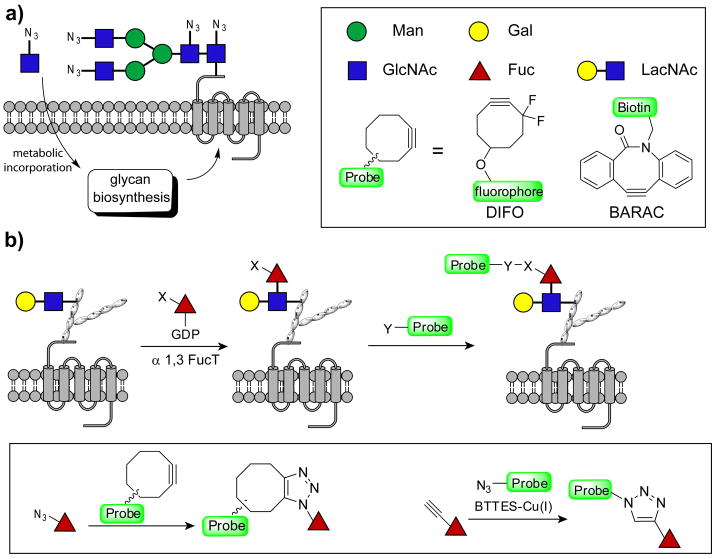
The glycome, the totality of glycans produced by a cell, is a dynamic indicator of the cell's physiology.[1] Changes in the glycome reflect a cell's developmental stage and the transformation state of a cell. Recently, imaging glycans in vivo has been enabled using a bioorthogonal chemical reporter strategy by treating cells or organisms with azide- or alkyne-tagged monosaccharide precursors.[2, 3] The modified monosaccharides, when taken up by cells, are activated in the cytoplasm to form nucleotide sugars, substrates of glycosyltransferases that generate complex glycans in the endoplasmic reticulum and Golgi. Once incorporated into cell surface glycoconjugates, the bioorthogonal chemical tags allow covalent conjugation with fluorescent probes for visualization,[2] or with affinity probes for enrichment and glycomic analysis.[4] This approach has been successfully used for the detection and imaging of mucin O-linked glycans,[2] sialylated[2] and fucosylated glycans,[5] and cytosolic O-GlcNAcylated proteins.[2] However, only monosaccharides are tracked by this strategy, and each monosaccharide is usually found on a plethora of glycans.[6] Higher order glycans, i.e. disaccharides or trisaccharides, of specific composition cannot be uniquely labeled by hijacking their biosynthetic pathways with unnatural monosaccharides (Figure 1a). Here we report a rapid and highly specific chemoenzymatic method for labeling cell surface glycans bearing a ubiquitous disaccharide—N-acetyllactosamine (LacNAc, Galβ1,4GlcNAc)—with biophysical probes for imaging or glycomic analysis.
Figure 1.
Labeling cell surface glycans with LacNAc using a two-step approach. a) Treating cells with unnatural monosaccharide precursors leads to general labeling of cell surface glycans containing that sugar. b) Specific labeling of glycans with LacNAc is achieved by an in situ fucosylation reaction with GDP-FucAz or GDP-FucAl followed by cyclooctyne-based copper-free click chemistry or BTTES-Cu(I)-catalyzed azide-alkyne cycloaddition.
LacNAc is widely distributed in most vertebrates, enveloped viruses, certain pathogenic bacteria and human parasites.[6] It is a universal component of complex and hybrid N-glycans as well as a few types of O-glycans and glycolipids.[6] Branched N- and O-glycans are modified in the trans Golgi by β(1,4)galactosyltransferases to generate the LacNAc disaccharide, which can be further elongated by β(1,3)N-acetylglucosaminyltransferases to ultimately form linear homopolymers of LacNAc of variable length, known as poly-LacNAc. The availability and localization of glycosyltransferases and donor substrates, i.e. UDP-GlcNAc and UDP-Gal, influences the biosynthesis and elongation of poly-LacNAc in the Golgi apparatus.[7] Terminal LacNAc may be capped by α-linked sialic acid, galactose or fucose added to the terminal galactose, or fucosylated on the internal N-acetylglucosamine residue to generate glycan epitopes such as Lewis X (Galβ1,4(Fucα1,3)GlcNAc, LeX) and sialyl Lewis X (Siaα2,3Galβ1,4(Fucα1,3)GlcNAc, sLeX). Many of these modifications are developmentally regulated.[8] Mice with homozygous knockout of β(1,4)galactosyltransferase-1, one of the β(1,4)galactosyltransferases that adds Gal to GlcNAc to form LacNAc, exhibit growth retardation and a markedly shortened life span.[8] Furthermore, cell-surface LacNAc levels are elevated in certain malignant tissues. For example, immunohistochemical analysis of normal mucosa and carcinoma of the human colorectum revealed a strong correlation between the level of cell-surface LacNAc and colorectal cancer.[9] LacNAc disaccharides are barely detectable in normal mucosa, but are markedly increased in carcinoma of the human colorectum. Thus, glycans with LacNAc constitute attractive targets for molecular imaging and potential biomarkers for cancer.
To detect glycans with LacNAc on the surface of live cells, we took advantage of a recently characterized recombinant H. pylori 26695 α(1,3)fucosyltransferase, which converts LacNAc to the LeX trisaccharide by specifically transferring a fucose residue from GDP-fucose (GDP-Fuc) to the 3-OH of the GlcNAc in the LacNAc unit.[10] Our studies showed that although this enzyme is highly specific for acceptor glycans possessing LacNAc, it has a relaxed specificity for the donor substrate GDP-Fuc—unnatural analogs bearing a wide variety of functional groups at the fucose C-5 position can be successfully transferred to LacNAc in vitro.[10] This observation echoed an earlier discovery by Palcic and coworkers that a human α(1,3)fucosyltransferase was promiscuous for C-5 modified GDP-Fuc analogues,[11] and suggested that LacNAc residues could be specifically labeled with an alkyne or azide tag via in situ fucosylation even in a complex cellular environment (Figure 1b). The labeled glycoconjugates could then be detected with azide- or cyclooctyne-functionalized probes via biocompatible copper-catalyzed azide-alkyne cycloadditon (CuAAC)[5] or copper-free click chemistry.[12] Here we use this strategy to detect the dynamic expression of LacNAc in glycans of mammalian cells, splenic lymphocytes and zebrafish embryos.
In order to perform live cell labeling, the α(1,3)fucosyltransferase had to be active under physiological conditions and added in concentrated form. However, storage of concentrated (>5mg/mL) wild-type H. pylori α(1,3)fucosyltransferase was complicated by dimerization followed by precipitation of the enzyme. Based on the X-ray crystal structure of a homologous α(1,3)fucosyltransferase from H. pylori (NCTC 11639),[13] we rationalized that cysteine 169 may be located on the surface of our enzyme and could thus be responsible for the observed protein dimerization. We created a C169S mutant of the wild-type fucosyltransferase (see Figure S1 in the Supporting Information) which maintained the high activity and selectivity of the parent enzyme towards LacNAc, as well as the promiscuity toward the GDP-Fuc analogs (see Table S1 and S2 in the Supporting Information). Furthermore, it could be stored at 4 °C over 3 months without loss of its catalytic activity. We termed the mutant α(1,3)FucT-M (M for monomer).
Lec2 cells, a Chinese hamster ovary (CHO) cell mutant, were chosen as a model system to explore the feasibility of cell-surface chemoenzymatic labeling of LacNAc due to its well-defined glycan complement.[14, 15] Lec2 cells have an inactive CMP-sialic acid Golgi transporter,[16] and consequently their cell surface complex/hybrid N-glycans mainly terminate in LacNAc residues. We treated Lec2 cells with GDP-Fuc and α(1,3)FucT-M in Hank's Buffered Salt Solution (HBSS) for 10 min at 37 °C. The treated cells were then stained with phycoerythrin (PE)-conjugated anti-Lex IgM and analyzed by flow cytometry (Figure 2a). Lec2 cells incubated with α(1,3)FucT-M showed a marked increase in PE fluorescence compared to untreated cells, confirming that cell surface LacNAc residues could be modified by this in situ fucosylation reaction. To examine if sialyl-LacNAc (sLacNAc) and internal LacNAc units could be labeled using this approach, we performed the same reaction with wide-type CHO cells and detected sLex and internal fucosylated LacNAc using anti-sLeX and anti-CD65s antibody VIM-2, respectively. Significant labeling was achieved in both cases, suggesting that sLacNAc and internal LacNAc units could be labeled using this strategy (Figure S2).
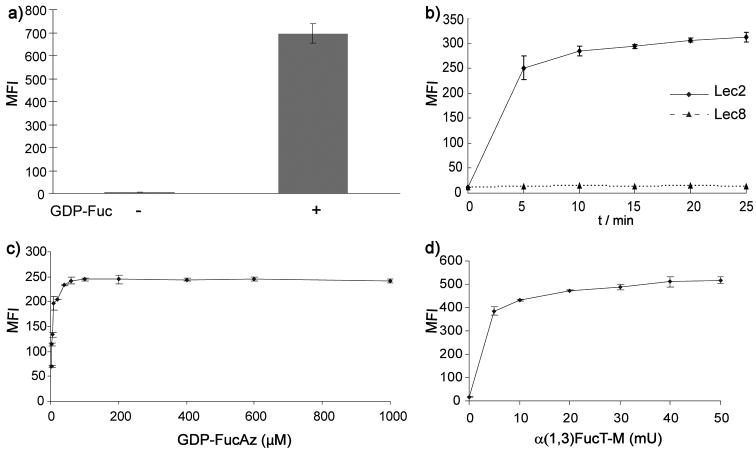
Figure 2.
Detection of cell surface glycans with LacNAc using flow cytometry. a) Lec2 CHO cells were treated with or without GDP-Fuc (500μM) in the presence of α(1,3)FucT-M for 10 min, and probed with PE-conjugated anti-LeX IgM. b) Time-dependent labeling of cell surface glycans with LacNAc. Lec2 and Lec8 CHO cells were treated with GDP-FucAz (500 μM) and α(1,3)FucT-M (30 mU) for 0–25 min. The labeled cells were then reacted with BARAC-biotin (10 μM) for 10 min and then probed with streptavidin-Alexa Fluor 488. c) and d) dose-dependent labeling of cell surface glycans with LacNAc. c) 30 mU α(1,3)FucT-M, labeling time for 10 min. d) 500 μM GDP-FucAz, labeling time for 10 min. Error bars represent the standard deviation of three replicates in one experiment.
To test the feasibility of incorporating 6-azido-fucose into cell-surface LacNAc residues using the same approach, we treated Lec2 cells in medium supplemented with the α(1,3)FucT-M (30 mU) and GDP-6-azidofucose (GDP-FucAz, 500 μM). After the in situ fucosylation, cells were reacted with biarylazacyclooctynone-biotin[17] (BARAC-biotin) via copper-free click chemistry and then stained with streptavidin-Alexa Fluor 488 for flow cytometry analysis. The GDP-FucAz-treated Lec2 cells showed robust azide-specific labeling with no significant background labeling compared to cells treated with GDP-Fuc. Remarkably, as low as 100 μM GDP-FucAz was required for a 10-min in situ fucosylation reaction to transfer a sufficient number of azide residues to the cell surface for detection by the second step click-labeling reaction (Figure 2b–d). Lec8 CHO cells, which do not express LacNAc due to a mutation in the UDP-Gal Golgi transporter,[18] were used as a negative control. The reduced fluorescence displayed by Lec8 cells compared to Lec2 cells showed that FucAz was selectively attached to cell-surface LacNAc in Lec2 (Figure 2b and Figure S3). Similarly, Lec2 cells could be labeled with alkynyl tags by treating them with α(1,3)FucT-M and GDP-6-alkynyl-fucose (GDP-FucAl), and detected via BTTES-promoted CuAAC, a biocompatible version of the canonical CuAAC (Figure S4).[5] The labeled LacNAc in CHO glycoproteins was shown to be primarily located in complex N-glycans.[15] Peptide: N-Glycosidase F (PNGase F)-treated and untreated control lysates were reacted with biotin-azide via the BTTES-mediated CuAAC, and probed with anti-biotin Western blot. As expected, PNGase F-treatment essentially abolished LacNAc-associated labeling (Figure S5).
To evaluate if the in situ fucosylation reaction causes any long term perturbation to treated cells, we labeled Lec2 cells with the GDP-Fuc analogs in the presence of α(1,3)FucT-M for 10 min, and cultured the treated cells for three days. Viable cells, based on Trypan Blue assay, were counted each day. Cells labeled with natural and unnatural fucose proliferated at similar rates as untreated cells (see Figure 6 in the Supporting Information). Taken together, this two-step chemoenzymatic method creates a nontoxic and highly efficient approach for labeling LacNAc disaccharides, setting the stage for use in the imaging of glycans with LacNAc on the surface of live cells.
To evaluate whether in situ fucosylation could selectively target LacNAc-bearing cells in a mixed cell population, we first labeled Lec2 CHO cells with cytosolic fluorescein as their identity marker. We then mixed Lec2 and Lec8 cells in a 1:5 ratio and cultured them on covered glass slides. After 3 days, we treated the cells with GDP-FucAz in the presence of α(1,3)FucT-M for 15 min, followed by reaction with Alexa Fluor 647 conjugated difluorinated cyclooctyne (DIFO-647, 25 min)[19] via copper-free click chemistry. Then we examined the labeled cells using fluorescence microscopy. Membrane-associated-647 fluorescence (red, arbitrary) was only observed for Lec2 cells (green), whereas no labeling could be detected for the neighboring Lec8 cells that were unstained (Figure 3a). Similar results were obtained using flow cytometry analysis (Figure S7).
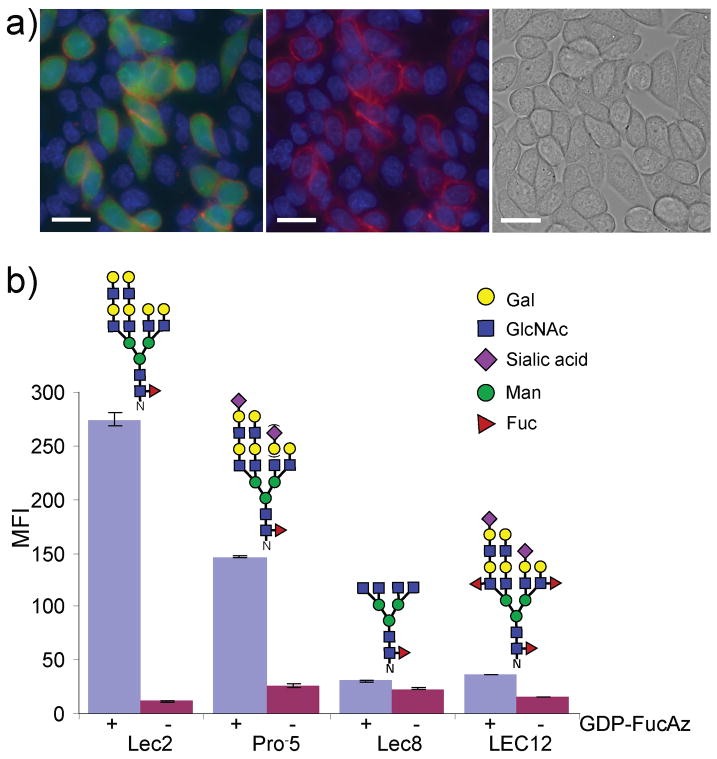
Figure 3.
Tracking of glycans with LacNAc on CHO glycosylation mutants. a) Imaging cell-surface glycans with LacNAc. Lec2 cells stained with CMFDA (fluorescein, green) and unstained Lec8 cells were mixed at a 1:5 ratio and were cultured for 3 days. The cells were treated with 500 μM GDP-FucAz and 30 mU α(1,3)FucT-M for 15 min, then labeled with 20 μM DIFO-647 for 25 min. The Alexa Fluor 647 image merged with the fluorescein and Hoechst 33342 images (left); the Alexa Fluor 647 image merged with the Hoechst 33342 image (middle); the bright field image (right). Scale bar: 20 μm. b) wild-type CHO, and mutant Lec2, Lec8, and LEC12 CHO cells were treated with 500 μM GDP-FucAz in the presence of 30 mU α(1,3)FucT-M for 10 min, then labeled with 10 μM BARAC-biotin for 10 min. The labeled cells were then probed with streptavidin-Alexa Fluor 488. Error bars represent the standard deviation of three replicates. The N-glycan shown above each cell line is representative of major N-glycans produced.[15]
The intrinsic expression patterns of glycan synthesizing enzymes play a major role in determining the quantity and distribution of cell surface LacNAc in individual cell types. The chemoenzymatic method described here should allow the determination of the cell surface LacNAc levels in closely related cell types. We chose to compare wild-type CHO cells and three CHO mutants, Lec2, Lec8, and LEC12, due to their well-characterized glycosylation patterns (Figure 3b).[14, 15] CHO cells were cultured in media under identical conditions for two days before being subjected to in situ fucosylation with α(1,3) FucT-M and GDP-FucAz. The fucosylated cells were then reacted with BARAC-biotin and stained with streptavidin-Alexa Fluor 488 for flow cytometry analysis. As shown in Figure 3b and Supporting Information Figure 8, Lec2 cells displayed the highest levels of fluorescence among the four CHO cell lines, followed by wild-type CHO and LEC12 cells, with Lec8 cells showing the weakest fluorescence signal. These observations reflect the different glycosylation complements of these cells.[15] Wild-type CHO cells have a similar spectrum of complex N-glycans to Lec2 mutants. However, they have an active CMP-sialic acid Golgi transporter, and thus a majority of their cell surface LacNAc units are capped with α(2,3)sialic acid.[14] The presence of bulky and charged sialic acid residues appears to slow down the fucosylation reaction significantly (Table S2), giving weaker labeling. In contrast to CHO and Lec2 cells, LEC12 CHO mutant cells express active α(1,3)fucosyltransferase IX[20] which converts many LacNAc units endogenously into Lex, leaving few unmodified LacNAc units on the cell surface as substrates of the in situ fucosylation reaction. Finally, Lec8 mutants express few cell surface LacNAc disaccharides due to a mutation in the UDP-Gal Golgi transporter, and therefore displayed the weakest labeling.
The pattern and quantity of glycans expressed by a cell correlates with its developmental stage or activation status.[8] To investigate whether the chemoenzymatic approach may be used to discriminate LacNAc levels on the surface of lymphoid cells with distinct activation status, we performed the in situ fucosylation reaction on splenocytes from wild-type mice. The treated splenocytes were then reacted with BARAC-biotin. Subsequently, the biotinylated or unbiotinylated (control) cells were probed with a fluorophore-coupled streptavidin, along with T and B cell-specific surface markers (CD4, CD8 and B220), and two well-established activation/memory markers, CD44 and CD62L, to discriminate between the activated and naïve cells (Figure 4). While control-treated cells did not exhibit any LacNAc-positive staining, the distinct lymphocyte populations were differentially stained, indicating that they express LacNAc at different levels or on different glycans. Most interestingly, cells exhibiting an activated/memory phenotype (CD44highCD62Llow, CD25+) exhibited greater fluorescence compared to their naïve counterparts (CD44lowCD62Lhigh, CD25-). Thus, our approach can serve to discriminate differentially activated cell subsets ex vivo (Figure 4 and Figure S9).
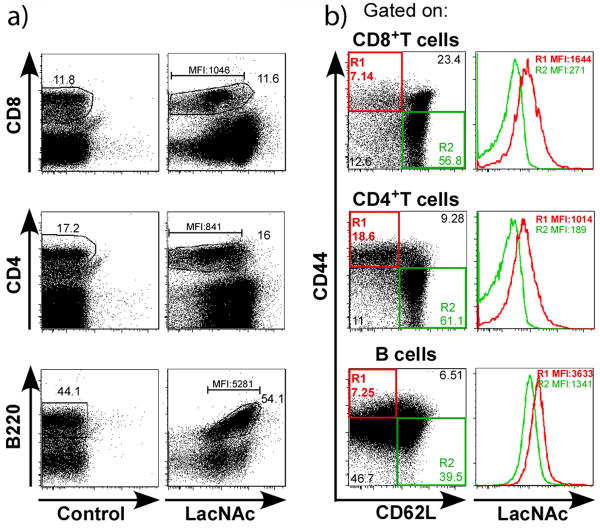
Figure 4.
Activated or memory lymphocytes express higher levels of LacNAc disaccharides. Spleen cells from wild type naïve mice were treated with 500 μM GDP-FucAz or no reagents (control) in the presence of 30 mU α(1,3) FucT-M for 20 min, and then reacted with BARAC-biotin. The biotinylated or unbiotinylated (control) cells were incubated with streptavidin-APC and a cocktail of antibodies that defines distinct populations of lymphocytes, e.g. T (anti-CD4, anti-CD8), B (anti-B220) lymphocytes, as well as activation markers (anti-CD44 and anti-CD62L). a) Dot plots show LacNAc-labeled cells (or controls) with different mAbs. b) Left panels show the cell-surface expression of CD44 and CD62L activation markers gated on each lymphocyte population, and right panels show the co-expression of LacNAc on activated (CD44highCD62Llow) or naïve (CD44lowCD62Lhigh) lymphocytes. Dot plots are representative of 9 individual mice and 4 independent experiments both on C57BL/6 (or BALB/c) genetic backgrounds.
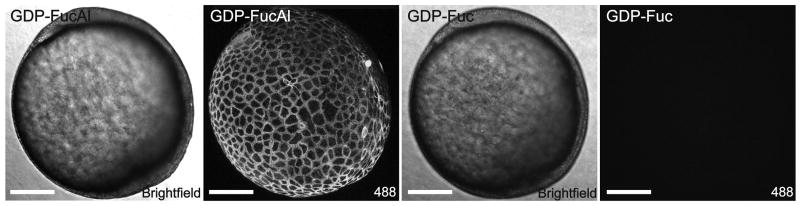
The development of a multicellular organism as it grows from a single zygote to a complex system of tissues is accompanied by complex changes of cell surface glycosylation. To test if the chemoenzymatic approach could be extended to image glycans bearing LacNAc in vivo, we took advantage of the rapid embryonic development and optical clarity of the zebrafish embryo as a vertebrate model system. At the tail bud stage (∼10 hpf), we reacted the embryos (n = 25) with GDP-FucAl in the presence of α(1,3)FucT-M. After a 10-min reaction, we rinsed the embryos and incubated them with Alexa Fluor 488-azide (100 μM) and the BTTES-Cu(I) catalyst to detect LacNAc-bearing glycans labeled by the alkyne. Immediately following a 5-min reaction, we were able to observe robust labeling of the treated embryos (Figure 5). Only background fluorescence was detected for control embryos treated with GDP-Fuc. After the reaction, we followed the development of the labeled embryos for four days, and we observed no developmental defects as compared to their untreated counterparts, suggesting that our labeling approach was well tolerated by the zebrafish embryos (Figure S10).
Figure 5.
In vivo imaging of glycans with LacNAc during zebrafish embryogenesis. Zebrafish embryos at tail bud stage were treated with 500μM GDP-FucAl or GDP-Fuc in the presence of 30 mU α(1,3)FucT-M for 10 min. The embryos were then reacted with Alexa Fluor 488-azide catalyzed by BTTES-Cu(I) for 5 min and imaged using confocal microscopy. Scale bar: 200 μm.
In conclusion, the two-step chemoenzymatic approach described here offers a practical and versatile method for site-specific chemical labeling of LacNAc-bearing glycans in live cells. A 10-min in situ fucosylation at 37 °C is typically sufficient to install enough azide or alkyne residues for the second step bioorthogonal click reaction. Furthermore, we discovered that the fucosyltransferase can accept donor substrates with functional groups larger than the azide introduced at the C-6 position of the fucose; we were able to transfer a fluorescein-conjugated GDP-Fuc analog to glycans with LacNAc on the surface of Lec2 cells directly (Figure S11), thereby alleviating the second step click reaction. We expect that further engineering of this fucosyltransferase will produce a versatile enzyme that is capable of accepting an even broader spectrum of unnatural GDP-Fuc analogs (e. g. biotin conjugates). Our fucosylation method also allowed probing of LacNAc levels on lymphocytes exhibiting different activation status as well as noninvasive imaging of LacNAc-bearing glycans in zebrafish embryos.
This method is far superior to the traditional lectin-based methods to detect LacNAc, in which detection and enrichment of LacNAc or poly-LacNAc-bearing glycans are realized using lectins such as Erythrina cristagalli (ECA) and Lycopersicon esculentum (LEA).[21] These lectins have comparatively low affinity and are not specific for LacNAc. For example, ECA binds to GalNAc and Gal residues[22] and LEA interacts with oligomers of β1,4–linked GlcNAc.[23] Furthermore, being of plant origin, these LacNAc-binding proteins are often toxic. Therefore, the utility of these lectins for labeling LacNAc in living systems is limited.[2] Finally, since azido or alkynyl-tags can be metabolically incorporated into cellular monosaccharides (e.g. sialic acids, N-acetylgalactosamine),[2, 24] exploiting both approaches would enable us to image multiple glycan types simultaneously in developing zebrafish and other living systems.
Supplementary Material
Footnotes
Dedicated to Professor K. Barry Sharpless on the occasion of his 70th birthday
This work was supported by the National Institutes of Health (R01GM093282 to P.W. and R01CA036434 to P.S.) and the Mizutani Foundation for Glycoscience (to P.W.). Y. L. was supported by the Office of Science, Office of Basic Energy Sciences, of the U. S. Department of Energy under contract No. DE-AC02-05 CH11231. We thank Prof. Carolyn Bertozzi for providing DIFO-647.
Supporting information for this article is available on the WWW under http://www.angewandte.org.
Contributor Information
Tianqing Zheng, Department of Biochemistry, Albert Einstein College of Medicine of Yeshiva University, 1300 Morris Park Ave, Bronx, NY 10461 (USA), Fax: (+1) 718-678-1022.
Dr. Hao Jiang, Department of Biochemistry, Albert Einstein College of Medicine of Yeshiva University, 1300 Morris Park Ave, Bronx, NY 10461 (USA), Fax: (+1) 718-678-1022
Dr. Marilyn Gros, Department of Microbiology and Immunology, Albert Einstein College of Medicine of Yeshiva University
Dr. David Soriano del Amo, Department of Biochemistry, Albert Einstein College of Medicine of Yeshiva University, 1300 Morris Park Ave, Bronx, NY 10461 (USA), Fax: (+1) 718-678-1022
Subha Sundaram, Department of Cell Biology, Albert Einstein College of Medicine of Yeshiva University.
Prof. Grégoire Lauvau, Department of Microbiology and Immunology, Albert Einstein College of Medicine of Yeshiva University
Prof. Florence Marlow, Department of Developmental and Molecular Biology, Albert Einstein College of Medicine of Yeshiva University
Dr. Yi Liu, The Molecular Foundry, Lawrence Berkeley National Laboratory, One Cyclotron Road, Berkeley, CA 94720 (USA)
Prof. Pamela Stanley, Department of Cell Biology, Albert Einstein College of Medicine of Yeshiva University
Prof. Peng Wu, Email: peng.wu@einstein.yu.edu, Department of Biochemistry, Albert Einstein College of Medicine of Yeshiva University, 1300 Morris Park Ave, Bronx, NY 10461 (USA), Fax: (+1) 718-678-1022.
References
- 1.Freeze HH. Nat Rev Genet. 2006;7:537–551. doi: 10.1038/nrg1894. [DOI] [PubMed] [Google Scholar]
- 2.Laughlin ST, Bertozzi CR. Proc Natl Acad Sci U S A. 2009;106:12–17. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baskin JM, Bertozzi CR. Qsar Comb Sci. 2007;26:1211–1219. [Google Scholar]
- 4.Hanson SR, Hsu TL, Weerapana E, Kishikawa K, Simon GM, Cravatt BF, Wong CH. J Am Chem Soc. 2007;129:7266–7267. doi: 10.1021/ja0724083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soriano del Amo D, Wang W, Jiang H, Besanceney C, Yan AC, Levy M, Liu Y, Marlow FL, Wu P. J Am Chem Soc. 2010;132:16893–16899. doi: 10.1021/ja106553e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Eseentials of Glycobiology. 2. Cold Spring Harbor; New York: 2008. [PubMed] [Google Scholar]
- 7.Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 8.Haltiwanger RS, Lowe JB. Annu Rev Biochem. 2004;73:491–537. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- 9.Ichikawa T, Nakayama J, Sakura N, Hashimoto T, Fukuda M, Fukuda MN, Taki T. J Histochem Cytochem. 1999;47:1593–1602. doi: 10.1177/002215549904701211. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Hu T, Frantom PA, Zheng T, Gerwe B, Del Amo DS, Garret S, Seidel RD, 3rd, Wu P. Proc Natl Acad Sci U S A. 2009;106:16096–16101. doi: 10.1073/pnas.0908248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srivastava G, Kaur KJ, Hindsgaul O, Palcic MM. J Biol Chem. 1992;267:22356–22361. [PubMed] [Google Scholar]
- 12.Jewett JC, Bertozzi CR. Chem Soc Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin SW, Yuan TM, Li JR, Lin CH. Biochemistry. 2006;45:8108–8116. doi: 10.1021/bi0601297. [DOI] [PubMed] [Google Scholar]
- 14.Patnaik SK, Stanley P. Methods Enzymol. 2006;416:159–182. doi: 10.1016/S0076-6879(06)16011-5. [DOI] [PubMed] [Google Scholar]
- 15.North SJ, Huang HH, Sundaram S, Jang-Lee J, Etienne AT, Trollope A, Chalabi S, Dell A, Stanley P, Haslam SM. J Biol Chem. 2010;285:5759–5775. doi: 10.1074/jbc.M109.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckhardt M, Gotza B, Gerardy-Schahn R. J Biol Chem. 1998;273:20189–20195. doi: 10.1074/jbc.273.32.20189. [DOI] [PubMed] [Google Scholar]
- 17.Jewett JC, Sletten EM, Bertozzi CR. J Am Chem Soc. 2010;132:3688–3690. doi: 10.1021/ja100014q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oelmann S, Stanley P, Gerardy-Schahn R. J Biol Chem. 2001;276:26291–26300. doi: 10.1074/jbc.M011124200. [DOI] [PubMed] [Google Scholar]
- 19.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Proc Natl Acad Sci U S A. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patnaik SK, Zhang A, Shi S, Stanley P. Arch Biochem Biophys. 2000;375:322–332. doi: 10.1006/abbi.1999.1693. [DOI] [PubMed] [Google Scholar]
- 21.Lanteri M, Giordanengo V, Hiraoka N, Fuzibet JG, Auberger P, Fukuda M, Baum LG, Lefebvre JC. Glycobiology. 2003;13:909–918. doi: 10.1093/glycob/cwg110. [DOI] [PubMed] [Google Scholar]
- 22.Lis H, Sharon N. Methods Enzymol. 1987;138:544–551. doi: 10.1016/0076-6879(87)38049-8. [DOI] [PubMed] [Google Scholar]
- 23.Nachbar MS, Oppenheim JD, Thomas JO. J Biol Chem. 1980;255:2056–2061. [PubMed] [Google Scholar]
- 24.Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.