Abstract
Nuclear factor of activated T cells (NFAT) proteins are Ca2+-regulated transcription factors that control gene expression in many cell types. NFAT proteins are heavily phosphorylated and reside in the cytoplasm of resting cells; when cells are stimulated by a rise in intracellular Ca2+, NFAT proteins are dephosphorylated by the Ca2+/calmodulin-dependent phosphatase calcineurin and translocate to the nucleus to activate target gene expression. Here we show that phosphorylated NFAT1 is present in a large cytoplasmic RNA-protein scaffold complex that contains a long intergenic noncoding RNA (lincRNA), NRON [noncoding (RNA) repressor of NFAT]; a scaffold protein, IQ motif containing GTPase activating protein (IQGAP); and three NFAT kinases, casein kinase 1, glycogen synthase kinase 3, and dual specificity tyrosine phosphorylation regulated kinase. Combined knockdown of NRON and IQGAP1 increased NFAT dephosphorylation and nuclear import exclusively after stimulation, without affecting the rate of NFAT rephosphorylation and nuclear export; and both NRON-depleted T cells and T cells from IQGAP1-deficient mice showed increased production of NFAT-dependent cytokines. Our results provide evidence that a complex of lincRNA and protein forms a scaffold for a latent transcription factor and its regulatory kinases, and support an emerging consensus that lincRNAs that bind transcriptional regulators have a similar scaffold function.
Keywords: calcium signaling, kinase scaffold, long noncoding RNA
Activation of nuclear factor of activated T cells (NFAT) proteins is regulated by the phosphorylation status of the NFAT regulatory domain, a conserved region of approximately 300 residues N terminal to the DNA-binding domain, which is necessary and sufficient for nuclear transport (1, 2). In resting cells, NFAT proteins are located in the cytoplasm, where they are heavily phosphorylated through synergistic actions of three different families of kinases, casein kinase 1 (CK1), glycogen synthase kinase 3 (GSK3), and dual specificity tyrosine phosphorylation regulated kinase (DYRK) (2–5). Phosphorylation results in masking of a nuclear localization sequence (NLS), exposure of a nuclear export sequence (NES), and cytoplasmic localization of NFAT (1, 2). When cells are stimulated to increase intracellular Ca2+ concentrations, the calmodulin-dependent phosphatase calcineurin is activated and dephosphorylates the NFAT regulatory domain, causing a conformational change that exposes the NLS and masks the NES (1, 2). This event results in nuclear accumulation of NFAT and transcription of NFAT target genes (1).
We previously found, by size-exclusion chromatography of hypotonic lysates from resting Cl.7W2 T cells, that fully phosphorylated NFAT1 migrated in an unexpectedly high-molecular weight complex with a substantial fraction of its inhibitory NFAT kinases CK1α and CK1ϵ (3). Neither calcineurin nor GSK3 was detected in this complex. Stimulation with Ca2+ ionophore caused CK1 to dissociate from the complex, with only a slight change in the elution properties of NFAT1 (3). These data suggested that NFAT1 and CK1 were part of a larger complex within the cytoplasm of resting cells, from which the kinase dissociated after stimulation.
To identify NFAT scaffold protein(s), we performed genome-wide RNAi screens for regulators of NFAT1 nuclear translocation in Drosophila S2R+ cells (4, 6, 7). A screen for candidates whose depletion resulted in nuclear accumulation of NFAT1 in resting cells yielded two well-known Drosophila scaffold proteins, Homer and Discs Large (4), known to affect Ca2+ signaling upstream of NFAT (8). Homer2 and Homer3 also directly interact with NFAT (9). We also performed a screen for candidates whose knockdown prevented NFAT1 nuclear translocation in stimulated cells (6, 7). This screen identified Drosophila Cul4, a component of a Skp, cullin, F-box E3 ligase complex whose closest human homologue is Cullin 4B (CUL4B), and two proteins involved in nuclear transport—Drosophila Fs(2)Ket, an importin-beta whose human homologue is karyopherin (importin) beta 1 (KPNB1), and Drosophila Cas, a protein that recycles importin-alpha to the cytoplasm, whose human homologue is chromosome segregation 1-like (CSE1L) (7).
Notably, KPNB1, CSE1L, and CUL4B have been shown to bind the long intergenic noncoding RNA (lincRNA) NRON [noncoding (RNA) repressor of NFAT] (10). Depletion of NRON increased NFAT accumulation and transcriptional activity in the nucleus (10). An NRON-interacting protein that could not have scored in Drosophila screens was IQ motif-containing GTPase-activating protein 1 (IQGAP1), a large scaffold protein represented in yeast and vertebrates but not Drosophila (11, 12). IQGAP proteins possess multiple protein interaction domains: a calponin homology domain that binds F actin; a WW domain that may bind proline-rich sequences or sequences containing phospho-serine or phospho-threonine; an IQ domain that binds the ubiquitous Ca2+ sensor calmodulin; a GTPase-activating protein-related domain (GRD) that binds Cdc42 and Rac; and a C-terminal domain that binds E cadherin, β-catenin, and adenomatous polyposis coli (APC) (11, 12). IQGAP1 also binds B-Raf, mitogen-activated protein kinase kinase, and ERK, as a scaffold for MAP kinases (11, 12).
In this study we investigated the relation between NFAT, the lincRNA NRON, and IQGAP. We find that phosphorylated NFAT1 is present with NRON and IQGAP1 in a cytoplasmic RNA-protein scaffold complex that also contains all three known NFAT inhibitory kinases, CK1, GSK3, and DYRK. Our data support the emerging hypothesis that lincRNAs may function as scaffolds for transcriptional regulators in the context of large RNA-protein complexes (13–23), reviewed in refs. 24–27.
Results
Size-exclusion chromatography demonstrated that IQGAP proteins and the lincRNA NRON comigrated with the high-molecular-weight NFAT complex (Fig. 1). The peak of NFAT1 was consistently shifted from the peak of IQGAP1 by approximately 2 fractions, consistent with the fact that IQGAP proteins form multiple associations in cells (11, 12). When cytoplasmic extracts of Jurkat T cells stably expressing low levels of HA-tagged NFAT1 were subjected to size-exclusion chromatography on a Superdex 200 column, IQGAP1 coeluted with NFAT1 in a complex of apparent molecular mass ca. 500 kDa (Fig. 1A). RT-PCR analysis of pooled column fractions detected NRON sequences in fractions that contained NFAT1 and IQGAP1 (fractions 54–60, Fig. 1B), but not in fractions lacking NFAT1 (fractions 74–80 and 87–93 with high and low protein content, respectively, Fig. 1B). Size-exclusion chromatography of hypotonic lysates of murine CD8+ T cells, activated with anti-CD3 and anti-CD28 antibodies 6 d previously and then expanded in Interleukin-2 (IL-2) (28, 29), confirmed that endogenous NFAT1 coeluted with IQGAP1, IQGAP2, calmodulin, and a substantial fraction (approximately 40%) of CK1ϵ (Fig. 1C).
Fig. 1.
NFAT1 coelutes with IQGAP and NRON in resting T cell lysates by size-exclusion chromatography. Hypotonic lysates from (A and B) HA-NFAT1 Jurkat T cells or (C) primary murine CD8+ T cells were fractionated on a Superdex 200 size-exclusion column. (A and C) Individual fractions were analyzed by SDS-PAGE and Western blotting for NFAT1, IQGAP1, IQGAP2, calmodulin (CaM), casein kinase epsilon (CK1ϵ), and calcineurin A (CnA). Column void volume (V) is indicated. (B) Pooled column fractions were analyzed for NRON sequences by RT-PCR.
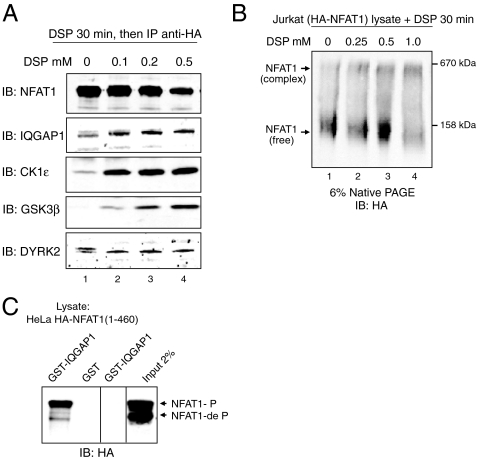
These results suggested the existence of a scaffolded complex containing NFAT1, NRON, IQGAP1, calmodulin, and at least one NFAT kinase, CK1ϵ. We performed cross-linking experiments to determine whether the complex was detectable in cells. HA-NFAT1 Jurkat cells were left untreated or treated with increasing concentrations of the cell-permeant, thiol-cleavable cross-linker Dithiobis[succinimidyl] propionate (DSP). HA-NFAT1 was immunoprecipitated from whole cell lysates, and immunoprecipitates were analyzed by immunoblotting after electrophoresis under reducing conditions. In the absence of cross-linking, IQGAP1 and the NFAT kinases DYRK2 and CK1ϵ coimmunoprecipitated with NFAT1, whereas GSK3β association was undetectable (Fig. 2A, lane 1); treatment with even the lowest concentration of DSP (0.1 mM) stabilized the interaction of NFAT1 with CK1ϵ and GSK3β, and to a lesser extent with IQGAP1 and DYRK2 (Fig. 2A, lanes 2–4). When lysates from HA-NFAT1 Jurkat cells were treated with DSP, NFAT1 shifted in its migration on native polyacrylamide gels from an apparent molecular mass of approximately 140 to 500–600 kDa (Fig. 2B), consistent with the data from size-exclusion chromatography (Fig. 1). In GST pulldown experiments, IQGAP1 interacted specifically with phosphorylated NFAT1(1-460)-GFP (Fig. 2C).
Fig. 2.
Association of NFAT1 with IQGAP1 and NFAT kinases demonstrated by protein cross-linking. (A) HA-NFAT1 Jurkat cells were left untreated (lane 1) or were treated with increasing concentrations of the thiol-cleavable cross-linker DSP (lanes 2–4). The cells were lysed, the lysates were immunoprecipitated (IP) with anti-HA antibody, and immunoprecipitates were resolved by denaturing SDS-PAGE. NFAT1, IQGAP1, and the NFAT kinases CK1ϵ, GSK3β, and DYRK2 were detected by immunoblotting (IB). (B) Whole cell lysates of HA-NFAT1 Jurkat cells were left untreated (lane 1) or treated with increasing concentrations of DSP (lanes 2–4) for 30 min, resolved by native PAGE, and immunoblotted (IB) for HA-NFAT1. (C) GST-IQGAP1 specifically binds phosphorylated NFAT1(1-460)-GFP. Phosphorylated and dephosphorylated forms are indicated.
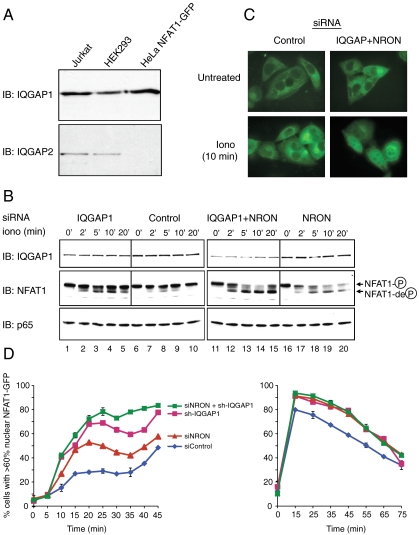
To examine the mechanism by which IQGAP1 and NRON regulate NFAT, HeLa cells stably expressing NFAT1(1-460)-GFP (4) were treated with siRNAs against IQGAP1 and/or NRON, and NFAT1 phosphorylation status and nuclear import kinetics were assessed (Fig. 3). HeLa cells were chosen because they express only IQGAP1, whereas Jurkat cells express IQGAP1 and IQGAP2 (Fig. 3A); because in contrast to T cells (30), their size and geometry are well suited for quantitative image analysis; and because siRNAs are taken up by > 80% of HeLa cells (4), but only by approximately 55% of Jurkat T cells after transient transfection (data not shown). NFAT1(1-460)-GFP from resting HeLa lysates migrated on size-exclusion columns as a high-molecular-weight complex, whose migration overlapped considerably with that of IQGAP1 (Fig. S1). Treatment with thapsigargin, which activates Ca2+ influx and consequent dephosphorylation of NFAT, led to a shift in the elution profile of both HA-NFAT1 and NFAT1(1-460)-GFP toward lower molecular weight, with no apparent change in the migration of IQGAP1 (Fig. S1). These results suggest that NFAT dissociates from IQGAP1 after stimulation, possibly as a result of the lower affinity of IQGAP1 for dephosphorylated NFAT (Fig. 2C). The elution profile of IQGAP1 is unaltered upon stimulation, consistent with its participation in multiple cellular protein complexes, including the immunological synapse in cytolytic T cells (11, 12, 31).
Fig. 3.
IQGAP1 and NRON depletion enhances NFAT1 dephosphorylation and nuclear import. (A) immunoblotting (IB) for IQGAP1 and IQGAP2 in HA-NFAT1 Jurkat, HEK293, and HeLa NFAT1(1-460)-GFP cells. (B and C) HeLa NFAT1(1-460)-GFP cells treated with control, IQGAP1, and/or NRON siRNAs were stimulated with 50 nM ionomycin (iono) and analyzed for (B) NFAT1 dephosphorylation by immunoblotting (phosphorylated and dephosphorylated NFAT1 are indicated) and (C) NFAT1(1-460)-GFP subcellular localization by fluorescence microscopy. (D, Left) Rate of nuclear import of NFAT1(1-460)-GFP in response to 250 nM thapsigargin (see Materials and Methods). (D, Right) HeLa NFAT1(1-460)-GFP cells were stimulated with 1 μM thapsigargin for 15 min to cause nuclear import of NFAT1(1-460)-GFP, and the rate of nuclear export of NFAT1(1-460)-GFP was quantified after addition of 50 nM CsA. Each data point represents an average of three independent values. Error bars denote standard deviation.
We observed a functional synergy between IQGAP1 and NRON in regulating NFAT (Fig. 3). First, combined treatment with siRNAs against IQGAP1 and NRON led to more efficient depletion of IQGAP1 protein than treatment with IQGAP1 siRNA alone (Fig. 3B, Top, compare lanes 11–15 with lanes 6–10 and lanes 1–5), suggesting that NRON stabilizes IQGAP1 in the NRON-IQGAP1 complex. Levels of p65/RelA were unaffected (Bottom). Second, siRNA-mediated knockdown of both IQGAP1 and NRON led to a substantial increase in NFAT dephosphorylation (Fig. 3B, Middle) and nuclear accumulation (Fig. 3C) in response to low concentrations of ionomycin, compared to cells treated with control siRNA or siRNAs against IQGAP1 or NRON alone. NFAT activation remained tightly regulated in cells with combined knockdown of IQGAP1 and NRON, without spontaneous dephosphorylation or nuclear import in the absence of stimulation (Fig. 3 B, lane 11, and C, Untreated).
We quantified the kinetics of NFAT1(1-460)-GFP nuclear transport using fluorescent images (30), scoring cells for nuclear NFAT using a cutoff of 60% colocalization with 4',6-diamidino-2-phenylindole (DAPI) staining. Import kinetics were followed after treating cells with relatively low concentrations (250 nM) of thapsigargin (Fig. 3D). The rate and magnitude of NFAT nuclear accumulation were substantially increased after depletion of IQGAP1, NRON, or both (Fig. 3D, Left). We also measured the kinetics of NFAT rephosphorylation and nuclear export: Cells were treated with thapsigargin (1 μM) to allow NFAT to accumulate rapidly in the nucleus, after which a low concentration of cyclosporin A (CsA) (50 nM) was added. Knockdown of IQGAP1, NRON, or both had no effect on the rate of nuclear export (Fig. 3D, Right), consistent with the fact that IQGAP1 is primarily cytoplasmic (31).
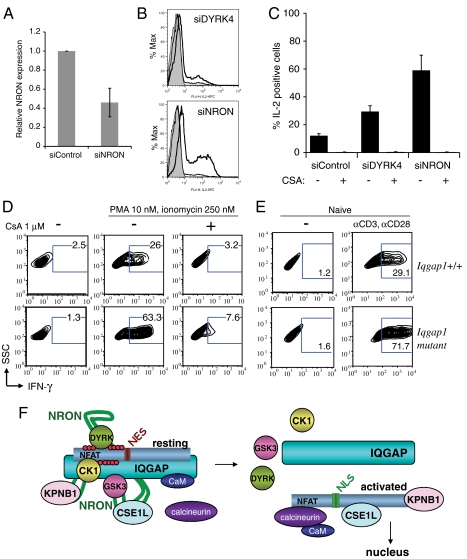
To examine the role of NRON in the regulation of NFAT-dependent genes, we transfected HA-NFAT1 Jurkat cells with a validated siRNA (10) to deplete NRON to levels ca. 45% of control (Fig. 4A), and analyzed IL-2 cytokine levels after phorbol-12-myristate-13-acetate (PMA) and ionomycin stimulation. Cells transfected with NRON siRNA showed a remarkable increase in IL-2-producing cells after stimulation, exceeding the moderate increase produced by treatment with siRNAs against a single NFAT kinase, DYRK4 (4) (Fig. 4B, bold lines, quantified in Fig. 4C). IL-2 production was sensitive to the calcineurin inhibitor cyclosporin A (Fig. 4C).
Fig. 4.
Increased expression of NFAT-dependent cytokines in NRON-depleted Jurkat cells and CD8+ T cells from Iqgap1 mutant mice. (A) HA-NFAT1 Jurkat cells were treated with control or NRON siRNAs and assessed for NRON expression by qRT-PCR. Results (average of three independent experiments) are normalized to β-actin and depicted relative to siControl. (B) siRNA-treated HA-NFAT1 Jurkat cells were analyzed for IL-2 production by intracellular staining and flow cytometry. Shaded histograms, unstimulated cells; thin lines, stimulated siControl-treated cells; thick lines, stimulated siDYRK4 or siNRON-treated cells (stimulation: PMA 50 nM, ionomycin 1 μM). As previously reported (4), only a small proportion of Jurkat cells transfected with control siRNA produce IL-2 upon stimulation. (C) Results from B were quantified and show the average and range of two independent experiments. (D) Primary CD8+ T cells from WT or IQGAP1-/- mice were isolated, differentiated, and restimulated with PMA (10 nM) and ionomycin (250 nM) for analysis of IFNγ production by intracellular staining and flow cytometry. (E) Naïve CD8+ T cells isolated from WT or IQGAP1-/- mice were isolated and IFNγ production was assessed in response to anti-CD3 (0.5 μg/mL) and anti-CD28 (1.0 μg/mL) stimulation. (F) Schematic representation of the NFAT1-IQGAP1-NRON-importin-kinase complex before and after stimulation. For details, see text.
To examine the effect of IQGAP1 deficiency on NFAT-mediated gene transcription, we examined T cells from mice with a deletion of exon 27 of Iqgap1 (32), which encodes part of the GRD motif (Fig. S2A). Mice with homozygous disruption of the Iqgap1 gene are viable and fertile, but show an increase in the frequency of late-onset spontaneous gastric hyperplasia (32). The mild phenotype of these mice could reflect functional redundancy among IQGAP1, IQGAP2, and IQGAP3; alternatively, it could reflect the fact that deletion of the distal GRD motif allows a truncated in-frame transcript to be translated. Indeed, RT-PCR analysis of RNA isolated from CD8+ T cells of Iqgap1 mutant mice confirmed the presence of Iqgap1 transcripts encoding the IQ motif adjacent to the deleted GRD region (Fig. S2B).
To determine whether T cells from Iqgap1 mutant mice showed dysregulated expression of NFAT-dependent cytokines, we analyzed CD8+ T cells which express the cytokine interferon-gamma (IFNγ), an established target gene of NFAT1 (1). IQGAP1 is abundant in the cytoplasm of CD8+ T cells, and a fraction shows regulated localization to the outer boundary of the cell–cell interface when cytolytic CD8+ T cells encounter target cells (31). CD8+ T cells were isolated from wild-type and Iqgap1 mutant mice and differentiated to yield functional cytolytic T cells (28, 29). The cells were restimulated for 6 h with PMA plus a limiting concentration of ionomycin (250 nM), and analyzed for IFNγ expression by intracellular staining and flow cytometry. Differentiated CD8+ T cells from Iqgap1 mutant mice did not produce IFNγ under resting conditions, but showed a CsA-sensitive increase in IFNγ production relative to cells from wild-type mice after stimulation with PMA plus ionomycin (Fig. 4D). Naïve CD8+ T cells isolated from Iqgap1 mutant mice also produced more IFNγ than wild-type cells in response to stimulation with anti-CD3 and anti-CD28 (Fig. 4E). These results are consistent with those obtained using HeLa cells, in which loss of IQGAP1 and NRON leads to increased NFAT dephosphorylation and nuclear translocation in stimulated cells (Fig. 3).
Discussion
In this study we provide evidence for the existence of a cytoplasmic RNA-protein complex containing inactive, phosphorylated NFAT1 and the three known NFAT1 kinases scaffolded by the lincRNA NRON and the scaffold protein IQGAP1. We show that (i) IQGAP1 interacts preferentially with phosphorylated NFAT1; (ii) IQGAP1 and NFAT kinases coimmunoprecipitate with NFAT1, especially after protein cross-linking; (iii) after cell stimulation, the peak of NFAT1 [and NFAT1(1-460)-GFP] in size-exclusion chromatography of cytoplasmic extracts shifts away from the peak of IQGAP1, to slightly lower apparent molecular weight; (iv) IQGAP1 protein levels are most effectively decreased by combined knockdown of both NRON and IQGAP1, compared to the relatively weak depletion achieved by IQGAP1 siRNA alone; (v) combined knockdown of NRON and IQGAP1 in HeLa cells enhances NFAT dephosphorylation and nuclear translocation in response to stimulation; and (vi) RNAi-mediated depletion of NRON in Jurkat cells results in a dramatic increase in IL-2 production, similar to that observed in primary T cells expressing a mutated IQGAP1. Together these data support a model (Fig. 4C) in which NRON stabilizes IQGAP protein in a high-molecular-weight complex containing NFAT, NRON, IQGAP1, and NFAT kinases in resting cells. Cell stimulation initiates a choreographed process in which NFAT associates with calcineurin, becomes dephosphorylated, and dissociates from IQGAP1 and NFAT kinases.
The cross-linking and coimmunoprecipitation data presented in this and an earlier study (3) document a physical linkage between NFAT1, IQGAP1, and NFAT kinases. Although it is possible that each of these proteins forms a separate complex with NFAT1, all of them are functional negative regulators of NFAT1, and all coelute with NFAT1 on size-exclusion chromatography at sizes considerably higher than their expected monomeric molecular masses, consistent with their presence in a single complex. The predicted size of a complex containing one molecule each of phosphorylated NFAT1 (110 kDa), IQGAP (180–190 kDa), CK1 (40–47 kDa), GSK (45–50 kDa), and DYRK (60–80 kDa) would be approximately 435–477 kDa, which falls into the observed range of elution on gel filtration, while still allowing for additional components such as RNA-binding proteins. NRON was previously reported to associate with KPNB1, CSE1L, and CUL4B (10); however, these proteins were not detected as NFAT-associated in either gel filtration or coimmunoprecipitation experiments, possibly because the proteins enter the complex only after activation. Our attempts to directly cross-link NRON RNA to proteins in the complex were not successful, likely because of very low NRON expression levels in cultured cells. We note that none of the proteins so far identified in the complex contains an obvious RNA-binding domain; moreover, we detect only a small change in the apparent size of the cytoplasmic NFAT complex upon NFAT dephosphorylation, suggesting that dissociation of NFAT from the scaffold complex upon NFAT dephosphorylation is accompanied by association with other components such as calcineurin, nuclear import proteins, and eventually, transcriptional regulators in the nucleus.
The repressive effect of NRON for NFAT was previously attributed to its ability to sequester proteins involved in nuclear transport, specifically KPNB1, an importin-beta, and CSE1L, which recycles importin-alpha from the nucleus to the cytoplasm (10). Our data show that the role of NRON is considerably more complex: It forms part of a large cytoplasmic RNA-protein scaffold for NFAT, which contains IQGAP proteins, calmodulin, and three distinct NFAT kinases, CK1, GSK3, and DYRK (see model in Fig. 4C). The scaffolded complex is stable, retaining its integrity in cell lysates and surviving the extended periods required for size-exclusion chromatography. The scaffold may have several functions: besides localizing NFAT adjacent to the maintenance kinases that promote its inactive state in the cytoplasm, it could hinder the access of calcineurin to NFAT in resting cells, and also serve as a reservoir for nuclear transport factors and for the calmodulin required to activate calcineurin in stimulated cells. In cells depleted of IQGAP1 or isolated from Iqgap1 mutant mice, basal activation of NFAT under resting conditions is not readily detected, most likely because calcineurin activity is very low under these conditions. In contrast, when cells are stimulated, calcineurin-mediated dephosphorylation of NFAT is accelerated and sustained compared to wild-type cells, presumably because NFAT is delocalized from rephosphorylation kinases.
Several transcriptional regulators have been shown to associate with lincRNAs (reviewed in refs. 24–27). In an early example, a lincRNA termed SRA (steroid receptor RNA activator) was shown to bind the AF2 activation domain of the progesterone receptor as well as the steroid receptor coactivator protein SRC-1, and to elute with SRC-1 in a high-molecular-weight complex from size-exclusion columns, in each case in the absence of ligand (13). Expression of SRA specifically increased hormone-dependent transcription driven by steroid receptors in reporter assays; conversely, antisense oligonucleotides to SRA diminished reporter expression (13). Similarly, the heat shock transcription factor 1 was shown to bind a complex of elongation factor eEF1A and the lincRNA HSR1 (14), and the homeodomain transcription factor Dlx-2 to bind the lincRNA Evf-2 (15). Remarkably, more than 20% of lincRNAs were found to bind proteins in the polycomb repressive complex 2 (PRC2) polycomb complex and other chromatin-modifying complexes (16). Specific, well-documented examples include HOTAIR, a lincRNA transcribed from the HOXC locus, which binds one or more proteins in the PRC2 complex, thereby affecting histone 3 lysine 27 (H3K27) methylation and silencing of the HOXD locus in trans (17, 18); HOTTIP, which binds the adapter protein WDR5, a component of the MLL H3K4 methyltransferase complex, and targets H3K4 trimethylation across the HOXA locus (19); an internal noncoding RNA, RepA, transcribed from the same locus as the larger lincRNA Xist responsible for X inactivation in female cells, which binds the Ezh2 H3K27 methyltransferase subunit of the PRC2 complex (20); Air, which binds the H3K9 histone methyltransferase G9a and recruits it to the promoter of paternally silenced genes in a narrow time window during embryonic development (21); lincRNA-p21, which is induced by p53 and feeds back to repress p53-mediated transcription (22); and lincRNA-RoR, which is upregulated by the pluripotency-associated transcription factors OCT4, SOX2, and NANOG and in turn promotes reprogramming of human fibroblasts to induced pluripotent stem cells (23). In several of these cases, the lincRNA was shown to localize with its associated transcriptional regulator to DNA, suggesting that it has a role in recruiting the transcriptional regulator to target genes (reviewed in refs. 24–27). However, in most cases, the exact mechanism of action of the noncoding RNA remains to be elucidated.
Based on our data, an attractive hypothesis is that lincRNAs and their protein partners modulate gene expression by serving as scaffolds for a variety of transcriptional regulators. As shown here for NRON, IQGAP1, and NFAT, one potential function of the scaffold complex is to promote efficient localization of the transcription factor to its regulators in the cytoplasm or its target genes in the nucleus. There is evidence that scaffold complexes can operate in diverse subcellular locations: lincRNAs are known to be key structural components of nuclear paraspeckles and are suspected to play a structural role in the cytokeratin network and the mitotic spindle (18). Further studies will be required to define mechanistically the functions of long noncoding RNAs, and potential scaffold complexes that incorporate such RNAs, in each experimental system.
Materials and Methods
Mice.
Iqgap1-null mice have been described (32). Mice were maintained in specific pathogen-free barrier facilities at Harvard Medical School, and were used in accordance with protocols approved by Immune Disease Institute, Brigham and Women’s Hospital, and Harvard Medical School animal care and use committees.
Size-Exclusion Chromatography.
Lysates were prepared by Dounce homogenization of HA-NFAT1 Jurkat T cells or mouse CD8+ T cells in hypotonic lysis buffer (3). Lysates were spun at 20,000 × g followed by a high-speed spin at 100,000 × g. Protein supernatant was loaded onto a Superdex 200 gel filtration column (Pharmacia).
RT-PCR.
Total RNA was extracted from column fractions or cells using TriZol reagent (Invitrogen), according to manufacturer’s instructions. Following DNase I treatment, cDNA was generated by random hexamer priming and SuperScript II (Invitrogen) reverse transcription, according to manufacturer’s instructions. PCR was performed for NRON or iqgap1 in a 50-μL volume containing 1X Taq buffer (New England BioLabs), 1.5 mM MgCl2, 0.2 mM dNTP, 1 pmol specific primer, 5 units of Taq DNA polymerase, and 1 μL room temperature product. Reaction mixtures were subjected to 28–32 amplification cycles, resolved on a 2% agarose gel, and visualized by ethidium bromide staining. Human NRON primers: ACCAGGCTGACTGTAGAATGG; GCAGAAAGGTCTCTGGACCTAC. Mouse Iqgap1 primers: CACCAAGCTGCAAGCCTGCTG; GGGTCCTCAGCATTGATGAGAGTC (IQ) and TGTGATCTTCACGCTGTACAACTATGC; AAGATCTGCCGGAGGGCATTCT (GRD). SYBR green (Roche) qRT-PCR was performed as described (4). Human NRON primers: ACGTTCCTT AATGTACGCCTTTGC; TTGGCCGTGTCCTGAGTCCTT; β-actin primers: TGAAGTGTGACG TGGACATC; GGAGGAGCAATGATCTTGAT.
Western Blotting.
Whole cell lysates were prepared, fractionated by SDS-PAGE, and immunoblotting was performed as described (4).
DSP Cross-Linking and Coimmunoprecipitation.
DSP (Pierce) was added to HA-NFAT1 Jurkat cells (5 × 107 cells per 500 μL PBS) for 30 min on ice, and whole cell lysates were prepared as described (4). Protein supernatants precleared with protein G sepharose (Amersham) were immunoprecipitated with 1 μg HA antibody (clone 12CA5) and protein G sepharose overnight at 4 °C. Immunoprecipitates were resolved by SDS-PAGE and Western blotting (4). For native PAGE, HA-NFAT1 Jurkat whole cell lysates (15 μg/μL) were DSP-treated for 30 min on ice. The reaction was stopped with 50 mM Tris•HCl pH 7.5 for 30 min on ice, and lysates were resolved by 6% PAGE (without SDS, DTT, or ß-mercaptoethanol) in the cold room. After transfer in Tris-Glycine, nitrocellulose membranes were soaked for 30 min in 20% ethanol followed by Western blotting. Negative control immunoprecipitations, including beads alone and isotype controls, were performed.
GST Pulldown.
GST-IQGAP1 was generated as described (33). Cells were lysed with Tritox-X lysis buffer and 1 mM CaCl2. Equal amounts of protein lysate were precleared by incubating for 1 h at 4 °C with glutathione-Sepharose. GST-IQGAP1 was added and samples were rotated at 4 °C for 3 h. After washing 5× with lysis buffer, beads were precipitated by centrifugation and dissolved in 30 μL sample buffer. Proteins were resolved by SDS-PAGE and Western blotting.
RNA Interference.
siRNA were purchased from Dharmacon. For HeLa, 0.8 × 106 cells were transfected with 60 nM control (4), human IQGAP1 (34), or human NRON siRNAs (10) using Lipofectamine 2000 (Invitrogen), according to manufacturer’s protocol. Cells were analyzed 5 d after transfection. For Jurkat, 1.0 × 106 cells were transfected with 60 nM control siRNA (4), human DYRK4 (4), or human NRON siRNAs (10) by NEON transfection (Invitrogen), according to manufacturer’s protocol. Cells were analyzed 2–3 d after transfection.
Quantification of Nuclear NFAT1(1-460)-GFP.
Cells seeded in black rim, clear bottom 96-well plates (Corning/Costar) were stimulated with thapsigargin (Sigma) at room temperature in complete growth media, fixed with 4% paraformaldehyde and stained with DAPI (Molecular Probes). Images were acquired on an ImageXpress Micro (Molecular Devices) using 10× magnification, and analyzed using the Translocation Application Module of MetaXpress software v.6.1 (Molecular Devices). Nuclear translocation was assessed by calculating the correlation of spatial fluorescence intensity between the GFP and DAPI compartments, with a cutoff of 60% for nuclear localization.
T Cell Differentiation.
HA-NFAT1 Jurkat T cells were maintained as described (4). CD8+ T cells were purified by magnetic bead negative selection (Dynal) from spleen and axillary, brachial and inguinal lymph nodes of C57BL/6 mice (6–8 wk old). T cell differentiation was induced as described (28, 29) for 6 d in 100 units per milliliter IL-2.
Intracellular Cytokine Staining.
Cells were stimulated 6 h with PMA (Calbiochem) and ionomycin (Calbiochem), or 24 h with anti-CD3 (clone 145.2C11) and anti-CD28 (clone 37.51) in plates coated with goat anti-hamster IgG (MP Biomedicals), with 2 μg/mL Brefeldin A (Sigma) added for the last 4 h of stimulation. Where indicated, 1 μM CsA (Calbiochem) was added to the culture 15 min before stimulation. Cells were stained as described (4) using APC-conjugated anti-human IL-2 or phycoerythrin-conjugated anti-mouse IFNγ (BD Bioscience), and analyzed on a FACSCalibur flow cytometer (Becton Dickinson) and FloJo software (Treestar).
Supplementary Material
Acknowledgments.
We thank Danya Machnes for technical support and members of the Rao lab for valuable discussions. This work was funded by National Institutes of Health (NIH) grants AI40127 and AI84167 (to A.R.), NIH Grant CA93645 (to D.B.S.) and NIH Grant DK62040 and a Research Scholar Grant from the American Cancer Society (to V.S.), a Fellowship from the Canadian Institutes of Health Research and a Special Fellowship from the Leukemia and Lymphoma Society (S.S.), a fellowship from the Lady Tata Memorial Fund (H.S.B.), and a postdoctoral training grant from the Joint Program in Transfusion Biology and Medicine, Children’s Hospital, Boston (S.O.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019711108/-/DCSupplemental.
References
- 1.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by Ca2+, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 2.Okamura H, et al. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell. 2000;6:539–550. doi: 10.1016/s1097-2765(00)00053-8. [DOI] [PubMed] [Google Scholar]
- 3.Okamura H, et al. A conserved docking motif for CK1 binding controls the nuclear localization of NFAT1. Mol Cell Biol. 2004;24:4184–4195. doi: 10.1128/MCB.24.10.4184-4195.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gwack Y, et al. A genome-wide Drosophila RNAi screen identifies DYRK-family kinases as regulators of NFAT. Nature. 2006;441:646–650. doi: 10.1038/nature04631. [DOI] [PubMed] [Google Scholar]
- 5.Arron JR, et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- 6.Feske S, et al. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 7.Gwack Y, et al. Biochemical and functional characterization of Orai proteins. J Biol Chem. 2007;282:16232–16243. doi: 10.1074/jbc.M609630200. [DOI] [PubMed] [Google Scholar]
- 8.Shaw AS, et al. Scaffold proteins and immune-cell signalling. Nat Rev Immunol. 2009;9:47–56. doi: 10.1038/nri2473. [DOI] [PubMed] [Google Scholar]
- 9.Huang GN, et al. NFAT binding and regulation of T cell activation by the cytoplasmic scaffolding Homer proteins. Science. 2008;319:476–481. doi: 10.1126/science.1151227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willingham AT, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 11.Briggs MW, Sacks DB. IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep. 2003;4:571–574. doi: 10.1038/sj.embor.embor867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White CD, Brown MD, Sacks DB. IQGAPs in cancer: A family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009;583:1817–1824. doi: 10.1016/j.febslet.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanz RB, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 14.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 15.Feng J, et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagano T, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 22.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loewer S, et al. Large intergenic noncoding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: An answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 25.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Cruz-Guilloty F, et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med. 2009;206:51–59. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pipkin ME, et al. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh-Hora M, et al. Dual functions for the endoplasmic reticulum Ca2+ sensors STIM1 and STIM2 in T cell activation and tolerance. Nat Immunol. 2008;9:432–443. doi: 10.1038/ni1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stinchcombe JC, Majorovits E, Bossi G, Fuller S, Griffiths GM. Centrosome polarization delivers secretory granules to the immunological synapse. Nature. 2006;443:462–465. doi: 10.1038/nature05071. [DOI] [PubMed] [Google Scholar]
- 32.Li S, Wang Q, Chakladar A, Bronson RT, Bernards A. Gastric hyperplasia in mice lacking the putative Cdc42 effector IQGAP1. Mol Cell Biol. 2000;20:697–701. doi: 10.1128/mcb.20.2.697-701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho YD, Joyal JL, Li Z, Sacks DB. IQGAP1 integrates Ca2+/calmodulin and Cdc42 signaling. J Biol Chem. 1999;274:464–470. doi: 10.1074/jbc.274.1.464. [DOI] [PubMed] [Google Scholar]
- 34.Mataraza JM, et al. IQGAP1 promotes cell motility and invasion. J Biol Chem. 2003;278:41237–44245. doi: 10.1074/jbc.M304838200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.