Abstract
An estimated 3% of the world's population is chronically infected with hepatitis C virus (HCV). Although HCV was discovered more than 20 y ago, its origin remains obscure largely because no closely related animal virus homolog has been identified; furthermore, efforts to understand HCV pathogenesis have been hampered by the absence of animal models other than chimpanzees for human disease. Here we report the identification in domestic dogs of a nonprimate hepacivirus. Comparative phylogenetic analysis of the canine hepacivirus (CHV) confirmed it to be the most genetically similar animal virus homolog of HCV. Bayesian Markov chains Monte Carlo and associated time to most recent common ancestor analyses suggest a mean recent divergence time of CHV and HCV clades within the past 500–1,000 y, well after the domestication of canines. The discovery of CHV may provide new insights into the origin and evolution of HCV and a tractable model system with which to probe the pathogenesis, prevention, and treatment of diseases caused by hepacivirus infection.
Keywords: flavivirus, zoonosis
Viral zoonoses account for up to 70% of human emerging infectious diseases; nonetheless, biological and epidemiological barriers to interspecies transmission are high (1, 2), and the majority of viruses that infect wildlife and domestic animals do not infect humans. Sustained contact between humans and other species increases the likelihood of the emergence of a virus adapted to infect and replicate in humans either directly or through intermediate hosts (2). Identification and characterization of animal virus homologs provide new insights into pathogenesis of human viruses, and, in some instances, models for investigating prevention and treatment of human disease must be pursued with related animal viruses (3–5). Comparative genetic analysis of closely related viruses can also identify genomic regions (RNA structures, amino acid motifs, and residues) important for virus receptor binding, entry, replication, immunity, and other biological functions (6–10).
Since its discovery about 20 y ago (11, 12), the origin of hepatitis C virus (HCV) remains obscure largely because a closely related animal virus homolog has not been identified (13, 14). Worldwide, 200 million people are chronically infected with HCV (13, 15) and are at risk for developing liver fibrosis, cirrhosis, and hepatocellular carcinoma. Given precedent with other blood-borne pathogens like HIV and hepatitis B virus, efforts to find homologs of HCV have focused on nonhuman primates. To date, these efforts have been unsuccessful (14). Dogs were domesticated as early as 8,000 BCE (16) and, as companion and working animals, occupy a unique niche at the human–animal interface. During an investigation to characterize respiratory viruses infecting domestic dogs, we identified several flavivirus-like sequences. Phylogenetic analysis of ∼6,500 nt of continuous genomic sequence revealed the presence of a virus genetically most similar to HCV, tentatively named canine hepacivirus (CHV).
HCV belongs to the genus Hepacivirus, one of the four genera in the family Flaviviridae (14). These viruses are classified in three established genera (Flavivirus, Pestivirus, and Hepacivirus) and one proposed genus, Pegivirus (14). Studies to understand the pathogenesis of HCV have been hampered by its restricted replication in humans or chimpanzees and, until recently, its inability to replicate in cell culture (17, 18). One alternative model is the distantly related GB virus B (GBV-B) that infects tamarins (Saguinus sp.) (19–21). However, GBV-B is highly divergent from HCV. Moreover, its elusive origins and ongoing uncertainty over whether tamarins are the natural host for GBV-B further restrict its value as a model system to study HCV pathogenesis (14). Here we report the discovery and unique genomic features of a hepacivirus that infects domestic dogs and is genetically most related to HCV. Interestingly, CHV was found in respiratory samples as well as in liver; because titers in liver specimens were low, it remains to be confirmed whether CHV is hepatotropic. Our results indicate that hepaciviruses are not restricted to primates and suggest the possibility that HCV may have been introduced in the human population through contact with canines or other nonprimate species.
Results
Discovery of CHV.
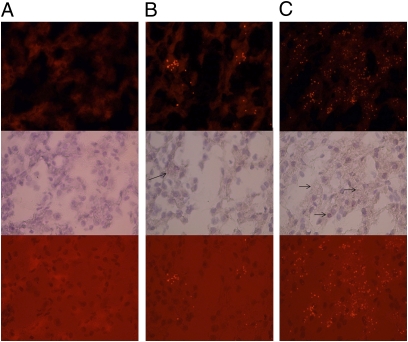
This study was undertaken to characterize the viral flora of companion animals. Respiratory samples of dogs associated with respiratory illness outbreaks were enriched for viral nucleic acids (22), randomly amplified, and subjected to unbiased high-throughput sequencing (23). Bioinformatic analysis of sequences at the predicted amino acid level revealed the presence of several sequences substantially similar to flaviviruses. Sequence fragments were mapped to prototypic flavivirus genomes, and gaps in genomic sequences were filled by PCR using specific and degenerate primers. Preliminary phylogenetic analysis of ∼6,500 nt of continuous genomic sequence revealed the presence of a unique virus most closely related to HCV, tentatively named CHV. Thereafter, specific primers targeting highly conserved helicase gene motifs in CHV were used in RT-PCR to screen samples from 33 dogs representing five different outbreaks of respiratory disease. Six of 9 animals in one outbreak and 3 of 5 in another were positive for CHV. Quantitative PCR assay yielded >107 copies of CHV RNA per nasal swab of most infected animals. Partial CHV sequences from the NS3 gene (399 nt) of viruses from different animals of the two outbreaks showed 99.2% sequence convergence (all substitutions occurred at synonymous sites). This high degree of genetic relatedness likely arose because both outbreaks occurred in same animal shelter facility within a period of 2 wk. To explore the epidemiology of CHV infection in dogs, we screened nasal swabs collected from 60 healthy pets. No healthy animals were found infected with CHV or its related variants. We also tested 19 liver and five lung samples from 19 unrelated dogs that had died of unexplained gastrointestinal illness. In five dogs (three from Michigan and two from Montana), we found CHV RNA in liver but not in lung tissue. Whereas respiratory samples contained >107 copies of CHV genomic RNA per 2 ng of total RNA, levels in liver samples were <103 copies of CHV genomic RNA in equivalent input. Blood, peripheral blood mononuclear cell, or other samples from these animals were not available for the study. The viral variants found in liver samples showed four synonymous and one nonsynonymous mutation compared with CHV (GenBank accession nos. JF744992 to JF744996). To test for the presence of CHV in liver by using an independent method, we used an in situ hybridization assay that revealed focal as well as dispersed infection of canine liver and presence of viral RNA predominantly in the cytoplasm of hepatocytes (Fig. 1). We have been unable to culture CHV in vitro using two continuous (Madin–Darby canine kidney and D17) and one primary (dog kidney primary) canine cell line. Although our findings are intriguing, further studies are necessary to determine the tissue tropism, pathogenic potential, and disease association of CHV.
Fig. 1.
In situ hybridization of CHV RNA in canine liver. (A) Uninfected liver. (B and C) Infected liver. Top, Middle, and Bottom represent fluorescent, bright-field, and superimposed images, respectively (bright red dots indicate probe bound to CHV genomic RNA; blue is hematoxylin counterstain).
Genomic Characterization of CHV.
The genome sequence of CHV was determined directly from a respiratory sample of one of the nine dogs with acute respiratory illness. The CHV genome comprises at least 9,195 nt (GenBank accession no. JF744991) and encodes a 2,942-aa polyprotein and a short 5′ UTR (Fig. 2A). We were unable to clone and sequence the complete 3′ UTR of CHV but note that the 3′ UTR of HCV was not cloned for several years after the initial identification of the virus (24, 25). Based on analogy to HCV and GBV-B viruses, which contain a poly-U tract upstream of 3′-terminal RNA structures (3′ UTR), we used a poly-A primer to initiate reverse transcription in efforts to clone and characterize the 3′ UTR of CHV. These efforts were unsuccessful after several attempts. However, cDNA construction using a poly-T primer resulted in early termination of CHV genome, indicating the presence of a poly-A stretch near the 3′ terminus. This finding was confirmed by using an adapter ligation approach similar to the one described for cloning the 3′ terminus of HCV (25). The sequence is submitted in GenBank under accession number JF744997. These results are unusual and will require further study using high-titer CHV samples.
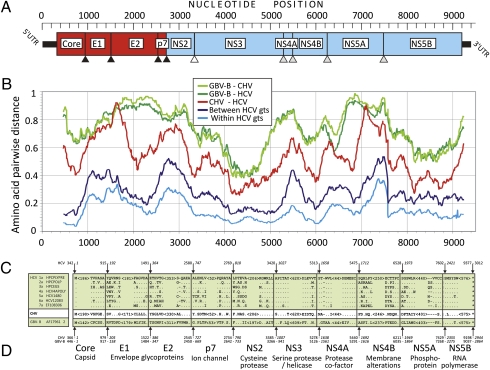
Fig. 2.
Structural and functional map of the CHV genome. (A) Structural protein cleavage is mediated by cellular signal peptidase (black triangle); NS2–NS3 cleavage is mediated by the NS2–NS3 autoprotease (white triangle); and cleavage of other nonstructural proteins is mediated by NS3–NS4A protease complex (gray triangles). (B) Amino acid sequence divergence scan of CHV polyprotein, HCV genotypes, and GBV-B. (C) Amino acid sequence of different viruses adjacent to predicted protease cleavage sites (10 aa on each side are shown). (D) Reported functional role of different proteins in the virus life cycle.
The G + C content of CHV (50.7%) is similar to GBV-B (50.6%), lower than HCV (55.9–58.4%) or pegiviruses (55.9–60.6%), and higher than pestiviruses (45.5–46.7%) and the majority of classical flaviviruses (38.4–54.9%). Similar to HCV and GBV-B, the CpG (and UpA) dinucleotides were underrepresented (72% of expected value based on G + C content) compared with flaviviruses and pestiviruses. Using a sliding window analysis, the degree of amino acid sequence divergence of CHV from other hepaciviruses was determined for the complete predicted polyprotein-coding region (Fig. 2). CHV is more similar to HCV than to GBV-B throughout the genome (including the 5′ UTRs) (Fig. 3). Furthermore, regions of greater diversity between HCV genotypes are also more divergent between HCV and CHV (E2 and 5′ end of nonstructural protein NS5A). Nonstructural proteins NS3 and NS5B of CHV have maximum amino acid identity to HCV (>55–65%), whereas E1 (the N-terminal half of E2), NS2, and the C terminus of NS5A have the least amino acid identity (<35–45%) (Fig. 2).
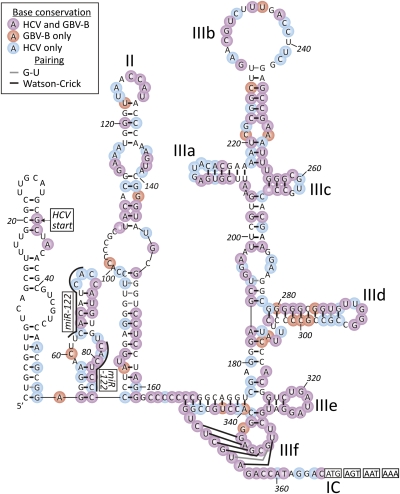
Fig. 3.
Sequence and secondary structure of CHV 5′ UTR. Bases conserved among different hepaciviruses are shown with different colored circles. The miR-122 binding sites and different internal ribosome entry site stems are labeled according to previously reported hepacivirus 5′ UTR structures (34).
In hepaciviruses, the structural and nonstructural proteins are typically generated by proteolytic cleavage by virus- and host-encoded proteinases. The hypothetical cleavage map of the CHV polyprotein was derived by alignment with representative sequences from the seven HCV genotypes (13). Similar to HCV, cleavage of the CHV polyprotein is predicted to create 10 viral proteins in a typical hepacivirus genomic organization (Fig. 2C). Signalase motifs (typically Ala-X-Ala preceding the cleavage site) are moderately conserved, as are the NS2–NS3 Zn-dependent cysteine protease and NS3–NS4A serine protease cleavage sites in the nonstructural polyprotein. Similar to HCV and GBV-B, the predicted CHV core protein is highly basic (pI = 11.4), consistent with an RNA binding/packaging function. Like HCV, the genome of CHV contains a ribosomal slippery sequence (A5nnA5) in the core protein-coding region, possibly for generation of a frame-shift [F protein or alternative reading frame protein (ARFP)] product (26, 27); however, the other two associated alternative reading frames in CHV are interrupted by stop codons and have no significant homology to HCV F proteins. GBV-B also lacks an F protein ortholog, a finding that suggests that the F protein has a unique role in the life cycle of HCV. Between HCV and GBV-B, we observed several long amino acid stretches in the E1E2 region with virtually no identifiable homology (>90% amino acid divergence). However, the E1E2 regions of CHV were readily aligned with HCV, with marked similarity in the C-terminal half of E2 (Fig. 2B). The translated E1 and E2 sequences contained 4 and 10 N-linked glycosylation sites similar to sites predicted for HCV (5 and 11, respectively) (ref. 28; Fig. S1) and greater than the number of sites in GBV-B (3 and 6, respectively) and members of the Pegivirus genus (e.g., 1 and 3 in GBV-A) (14). The ectodomain of E2 envelope protein of all HCV genotypes contain 18 highly conserved cysteine residues that form nine disulphide bonds crucial for proper 3D folding of the HCV structural protein (29). Interestingly, CHV E2 contained 14 of the 18 highly conserved cysteine residues known to form disulfide bridges in HCV E2, whereas GBV-B lacked these cysteines (Fig. S1). The similar genome organization, presence of conserved protein motifs, processing sites, and the overall high sequence homology between CHV and the other hepaciviruses (as determined with the National Center for Biotechnology Information Conserved Domains Database) enables a robust prediction of the location and function of all CHV-encoded proteins (Fig. 2D) (18, 30).
Secondary RNA Structures in the CHV Genome.
The 5′ UTR of CHV has 366 nt, which is more similar in length to HCV (341 nt) than to GBV-B (445 nt). Standard ClustalW-based sequence alignment demonstrated 66% and 57% nucleotide identity with HCV and GBV-B, respectively. The secondary structure of the CHV 5′ UTR RNA was predicted by using a simple thermodynamic folding energy minimization algorithm (MFOLD) (Fig. 3) to reveal four stem-loops with moderate/high Pnum values (measure of the robustness of the predicted paired/unpaired state at each base position), two of which corresponded in position and shape to previously designated stem-loops II and III in the type IV internal ribosome entry sites of HCV, GBV-B, pestiviruses, and a few genera within the picornavirus family (31). Sequence conservation among CHV, HCV, and GBV-B is most apparent in the pseudoknot region (the IIIf, IIIe, and IIIc stem-loops), whereas other regions (large parts of stem-loop II, the IIIb terminal loop, IIIa, and IIId) show conservation only in the predicted RNA secondary structure (multiple covariant sites or nonhomologous base pairings). Substantial structural differences and lack of sequence homology are most evident at the 5′ end of the 5′ UTR between CHV (and HCV) and the much longer equivalent region in GBV-B (10). Furthermore, the first stem-loop predicted for CHV is distinct from stem-loop I of HCV; indeed, based on the alignment, the 5′ base of the HCV genome falls within the 3′ half of the CHV structure (Fig. 3). Intriguingly, a second stem-loop of CHV is formed by a stretch of 30 nt (positions 55–84) with evident homology to HCV (and, to a lesser extent, to GBV-B) (Fig. 3). However, in HCV, this region is predicted to be unpaired and contains binding sites for the human microRNA, miR-122 (32, 33). Whether the stem-loop evident in CHV precludes equivalent binding of the canine homolog of miR-122 (and indeed whether sequence variability in the first seed match is compatible with binding) remains to be determined (Fig. 3). A scan of the dog microRNA registry failed to identify any high-probability alternative seed matches in the CHV 5′ UTR. Finally, the predicted CHV 5′ UTR structure lacks the stem-loop around the polyprotein initiation codon found in HCV and GBV-B (but not in pestiviruses). The translational control mechanisms proposed for HCV mediated by this pairing (34) are thus unlikely to apply in CHV.
RNA secondary structure within coding regions of the genome was characterized by extensive internal base pairing. For example, MFOLD analysis of the last 540 nt of the coding region of the CHV genome (corresponding to the region containing a cis-replicating element in HCV) predicts three stable stem-loop structures (each spanning 120–145 nt) with long duplex regions (Fig. S2). RNA viruses containing genome-scale ordered RNA structure (GORS) with high mean folding energies (MFEs) are more likely to cause persistent infection (35). The CHV genome sequence was analyzed for evidence of GORS by comparing folding energies of consecutive fragments of nucleotide sequence with random sequence-order controls (35). The MFE difference value of the CHV genome at 13.8% is higher than that of HCV (7.8–9.5%), GBV-B (9.5%), or pegiviruses (10.3–14.4%). Based on the characteristics of other RNA viruses with GORS, these observations predict that CHV infections may be persistent in its natural hosts.
Phylogenetic and Evolutionary Analysis of CHV.
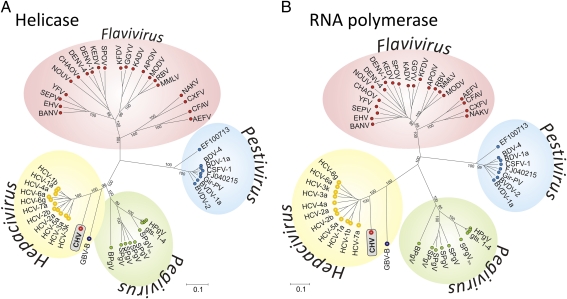
CHV was phylogenetically classified by determining its genetic relatedness to representative viruses of different genera of Flaviviridae. These viruses are classified in three established genera (Flavivirus, Pestivirus, and Hepacivirus) and one proposed genus, Pegivirus (14). Comparative phylogenetic analysis of conserved regions in the predicted helicase (NS3) and RNA-dependent RNA polymerase (RdRp or NS5B) regions were congruent with CHV consistently closest and equidistant from the seven HCV genotypes (Fig. 4). These clusters were consistent with the established taxonomic groups and supported by bootstrap values of above 90% (of 1,000 replicates) (36). The phylogenetic position of CHV relative to HCV and GBV-B is consistent with their pairwise distances (Table S1). The dissimilarity in S region sequences between GBV-B and other hepaciviruses is reflected in the virtual absence of sequence similarity of amino acid sequences after alignment (11–12%) compared with 36% amino acid similarity between CHV and HCV. The degree of sequence divergence between HCV and CHV is almost the same as that observed between GBV-C (a human virus) and GBV-A (found in chimpanzees and New World primates) (Fig. 4). Considering the results of phylogenetic analyses, pairwise protein distances, and identification of CHV in a different natural host, we suggest classification of CHV as a prototype new virus species in the genus Hepacivirus.
Fig. 4.
Phylogenetic analysis of conserved regions in the helicase (motifs I–VI) (A) and RdRp (B) genes of CHV aligned with representative members of the Hepacivirus, Pegivirus (GBV viruses A, C, and D), Pestivirus, and Flavivirus genera. Translated amino acid sequences were aligned with the program ClustalW. Trees were constructed by neighbor joining of pairwise amino acid distances with the program MEGA5 (according to the distance scale provided). Bootstrap resampling was used to determine robustness of branches; values of ≥70% (from 1,000 replicates) are shown. Regions compared corresponded to positions 3697–4477 (helicase domain of NS3) and 7705–8550 (RdRp in NS5B; numbered according to the AF011751 HCV genotype 1a reference sequence). A listing of virus abbreviations and original accession numbers for each sequence are provided in Table S2.
To determine the evolutionary relation between CHV and other hepaciviruses, a Bayesian Markov chains Monte Carlo (MCMC) estimation of the time to most recent common ancestor (TMRCA) for the HCV genotypes, GBV-B, and CHV was performed by using an external rate calibration based on the evolutionary rates estimated for (i) HCV subtypes 1a and 1b (37) and (ii) HCV subtype 6 (38). The mean estimated TMRCA for the Hepacivirus clade and CHV is 341 y before present (ybp) [95% highest posterior density (HPD) = 69–705 ybp] based on the HCV subtype 1 calibration and 1,680 ybp (521–3,291 ybp) based on the HCV subtype 6 calibration (Fig. S4). Thus, we estimate that the shared common ancestor between CHV and the HCV genotypes probably existed between 500 and 1,000 ybp. However, this should only be regarded as a minimum estimate given the difficulties associated with extrapolating short-term substitution rates to longer evolutionary periods (39, 40). We estimated the TMRCA between CHV and HCV using a substitution rate previously used to infer the divergence times within HCV genotypes/subtypes (37, 38). However, whether these rates can be reliably applied to much more divergent CHV and HCV sequences is unclear. Observation of time dependency in substitution rates in host genes (41) and recent evidence from endogenous viral elements for substantially slower long-term substitution rates in a variety of animal viruses argue against simple extrapolation of substitution rates measured over short observation periods (42–45).
Discussion
Molecular characterization of CHV indicates that it is the most genetically related homolog of HCV. Viral structural proteins typically contain major determinants of viral immunogenicity and host/cell tropism. The envelope protein E2 of HCV is among the most variable portions of its genome, yet it has remarkable sequence similarity with CHV. Moreover, the number and position of cysteine residues in E2 protein of CHV indicate that even the tertiary structure of CHV is likely to be more similar to HCV than to other genetically related viruses (29). However, there are notable differences between CHV and HCV that may have biological significance. Most strikingly, the potential occlusion of the binding site of miR-122 in the CHV 5′ UTR and the absence of microRNA sequences in the dog genome capable of binding to the equivalent site in CHV suggest that the interaction, which enhances the replication of HCV in human liver (32, 33), may not be needed in CHV infections. It remains to be determined whether the unique stem-loop I of CHV allows it to replicate in a manner independent of miR-122 or influences tissue tropism. Although reverse-genetic experiments wherein genomic regions are swapped between HCV and CHV will be required to address these questions, we note that, although CHV is found at high levels in respiratory samples of infected animals, only low or undetectable levels of HCV RNA are typically detected in respiratory samples from HCV-infected humans (46). A significant difference in life span of humans and canines can also affect the disease pattern caused by genetically related viruses. The availability of CHV genome and its comparative genetic analysis with HCV genotypes is therefore likely to advance our understanding of the role genetic elements and proteins play in the viral life cycle. Moreover, the sequence data presented here will help in designing reagents necessary to further explore the biology, pathogenesis, and tissue tropism of CHV and its variants.
The limited genetic diversity observed among CHV variants is atypical for RNA viruses including HCV and is likely attributable to the study animals being in close contact (same disease outbreak) or the highly specific PCR assay used in this study. We expect that the use of broadly reactive reagents (e.g., degenerate primers) and samples from unrelated dogs from different geographies will result in identification of many diverse CHV-related viruses. Without further information on the distribution of HCV-related viruses in other mammalian species, it is too early to draw conclusions on the evolutionary events underlying their distribution in humans and dogs and their apparent absence in nonhuman primates. Indeed, there is virtually no information on the existence of HCV-like viruses in other mammalian orders, which have probably remained untested because of a primate focus in screening paradigms. Hepaciviruses may be widely distributed among different mammalian species, perhaps highly host-specific, effectively transmitted by nonparenteral routes and largely nonpathogenic (as appears to be the case for pegiviruses in primates) (35). An alternative scenario is one where hepaciviruses are primarily canine viruses and HCV in humans arose zoonotically from contact from dogs or other related members of carnivore mammalian order that harbor these types of viruses. A zoonotic origin for HCV and lack of host adaptation would explain its high degree of pathogenicity in humans, inefficient transmission by nonparenteral routes, and apparent absence of HCV homolog in nonhuman primates. However, the miR-122 interaction appears to be human-specific and likely represents a virus/host coadaptation, unlikely responsible for a recent zoonosis.
Although its hepatotropism and ability to establish persistence remain to be determined, the presence of CHV in hepatocytes is reminiscent of HCV infections in humans. Furthermore, the presence of GORS in CHV is consistent with persistence in other viral systems. Irrespective of its evolutionary origins, the discovery of CHV provides the exciting prospect of a unique experimental model for HCV infections in humans and opens future avenues for research into the pathogenesis, prevention, and treatment of hepacivirus infections.
Materials and Methods
Samples and High-Throughput Sequencing.
Respiratory samples (nasal swabs) were collected in 3 mL of MEM from affected dogs in five respiratory disease outbreaks in four shelters (one each in Texas and Utah and three in Pennsylvania). Postmortem lung and liver samples were from euthanized dogs in clinics in Montana and Missouri. Centrifuged respiratory sample (140 μL) was filtered through a 0.45-μm filter to remove eukaryotic and bacterial-sized particles. The filtrate was then treated with nucleases to digest non-particle-protected nucleic acid. RNA from filtered (0.45-μm) respiratory samples or tissue homogenates was treated with DNase before random amplification and pyrosequencing (22, 47). After assembly (Newbler v2.3; 454 Life Sciences), sequence contigs and singletons were analyzed by using BlastX against a National Center for Biotechnology Information nonredundant protein database (47).
Genome Sequencing and Phylogenetic and Evolutionary Analyses.
Details are provided in SI Materials and Methods.
RNA Structure and GORS Predictions.
Independent of phylogenetic information, the secondary structure of the CHCV 5′ UTR RNA was modeled with MFOLD. Labeling of the predicted structures in the 5′ UTR followed numbering used for reported homologous structures in HCV and GBV-B (10, 34). The CHV genome sequence was analyzed for evidence of GORS by comparing folding energies of consecutive fragments of nucleotide sequence with random sequence-order controls (35). MFEs of CHCV were calculated by using default setting in the program ZIPFOLD. MFE results were expressed as MFE differences, i.e., the percentage difference between the MFE of the native sequence from that of the mean value of the 50 sequence order–randomized controls.
In Situ Hybridization.
For in situ hybridization assay, multiple branched-chain probe amplifiers labeled with alkaline phosphatase (Panomics/Affymetrix) were used against CHV genomic RNA (nucleotides 840–2040 of CHV genome corresponding to the coding region for partial core, envelope glycoprotein E1/E2, and partial NS1 protein). Liver sections (10 μm) were fixed with 4% formaldehyde at 4 °C overnight, dehydrated, permeabilized, and stained with Fast Red substrate (QG ViewRNA Chromogenic Signal Amplification Kit; Panomics/Affymetrix) for light and florescent microscopy.
Supplementary Material
Acknowledgments
We thank Natasha Mehta, Jan Medina, and Ahmed Sandhu for excellent technical assistance. We are grateful to Roche 454 Life Sciences for high-throughput sequencing support. This work was supported by National Institutes of Health Grants AI090196, AI079231, AI57158, AI070411, AI090055, AI072613, and EY017404; the Department of Defense; US Department of Agriculture Grant 58-1275-7-370; the Greenberg Medical Research Institute; the Starr Foundation; German Academy of Sciences Leopoldina Grant LPDS-2009-9; and International Human Frontier Science Program Organization Grant LT000048/2009-L.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JF744991 and JF744997).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101794108/-/DCSupplemental.
References
- 1.Parrish CR, Kawaoka Y. The origins of new pandemic viruses: The acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu Rev Microbiol. 2005;59:553–586. doi: 10.1146/annurev.micro.59.030804.121059. [DOI] [PubMed] [Google Scholar]
- 2.Parrish CR, et al. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hatziioannou T, et al. A macaque model of HIV-1 infection. Proc Natl Acad Sci USA. 2009;106:4425–4429. doi: 10.1073/pnas.0812587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai CC, et al. Prevention of SIV infection in macaques by (R)-9-(2-phosphonylmethoxypropyl)adenine. Science. 1995;270:1197–1199. doi: 10.1126/science.270.5239.1197. [DOI] [PubMed] [Google Scholar]
- 5.Van Rompay KK. Evaluation of antiretrovirals in animal models of HIV infection. Antiviral Res. 2010;85:159–175. doi: 10.1016/j.antiviral.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Kapoor A, et al. Multiple novel astrovirus species in human stool. J Gen Virol. 2009;90:2965–2972. doi: 10.1099/vir.0.014449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapoor A, et al. Identification and characterization of a new bocavirus species in gorillas. PLoS ONE. 2010;5:e11948. doi: 10.1371/journal.pone.0011948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapoor A, et al. A highly divergent picornavirus in a marine mammal. J Virol. 2008;82:311–320. doi: 10.1128/JVI.01240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klatt NR, et al. CD8+ lymphocytes control viral replication in SIVmac239-infected rhesus macaques without decreasing the lifespan of productively infected cells. PLoS Pathog. 2010;6:e1000747. doi: 10.1371/journal.ppat.1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rijnbrand R, Abell G, Lemon SM. Mutational analysis of the GB virus B internal ribosome entry site. J Virol. 2000;74:773–783. doi: 10.1128/jvi.74.2.773-783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alter HJ. Discovery of the non-A, non-B hepatitis virus: The end of the beginning or the beginning of the end. Transfus Med Rev. 1989;3:77–81. doi: 10.1016/s0887-7963(89)70071-7. [DOI] [PubMed] [Google Scholar]
- 12.Choo QL, et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 13.Simmonds P. Genetic diversity and evolution of hepatitis C virus—15 years on. J Gen Virol. 2004;85:3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- 14.Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P. The GB viruses: A review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J Gen Virol. 2011;92:233–246. doi: 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ray Kim W. Global epidemiology and burden of hepatitis C. Microbes Infect. 2002;4:1219–1225. doi: 10.1016/s1286-4579(02)01649-0. [DOI] [PubMed] [Google Scholar]
- 16.Protsch R, Berger R. Earliest radiocarbon dates for domesticated animals: Europe is added to the Near East as another early center of domestication. Science. 1973;179:235–239. doi: 10.1126/science.179.4070.235. [DOI] [PubMed] [Google Scholar]
- 17.Lohmann V, et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 18.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 19.Nam JH, et al. In vivo analysis of the 3′ untranslated region of GB virus B after in vitro mutagenesis of an infectious cDNA clone: Persistent infection in a transfected tamarin. J Virol. 2004;78:9389–9399. doi: 10.1128/JVI.78.17.9389-9399.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bukh J, Apgar CL, Govindarajan S, Purcell RH. Host range studies of GB virus-B hepatitis agent, the closest relative of hepatitis C virus, in New World monkeys and chimpanzees. J Med Virol. 2001;65:694–697. doi: 10.1002/jmv.2092. [DOI] [PubMed] [Google Scholar]
- 21.Bukh J, Apgar CL, Yanagi M. Toward a surrogate model for hepatitis C virus: An infectious molecular clone of the GB virus-B hepatitis agent. Virology. 1999;262:470–478. doi: 10.1006/viro.1999.9941. [DOI] [PubMed] [Google Scholar]
- 22.Kapoor A, et al. A highly prevalent and genetically diversified Picornaviridae genus in South Asian children. Proc Natl Acad Sci USA. 2008;105:20482–20487. doi: 10.1073/pnas.0807979105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Epstein JH, et al. Identification of GBV-D, a novel GB-like flavivirus from old world frugivorous bats (Pteropus giganteus) in Bangladesh. PLoS Pathog. 2010;6:e1000972. doi: 10.1371/journal.ppat.1000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka T, Kato N, Cho MJ, Shimotohno K. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem Biophys Res Commun. 1995;215:744–749. doi: 10.1006/bbrc.1995.2526. [DOI] [PubMed] [Google Scholar]
- 25.Kolykhalov AA, Feinstone SM, Rice CM. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Branch AD, Stump DD, Gutierrez JA, Eng F, Walewski JL. The hepatitis C virus alternate reading frame (ARF) and its family of novel products: The alternate reading frame protein/F-protein, the double-frameshift protein, and others. Semin Liver Dis. 2005;25:105–117. doi: 10.1055/s-2005-864786. [DOI] [PubMed] [Google Scholar]
- 27.Xu Z, et al. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 2001;20:3840–3848. doi: 10.1093/emboj/20.14.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohr EL, Stapleton JT. GB virus type C interactions with HIV: The role of envelope glycoproteins. J Viral Hepat. 2009;16:757–768. doi: 10.1111/j.1365-2893.2009.01194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krey T, et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 2010;6:e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 31.Kieft JS. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci. 2008;33:274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jangra RK, Yi M, Lemon SM. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol. 2010;84:6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 34.Honda M, Brown EA, Lemon SM. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 35.Simmonds P, Tuplin A, Evans DJ. Detection of genome-scale ordered RNA structure (GORS) in genomes of positive-stranded RNA viruses: Implications for virus evolution and host persistence. RNA. 2004;10:1337–1351. doi: 10.1261/rna.7640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 37.Magiorkinis G, et al. The global spread of hepatitis C virus 1a and 1b: A phylodynamic and phylogeographic analysis. PLoS Med. 2009;6:e1000198. doi: 10.1371/journal.pmed.1000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pybus OG, et al. Genetic history of hepatitis C virus in East Asia. J Virol. 2009;83:1071–1082. doi: 10.1128/JVI.01501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keckesova Z, Ylinen LM, Towers GJ, Gifford RJ, Katzourakis A. Identification of a RELIK orthologue in the European hare (Lepus europaeus) reveals a minimum age of 12 million years for the lagomorph lentiviruses. Virology. 2009;384:7–11. doi: 10.1016/j.virol.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Worobey M, et al. Island biogeography reveals the deep history of SIV. Science. 2010;329:1487. doi: 10.1126/science.1193550. [DOI] [PubMed] [Google Scholar]
- 41.Ho SY, Phillips MJ, Cooper A, Drummond AJ. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol Biol Evol. 2005;22:1561–1568. doi: 10.1093/molbev/msi145. [DOI] [PubMed] [Google Scholar]
- 42.Kapoor A, Simmonds P, Lipkin WI. Discovery and characterization of mammalian endogenous parvoviruses. J Virol. 2010;84:12628–12635. doi: 10.1128/JVI.01732-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belyi VA, Levine AJ, Skalka AM. Unexpected inheritance: Multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 2010;6:e1001030. doi: 10.1371/journal.ppat.1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horie M, et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature. 2010;463:84–87. doi: 10.1038/nature08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet. 2010;6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferreiro MC, Dios PD, Scully C. Transmission of hepatitis C virus by saliva? Oral Dis. 2005;11:230–235. doi: 10.1111/j.1601-0825.2005.01076.x. [DOI] [PubMed] [Google Scholar]
- 47.Victoria JG, et al. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol. 2009;83:4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.