Abstract
Objective
Our goal was to evaluate the influence of quality control (QC) decisions using two genotype calling algorithms, CRLMM and Birdseed, designed for the Affymetrix SNP Array 6.0.
Methods
Various QC options were tried using the two algorithms and comparisons were made on subject and call rate and on association results using two data sets.
Results
For Birdseed, we recommend using the contrast QC instead of QC call rate for sample QC. For CRLMM, we recommend using the signal-to-noise rate ≥4 for sample QC and a posterior probability of 90% for genotype accuracy. For both algorithms, we recommend calling the genotype separately for each plate, and dropping SNPs with a lower call rate (<95%) before evaluating samples with lower call rates. To investigate whether the genotype calls from the two algorithms impacted the genome-wide association results, we performed association analysis using data from the GENOA cohort; we observed that the number of significant SNPs were similar using either CRLMM or Birdseed.
Conclusions
Using our suggested workflow both algorithms performed similarly; however, fewer samples were removed and CRLMM took half the time to run our 854 study samples (4.2 h) compared to Birdseed (8.4 h).
Key Words: Genotype call, Birdseed, CRLMM, Quality control decisions, Association
Background
With the advances of technology, several DNA analysis arrays have been developed to measure variation in single nucleotide polymorphisms (SNPs) and copy number variation (CNV). In this paper, we focused on the Affymetrix Genome-Wide Human SNP Array 6.0 (Affy SNP 6.0) that features 1.8 million genetic markers, including more than 906,600 SNPs and more than 946,000 additional probes for the detection of CNV. This chip was an updated version of the previous two-chip design based on the NspI and StyI restriction enzymes.
There are two genotype calling algorithms (GCAs) broadly used for the Affy SNP 6.0: Birdseed and CRLMM. Birdseed is an evolution of the Bayesian Robust Linear Model with Mahalanobis distance classifier (BRLMM) GCA [1,2] and is the default algorithm used by Affymetrix [3]. It performs a multiple-chip analysis to estimate signal intensity for each allele of each SNP, fitting probe-specific effects to increase precision (similar to the BRLMM-P algorithm) [2]. The Birdseed algorithm then makes genotype calls by fitting a Gaussian mixture model in the two-dimensional A allele intensity signal versus B allele intensity signal space, using SNP-specific models to increase performance. Carvalho et al. [4] observed that this algorithm did not perform well for data from different laboratories and developed a new algorithm named CRLMM (corrected RLMM) to correct for various batch-related effects. Similarly, Lin et al. [5] observed, using HapMap samples, that Birdseed had similar limitations to BRLMM in cross-laboratory stability of their calls. They also found CRLMM provided confidence measures that better correlated with observed accuracy.
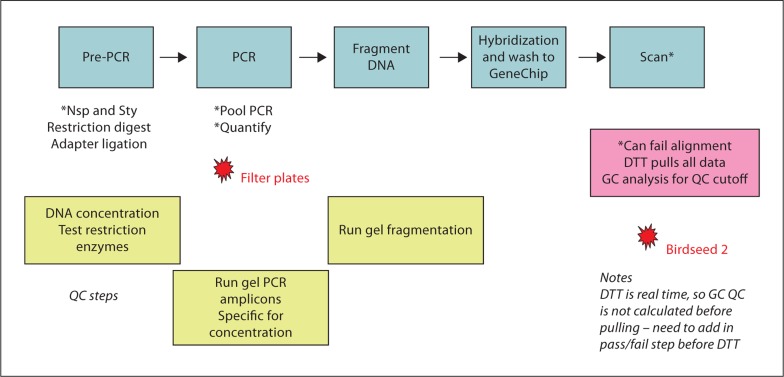
As with many new technologies, the Affymetrix analysis workflow is still a work in progress [6,7,8,9,10]. In the laboratory, a sequence of time-consuming steps must be performed taking one lab technician five days to process a sample (fig. 1). In our experience, this platform is very sensitive to sample DNA quality such as concentration, buffer solution, and DNA degradation, furthering the challenges within the laboratory. In addition to sample quality problems, we experienced issues with hardware such as filter plate failures during the PCR step and alignment problems during the scan process, further delaying the sample processing. Besides challenges in the laboratory, the recommended analysis software has undergone several revisions. Initially the Birdseed-v1 algorithm was used and one year later this was updated to Birdseed-v2 [9,10] to fix several bugs. Additionally, the quality control (QC) step used to identify poor quality samples was modified in February 2008 by Affymetrix [8]. The QC call rate (QCR) was used first, identifying samples with values less than 0.86 as ‘failures’ that needed to be repeated [7]. However, QCR was shown to be of limited value for predicting genotyping performance in problematic samples [11]. Thus, the current recommended algorithm is the contrast QC (CQC), where values less than 0.40 are considered failures [8,11]. Although the QCR is no longer the recommended QC step, it is still being used by some genotyping facilities.
Fig. 1.
Description of the Affymetrix SNP Array 6.0 processing. This workflow details the steps required for a single individual performing the process. The QC steps are essential: verifying restriction enzyme activity and DNA concentration (by pico green); verifying PCR by gel analysis, and lastly verifying optimal fragmentation. Any sample that does not pass these steps is not applied to the Affymetrix array. The two starred points (filter plates and Birdseed 2) indicate the unreliability of filter plates recommended by Affymetrix and Birdseed 2 that cannot be recalled.
Thus, our objective was to evaluate the influence of QC decisions for the Affymetrix SNP Array 6.0 using two GCAs, CRLMM and Birdseed v2, and determine whether these decisions had an effect on association analysis results.
Methods
Data
Two data sets were used in our evaluation. The first data set included samples from 854 non-Hispanic white adults enrolled in the Genetic Epidemiology Network of Arteriopathy (GENOA) study of the Family Blood Pressure Program (FBPP), a study designed to identify genetic determinants of hypertension in multiple ethnic groups [12]. The second data set included samples from 746 non-Hispanic white adults enrolled in the Genetic Epidemiology study of Chronic Lymphocytic Leukemia (CLL) [13]. These samples were genotyped at the same time at the Genotyping Shared Resource of the Mayo Clinic Technology Center using Affymetrix SNP Array 6.0.
Affy SNP 6.0
There are 934,968 SNPs on the 6.0 chip. Of these, 3,022 are duplicates and were included purely for QC purposes (2,906 of these have rsIDs). An additional 25,346 SNPs have been excluded from the officially released SNPs due to poor performance. The default extraction of the SNPs using most software packages yielded only 909,622 SNPs. Once the duplicates were excluded, 906,600 SNPs remained for analysis (table 1).
Table 1.
Distribution of the 906,600 SNPs
| 1 | 71,312 | 14 | 28,065 |
| 2 | 73,937 | 15 | 26,074 |
| 3 | 60,685 | 16 | 27,716 |
| 4 | 55,995 | 17 | 20,658 |
| 5 | 56,416 | 18 | 26,529 |
| 6 | 56,271 | 19 | 11,929 |
| 7 | 47,056 | 20 | 22,843 |
| 8 | 48,608 | 21 | 12,579 |
| 9 | 41,442 | 22 | 11,537 |
| 10 | 48,195 | X | 36,886 |
| 11 | 44,539 | Y | 258 |
| 12 | 42,555 | MT | 119 |
| 13 | 34,283 | Unknown | 113 |
Birdseed Algorithm
Birdseed-v2 uses the expectation-maximization (EM) algorithm to derive a maximum likelihood fit of a 2-dimensional Gaussian mixture model in the A versus B allele space. A key difference between Birdseed-v1 and Birdseed-v2 is that the former uses SNP-specific models or priors only as an initial condition from which the EM fit is free to wander; on rare occasions this allows for mislabeling of the clusters. For Birdseed-v2 the SNP-specific priors are used not only as initial conditions for EM, but are incorporated into the likelihood as Bayesian priors. This constrains the extent to which the EM fit can wander off. Correctly labeling SNP clusters, whose centers have shifted relative to the priors, is problematic for both Birdseed versions. However, given the additional constraint on the EM fit, Birdseed-v2 is more likely than Birdseed-v1 to either correctly label the clusters or set genotypes to no-calls [8]. The algorithm by default uses minor allele frequency (MAF) values from the Caucasian HapMap data as ‘seeds’. It is also possible to specify your own ‘seeds’ based on other data sources. Each call is given a confidence score that varies from 0 (best) to 1 (worst). Confidence is calculated as Confidence = 80% × E1 + 20% × E2, where E1 is the posterior probability (PP) ratio between the 2nd closest peak to the closest peak, and E2 is the deviation penalty from the closest peak. From the confidence we can stipulate a confidence score threshold, which is the maximum score at which the Birdseed algorithm will make a genotype call. Larger values of the confidence threshold indicate less certain calls. Calls with confidence scores above the threshold are assigned a no-call. The confidence score threshold default is 0.5.
There are two QC assessments available for use prior to executing the Birdseed calling algorithm: QCR and CQC. The QCR uses a dynamic model QCR analysis of a subset of 3,022 duplicate SNPs following chip scanning. However, the QCR was shown to be dependent on sample quality, and it did not measure the degree to which the perfect match intensities of all SNPs cluster by genotype. In high quality samples, the A and B allele probe intensities display three clusters for AA, AB, and BB genotypes. For poor quality samples, these clusters will merge. Since the genotype calling is based on cluster resolution, this QC algorithm was not ideal. The CQC algorithm was developed to better track the genotyping call rates yielded by the Birdseed algorithm. This algorithm measures the separation of allele intensities into three clusters in a single dimension generated by the projection of the two-dimensional allele intensity space. A detailed description of these two QC algorithms is described in the Affymetrix White Paper [10]. According to the initial Affymetrix SNP 6.0 workflow, re-processing was recommended for samples with a QCR <0.86 [6]. The current Affymetrix recommendation is to re-process samples with CQC <0.4 [9,10].
CRLMM Algorithm
CRLMM uses a supervised learning approach based on a two-stage hierarchical model [4]. Unlike other algorithms, CRLMM models M = log2 (IA) – log2 (IB) instead of the intensity pair, where IA and IB are the intensity of alleles A and B, respectively. This choice makes CRLMM more robust to probe effects [Carvalho, unpubl. data]. To account for the dependence of M on the overall intensity S, defined as S = [log2(IA) + log2(IB)]/2, Carvalho et al. [4] fitted splines using a mixture model and corrected the bias using the fitted curves. Then, for a given SNP, the distribution of M, conditioned on genotype, is modeled as Gaussian. To account for the remaining probe effect, each SNP has a different mean and standard deviation which are estimated from HapMap samples. Because of its design, HapMap provides few rare genotypes in some SNPs, so Carvalho et al. [4] used an empirical Bayes approach in which the means, conditioned on genotype, follows a multivariate normal distribution and the variances have a scale-inverse χ2 distribution. This approach permits CRLMM to borrow strength from other SNPs. To make calls, CRLMM treats the estimated parameters as known and computes PPs for each genotype given the observed log-ratio M. The posteriors are then used as a confidence measure. Lin et al. [5] found that the confidence measures provided by CRLMM were not optimal and proposed an ad-hoc adjustment based on a training approach. These are the confidence measures currently implemented in CRLMM. There are other measures such as SNPQC and SNR. SNPQC is a quality score for each SNP, and a SNPQC value >0.25 indicates a SNP with very accurate genotype call. SNR is a signal-to-noise ratio quantity, and when it is close to zero it is impossible to distinguish between homozygous and heterozygous clusters [4]. By default, CRLMM considers SNPs with SNR >5.0 to be of excellent quality.
The CRLMM workflow requires the user to make several decisions about which samples and genotype calls are going to be trusted. The impact of choosing different cut-off values for the sample quality scores, SNR, and for the PP should be investigated for each study. In our investigation we tried the following:
– SNR cutoffs of 3, 4, and 5
– PPs of 75, 80, 85, 90, and 95%.
Additional Decisions
Our recommended workflow for Birdseed and CRLMM algorithms is shown in table 2. However, there are additional decisions that need to be made by the investigator which can have a large influence on the final results. After applying the appropriate QC algorithm, samples can be called separately for each plate (individual) or using all samples together (pooled). The current workflows leave this decision up to the investigator. Additionally, order matters when determining call rates for SNPs and subjects. We investigated the impact of dropping SNPs with call rates <95%, then dropping subjects with call rates <95% or reversing the order and dropping subjects first.
Table 2.
Suggested workflow for Birdseed and CRLMM
| Birdseed | CRLMM | Action |
|---|---|---|
| QC contrast ≥ 0.4 | SNR ≥ 4.0 | Remove low quality samples |
| Confidence measure < 5 | PP < 0.90 | Set unreliable genotype calls to missing |
| SNP call rate ≥ 95% | SNP call rate ≥ 95% | Remove SNPs with a low call rate |
| Sample call rate ≥ 95% | Sample call rate ≥ 95% | Remove samples with a low call rate |
Statistical Analysis
For the association analysis we used the quantitative trait, estimated glomerular filtration rate (eGFR), adjusting for age and sex, available only in the GENOA data set, and ran a mixed effects linear model taking into account the familial relationship among the sibships using the R library multic (http://mayoresearch.mayo.edu/mayo/research/biostat/splusfunctions.cfm) for the association analysis. We used PLINK for data management, for calculating identify-by-state analysis to confirm familial relationships and to identify hidden familial relationships, and for performing population stratification analysis [14].
Results
Comparison of the Birdseed QC Measures
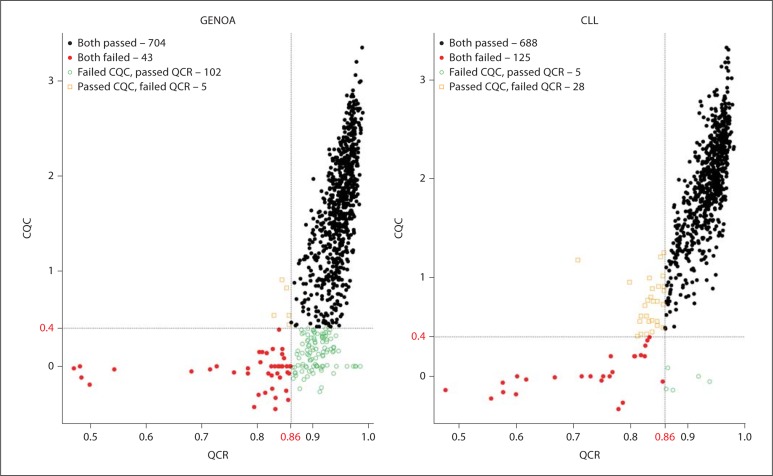
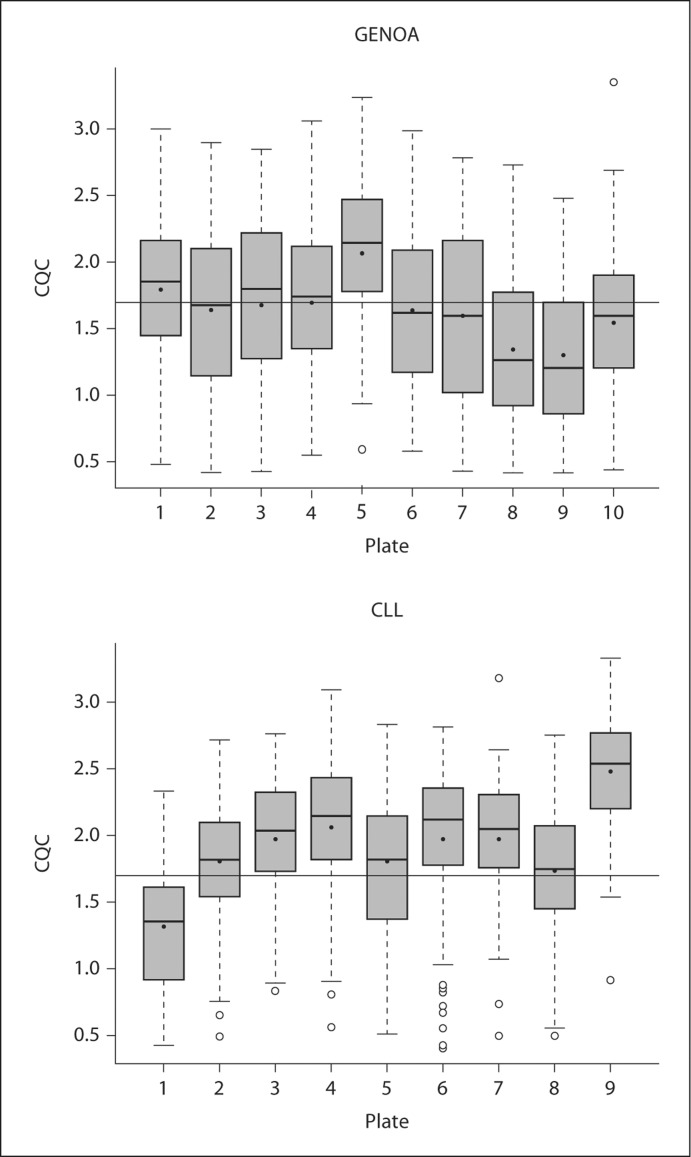
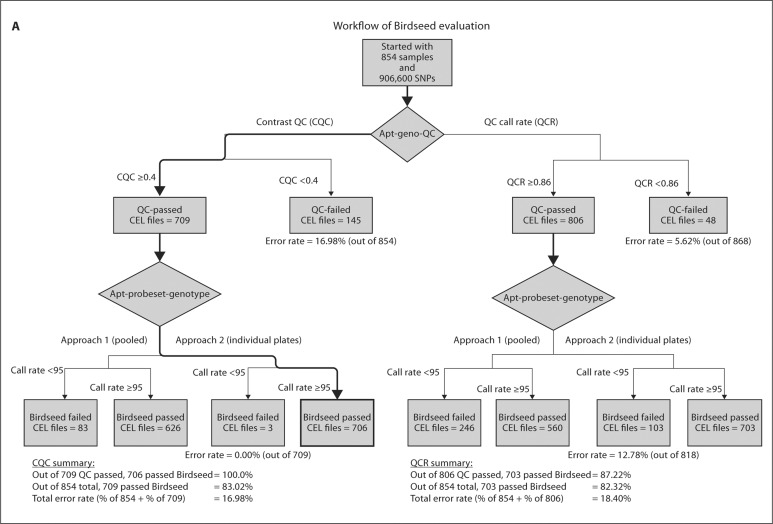
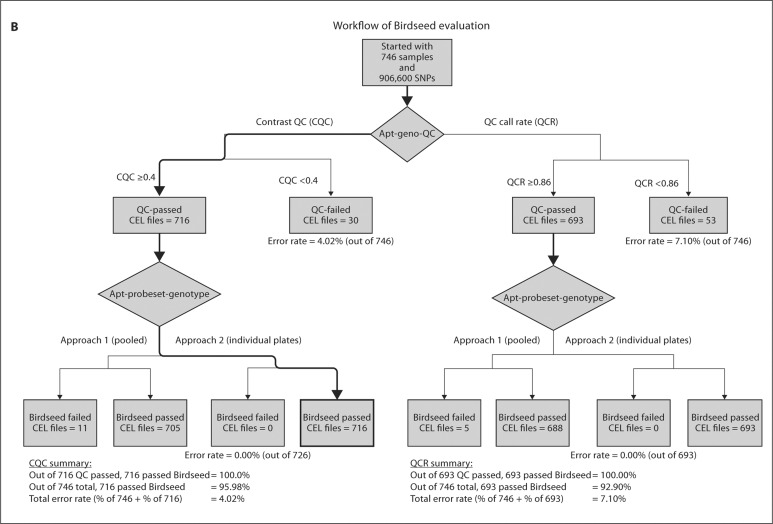
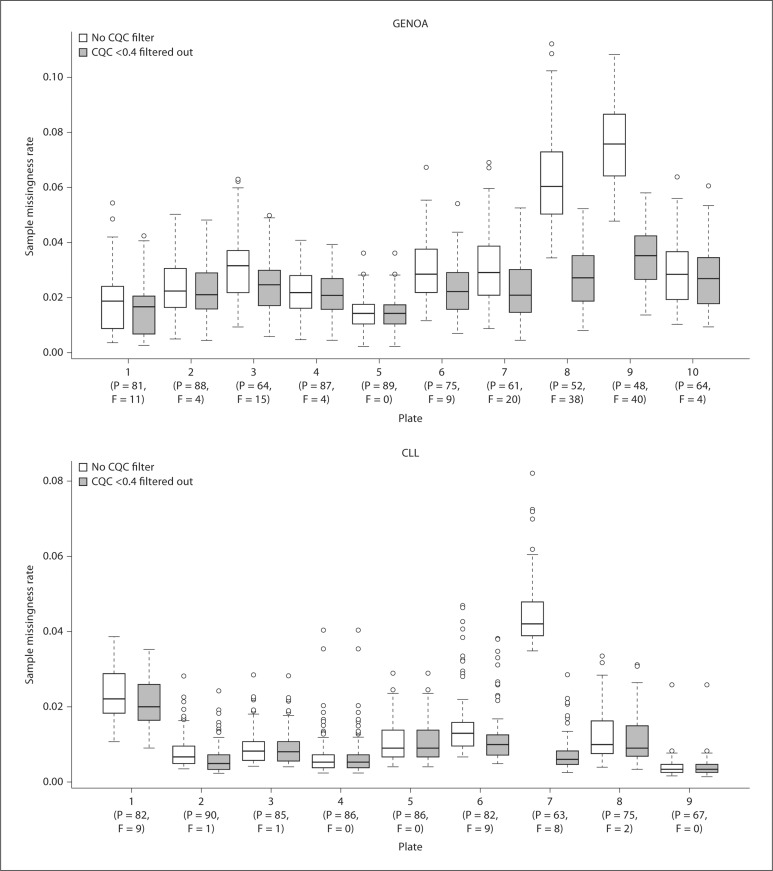
The purpose of QCR and CQC is to identify samples that are of poor quality and might negatively influence the Birdseed calling of other subjects. Figure 2 compares the QCR and CQC values for each sample for the GENOA and CLL cohorts, where 94 and 93% of the samples passed QCR and 83 and 96% passed CQC, respectively. Additionally, the most recent Affy SNP 6.0 user guide [10] suggests that plates should be flagged as potentially problematic if less than 90% of a given plate passes the CQC threshold or if the average CQC of those samples that passed is less than 1.7. For the GENOA samples, the mean and median CQC over all the plates were 1.67 and 1.72, respectively, and for the CLL samples, the mean and median CQC over all the plates were 1.81 and 1.91, respectively. As shown in figure 3, these values can vary significantly by plate. Based on this recommendation, 60% of the GENOA plates were flagged as opposed to 11% of the CLL plates. Despite this recommendation we did not exclude these plates from our evaluation because we were interested in assessing how the performance of these algorithms impacts the performance of the Birdseed calling algorithm. As was observed by Affymetrix White Paper [10], we also observed that although more samples passed the initial screening using QCR, more samples ultimately passed Birdseed using CQC suggesting that the ‘bad’ samples perhaps contaminated the results. Of the GENOA and CLL samples that passed QCR (n = 806 and 693, fig. 2A, B), 703 (87.2%, fig. 4A) and 693 (93%, fig. 4B) passed Birdseed (sample call rate ≥0.95). On the other hand, of the 709 GENOA samples that passed CQC in figure 2A, 706 passed Birdseed (99.5%, fig. 4A), and of the 716 CLL samples that passed CQC in figure 2B, 716 passed Birdseed (100%, fig. 4B). Thus, the samples that passed QCR but failed CQC generally failed the Birdseed algorithm. We also observed that these ‘bad’ samples influenced the Birdseed calling rate of the ‘good’ samples if the prior filtering (CQC ≥0.4) was not applied (fig. 5: plates 8 and 9, GENOA; plate 7, CLL).
Fig. 2.
Comparison between the two Affymetrix QC measures: QCR and CQC for the GENOA and CLL samples. The x-axis displays the QCR with its cut-off value of 0.86. The y-axis displays the CQC with its cut-off value of 0.4. For GENOA, 48 samples failed QCR and 102 samples failed CQC. For CLL, 30 samples failed QCR and 53 samples failed CQC.
Fig. 3.
CQC distribution across plates using box-plots for the GENOA and CLL samples. The x-axis displays the summary statistics for each plate including the average CQC value of 1.7 (straight line).
Fig. 4.
Description of the Affymetrix workflow for the two QC measures: QCR and CQC. A GENOA samples. B CLL samples.
Fig. 5.
Influence of a CQC filter in the Birdseed genotype calling for the GENOA and CLL samples. A higher sample missingness rate is observed when no CQC filter is applied, particularly for plates 8 and 9 for GENOA samples and plate 7 for CLL samples. P = Passed; F = failed.
Pooled versus Individual Plate Calling Using Birdseed
The impact of pooling samples was evident in the GENOA samples but not in the CLL samples, regardless of which options were tried for the GENOA samples, and we ultimately passed more GENOA samples running the Birdseed algorithm per plate rather than pooled. The influence of the QC algorithms was also evident when comparing calling using pooled or individual plates. The increased failure rate for the pooled samples was considerably higher after using the CQC in the GENOA samples compared to the CLL samples. More failures were observed from samples using QCR compared to CQC for pooled plates (31 vs. 12% for GENOA; 1.5 vs. 0.7% for CLL) and for individual plates (13 vs. 0.4% for GENOA; 0 and 0% for CLL). Based on these results, we recommend running the Birdseed algorithm separately for each plate if there are underperforming plates as we observed in GENOA (fig. 4A).
Comparison of the CRLMM Options
CRLMM also provides a QC measure for samples (SNR) to assess sample quality. We tried multiple SNR thresholds, and observed that there were overall more samples failing under the default SNR value of 5 (table 3). Thus, we concluded that a SNR threshold of 5, though recommended, may be too conservative and in our data a SNR threshold of 4 appeared to be more appropriate (table 3). Using this SNR threshold of 4, we investigated the appropriate threshold for the PP that determined an acceptable sample call rate and SNP call rate. We observed that a PP of 90% yielded an acceptable sample call rate and SNP call rate (table 4). Defining what is acceptable for sample and SNP call rates is left to the investigators. In our case, this decision was made after balancing the number of samples and SNPs we would keep and how comparable these numbers were with the ones obtained with Birdseed. We also observed similar results using the CLL samples (results not shown).
Table 3.
CRLMM evaluation for SNR thresholds under pooled and individual plate calling for 854 samples and 906,600 SNPs
| SNR threshold | Plate calling | Samples passed after SNR, n (%) | Samples passed after CRLMM*, n (%) | SNP passed, n (%) |
|---|---|---|---|---|
| 3 | individual | 836 (98) | 812 (95) | 679,968 (75) |
| 3 | pooled | 836 (98) | 808 (95) | 679,985 (75) |
| 4 | individual | 805 (94) | 799 (94) | 692,011 (76) |
| 4 | pooled | 805 (94) | 798 (93) | 693,008 (76) |
| 5 | individual | 703 (82) | 703 (82) | 712,076 (79) |
| 5 | pooled | 703 (82) | 703 (82) | 711,985 (79) |
Using the PP threshold of 0.90 and ‘drop SNP first’ filter.
Table 4.
CRLMM evaluation for PP using a SNR threshold of 4 under individual plate calling and ‘drop SNP first’ filter for 805 samples and 906,600 SNPs
| PP threshold | Samples passed, n (%) | SNP passed, n (%) |
|---|---|---|
| 0.95 | 794 (93) | 639,107 (70) |
| 0.90 | 799 (94) | 692,011 (76) |
| 0.85 | 805 (94) | 728,910 (80) |
| 0.80 | 805 (94) | 754,930 (83) |
| 0.75 | 805 (94) | 773,633 (85) |
Pooled versus Individual Plate Calling Using CRLMM
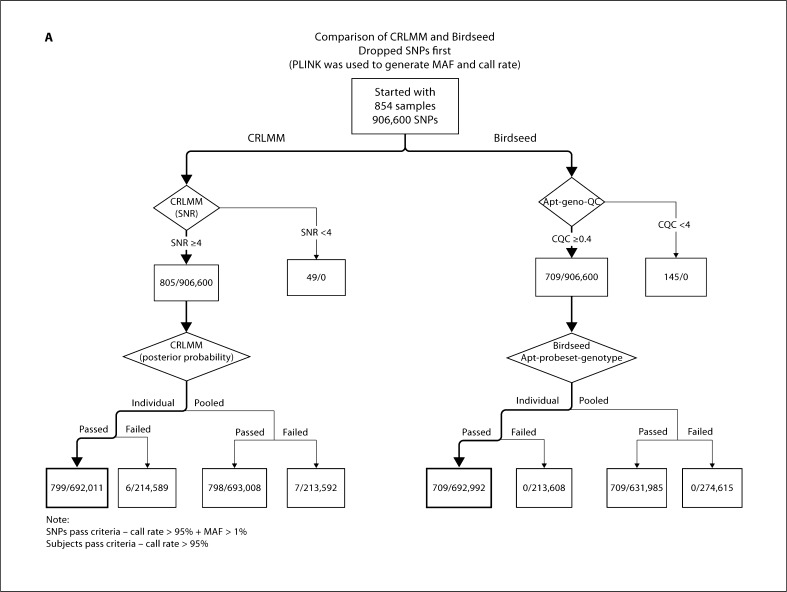
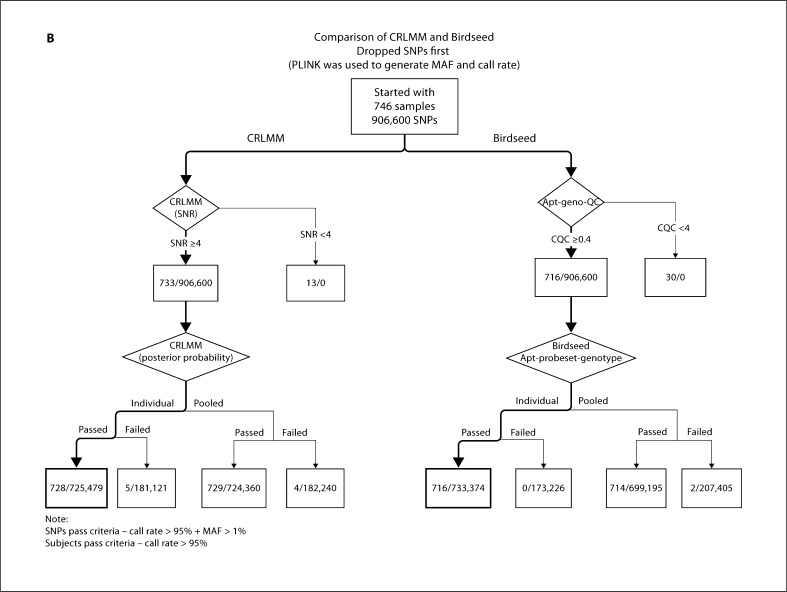
Since the impact of pooling was evident in the GENOA samples using Birdseed, we decided to perform a similar analysis using CRLMM to see whether we observed a similar effect. The number of samples with an acceptable call rate was similar using either pooled or individual plate calling. Using our suggested threshold of 90% for PP and 4 for SNR, 94% of the samples and 76% of the SNPs passed under individual plate calling compared to 93% of the samples and 76% of the SNPs that passed under pooled plate calling (table 3, fig. 6A). Using the same thresholds for PP and SNR in the CLL samples, we observed that 98% of the samples and 80% of the SNPs passed with either individual or pooled plate calling (fig. 6B). Based on these results, we observed that pooled plate calling was more susceptible to underperforming plates than individual plate calling. Thus we recommend running the CRLMM algorithm separately for each plate since it is less susceptible to underperforming plates and does not appear to negatively influence the genotype calling.
Fig. 6.
Comparison between CRLMM and Birdseed algorithms dropping SNP first. A GENOA samples. B CLL samples.
Identifying Poor Quality SNPs and Samples
Regardless of the calling algorithm, the order of investigating call rates of samples and SNPs needs serious consideration. For the GENOA samples, when we dropped samples first using our suggested workflow for Birdseed, 17% of the samples and 33% of the SNPs were dropped (online suppl. fig. 1, for all online suppl. material, see www.karger.com/doi/10.1159/000328843); using our suggested workflow for CRLMM, 11% of the samples and 22% of the SNPs were dropped (online suppl. fig. 1). When we dropped SNPs first using our suggested workflow for Birdseed, 17% of the samples and 33% of the SNPs were dropped (fig. 6A); using our suggested workflow for CRLMM, 6% of the samples and 24% of the SNPs were dropped (fig. 6A). These 6% samples were among the 11% of samples dropped using CRLMM when samples were dropped first, indicating that by removing underperforming SNPs first, we can retain more samples for analysis. For the CLL samples, when we dropped samples first using our suggested workflow for Birdseed, 4% of the samples and 19% of the SNPs were dropped (online suppl. fig. 2); using our suggested workflow for CRLMM, 3% of the samples and 20% of the SNPs were dropped (online suppl. fig. 2). When we dropped SNPs first, using our suggested workflow for Birdseed, 4% of the samples and 19% of the SNPs were dropped (fig. 6B); using our suggested workflow for CRLMM, 2% of the samples and 20% of the SNPs were dropped (fig. 6B). Based on our results, we recommend dropping SNPs first rather than samples, since fewer samples are dropped and dropped SNPs can be imputed [15]. The same recommendation was also suggested recently by another genome-wide association QC study [16].
Comparison of Birdseed and CRLMM in Practice
To perform this comparison, we ran a genome-wide association analysis using the GENOA cohort and the continuous trait eGFR as the outcome variable. Samples were analyzed using the genotyping algorithms by plate using our recommended workflow (fig. 6A). Using the samples and SNPs that passed both algorithms, we also observed similar genotypes (99.8% concordance rate).
The final data sets using CRLMM and Birdseed consisted of 799 (93.6%) and 709 (83.0%) samples, respectively. From those samples, 709 (83.0%) passed both algorithms, 55 (6.4%) failed both, and 90 (10.5%) failed Birdseed. We observed that the genomic inflation factor λ was the same for both algorithms (1.019), and the number of significant SNPs using either calling algorithm was similar for p values more significant than 10–3; however, for more stringent significant levels, more SNPs were observed using Birdseed (fig. 7A, B, table 5, online suppl. tables 1–5). Furthermore, the most significant SNP identified by one algorithm was not always revealed by the other (fig. 7A, B, online suppl. fig. 3–5). For instance, the most significant SNP (rs100488947 on chromosome 4) identified by CRLMM (p value = 2.35 × 10–7) was seen by Birdseed at a slightly lower level of significance (p value = 7.42 × 10–6). One sample did not have information for this particular SNP. Its sample size, MAF, Hardy-Weinberg equilibrium p value and sample call rate were 709 (798), 0.026 (0.028), 1.0 (0.125), and 1.0 (0.999) for Birdseed (and CRLMM), respectively.
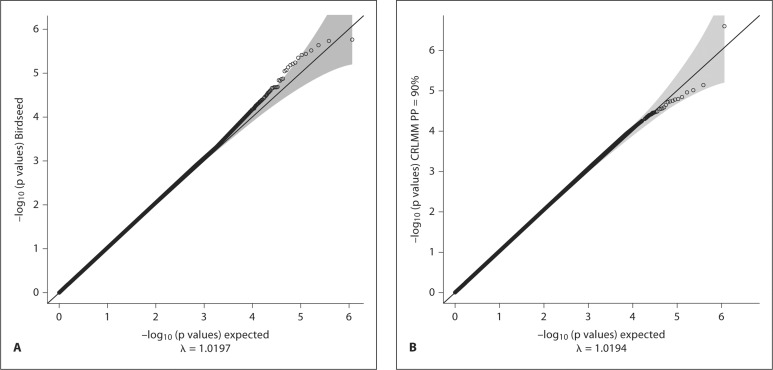
Fig. 7.
Q-Q plots for the association analysis p values using GENOA samples with genomic inflation factor λ. A Birdseed GCA. B CRLMM GCA.
Table 5.
Evaluation of the significant SNPs using either CRLMM or Birdseed calling algorithms from a total of 638,947 SNPs on chromosomes 1–22 that passed the process of QC
| p value cutoff | Birdseed only*, n (%) | CRLMM only*, n (%) | Both, n (%) |
|---|---|---|---|
| 0.1 | 19,692 (3.38) | 19,833 (3.40) | 40,408 (6.93) |
| 0.01 | 2,886 (0.49) | 2,819 (0.48) | 3,476 (0.59) |
| 0.001 | 328 (0.06) | 389 (0.07) | 323 (0.06) |
| 1.00−4 | 56 (0.01) | 27 (0.005) | 25 (0.004) |
| 1.00−5 | 12 (0.002) | 2 (0.0003) | 1 (0.0002) |
| 1.00−6 | 0 (0) | 1 (0.0002) | 0 (0) |
The genomic inflation factor λ was 1.019 for both calling algorithms.
Discussion and Conclusion
According to the most recent Affy SNP 6.0 user guide [10], data sets should be identified as being potentially problematic if less than 90% of the samples pass the CQC threshold or if the average CQC of the remaining samples is less than 1.7. The GENOA data did not fulfill the first condition and was close to the cut-off value for the second condition due to an abysmal CQC of more than 40% in two plates. Even with these potential problems, we ultimately were able to identify significant genome-wide association study results. We also observed that CQC is a more reliable measure than QCR, which is more sensitive to problematic samples as were observed in the GENOA data. In the GENOA data, 99.5% of the samples that passed CQC also passed Birdseed compared to 87.2% using QCR, whereas 100% of the CLL samples passed using either CQC or QCR. Our results have also shown, using both cohorts, that more samples passed if Birdseed was performed by individual plate instead of pooling the plates. Finally, run time for each algorithm may be another consideration; it took CRLMM 4.2 h (3 h) and Birdseed 8.4 h (8.4 h) to run 854 GENOA samples by plate (run time for pooled data).
We created guidelines based on two data sets; however, given the sensitivity of these algorithms we suggest examining each project carefully. Recently there have been several articles published discussing similar problems, focusing either on statistical issues of genome-wide association [17] or the usefulness of QC measures when using high-throughput genotyping from existing public databases [18]. Several factors may influence call rates such as mixtures of populations within a plate or unbalanced numbers of cases and controls in a plate that can result in bad genotype clustering [17,18]. These factors did not apply in either of our two data sets since the phenotypes for GENOA were continuous, and the primary phenotype for the CLL study, although discrete, was controlled by randomization of the samples to each plate. Additionally, our samples were all white non-Hispanics which was also confirmed by population stratification analysis implemented in PLINK [14]. Furthermore, each of our plates had a minimum of 15 females as required by Affymetrix – Best Practices SNP 6.0 Workflow [7,8,9]. We have performed our evaluation on two data sets that were genotyped at the same time in the same platform and using the same genotyping center. By comparing these two studies we observed that QC measures are important to detect problematic samples. If the samples are not appropriately filtered, it can influence the sample and SNP call rates, regardless of which algorithm is used. We believe that evaluating these two GCAs in a real set-up can provide a useful workflow for other researchers. The factors that influence genotyping are difficult to capture in a realistic simulation of the intensity files and are outside the scope of this paper. Additionally, real data has issues not seen by simply using the HapMap samples, and previous validation of GCAs has used the intensity files of HapMap data and not simulated data [1,4,5].
In summary, we have shown that the overall performance of the Birdseed genotyping process varies by choice of the two pre-cluster sample QC checks (QCR and CQC) and the number of samples used to call the genotypes (by plate versus using all samples). We have summarized study results using the two GCAs Birdseed and CRLMM, investigating the impact of several options within the CRLMM function and comparing advantages and disadvantages of the two algorithms. We have performed a genome-wide association analysis in the GENOA samples using the genotype calls from those two algorithms and observed similar association results when the level of significance was more significant than 10–3, but not when the level of significance was below 10–3 where more significant SNPs were picked up by Birdseed; we could not evaluate the number of false positives since truth was unknown. Finally, we conclude that it is useful to validate findings using either another platform or GCA if available, and to look at SNP genotype clustering plots if intensity data are available.
Supplementary Material
Supplemental materials
Acknowledgements
We thank DeLaine Anderson for her technical assistance with the manuscript and Benilton Carvalho for his helpful comments about CRLMM. The Genotyping Shared Resource of the Mayo Clinic Technology Center did all the genome-wide association genotyping described in this paper. This paper was supported by the National Institutes of Health research grant R01 HL87660 (to M.d.A. and S.L.R.K.) and National Cancer Institute grant U01 CA118444 (to S.L.S.).
References
- 1.Rabbee N, Speed TP. A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics. 2006;22:7–12. doi: 10.1093/bioinformatics/bti741. [DOI] [PubMed] [Google Scholar]
- 2.BRLMM-P: a genotype calling method for the SNP 5.0 array. http://www.affymetrix.com/support/technical/whitepapers/brlmmp_whitepaper.pdf
- 3.Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, Hubbell E, Veitch J, Collins PJ, Darvishi K, Lee C, Nizzari MM, Gabriel SB, Purcell S, Daly MJ, Altshuler D. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carvalho B, Bengtsson H, Speed TP, Irizarry RA. Exploration, normalization, and genotype calls of high-density oligonucleotide SNP array data. Biostatistics. 2007;8:485–499. doi: 10.1093/biostatistics/kxl042. [DOI] [PubMed] [Google Scholar]
- 5.Lin S, Carvalho B, Cutler D, Arking D, Chakravarti A, Irizarry RA. Validation and extension of an empirical Bayes method for SNP calling on Affymetrix microarrays. Genome Biol. 2008;9:r63. doi: 10.1186/gb-2008-9-4-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genotyping console 1.0 user manual: Affymetrix; 2007. http://www.affymetrix.com/support/downloads/manuals/genomewidesnp6_manual.pdf
- 7.Affymetrix® genotyping console 2.1 user manual: appendix F, Affymetrix; 2008(a). http://www.affymetrix.com/support/downloads/manuals/genomewidesnp6_manual.pdf
- 8.Affymetrix® genotyping console 3.0 user manual: appendix F, Affymetrix; 2008(b). http://www.affymetrix.com/support/downloads/manuals/genomewidesnp6_manual.pdf
- 9.Affymetrix® genome-wide human SNP Nsp/Sty 6.0 user guide: pp 288–289, Affymetrix; 2008(c). http://www.affymetrix.com/support/downloads/manuals/genomewidesnp6_manual.pdf
- 10.Affymetrix manual: apt-probeset-genotype (apt-1.10.2), Affymetrix 2009. http://www.affymetrix.com/support/downloads/manuals/genomewidesnp6_manual.pdf
- 11.Quality control assessment in genotyping console™. Affymetrix White Paper. September 2008.
- 12.The FBPP Investigators: Multi-center genetic study of hypertension: the Family Blood Pressure Program (FBPP) Hypertension. 2002;39:3–9. doi: 10.1161/hy1201.100415. [DOI] [PubMed] [Google Scholar]
- 13.Slager SL, Rabe KG, Achenbach SJ, Vachon CM, Goldin LR, Strom SS, Lanasa MC, Spector LG, Rassenti LZ, Leis JF, Camp NJ, Glenn M, Kay NE, Cunningham JM, Hanson CA, Marti GE, Weinberg JB, Morrison VA, Link BK, Call TG, Caporaso NE, Cerhan JR. Genome-wide association study identifies a novel susceptibility locus at 6p21.3 among familial CLL. Blood. 2011;117:1911–1916. doi: 10.1182/blood-2010-09-308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Willer C, Serena Sanna S, Abecasis G. Genotype imputation. Annu Rev GenomicsHum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turner S, Armstrong LL., Bradford Y, Carlson CS., Crawford DC, Crenshaw AT, de Andrade M, Doheny KF, Haines JL, Hayes G, et al: Quality control procedures for genome-wide association studies. Curr Protoc Hum Genet DOI: 10.1002/0471142905.hg0119s68. [DOI] [PMC free article] [PubMed]
- 17.Ziegler A, Koenig IR, Thompson JR. Biostatistical aspects of genome-wide association studies. Biom J. 2008;50:8–28. doi: 10.1002/bimj.200710398. [DOI] [PubMed] [Google Scholar]
- 18.Pluzhnikov A, Below JE, Konkashbaev A, Tikhomirov A, Kistner-Griffin E, Roe CA, Nicolae DL, Cox NJ. Spoiling the whole bunch: quality control aimed at preserving the integrity of high-throughput genotyping. Am J Hum Genet. 2010;87:123–128. doi: 10.1016/j.ajhg.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials