Abstract
The incurability of malignant glioblastomas is mainly attributed to their highly invasive nature coupled with resistance to chemo- and radiation therapy. Because invasiveness is partially dictated by the proteins these tumors secrete we used SILAC to characterize the secretomes of four glioblastoma cell lines (LN18, T98, U118 and U87). Although U87 and U118 cells both secreted high levels of well-known invasion promoting proteins, a Matrigel invasion assay showed U87 cells to be eight times more invasive than U118 cells, suggesting that additional proteins secreted by U87 cells may contribute to the highly invasive phenotype. Indeed, we identified a number of proteins highly or exclusively expressed by U87 cells as compared to the less invasive cell lines. The most striking of these include ADAM9, ADAM10, cathepsin B, cathepsin L1, osteopontin, neuropilin-1, semaphorin-7A, suprabasin and chitinase-3-like protein 1. U87 cells also expressed significantly low levels of some cell adhesion proteins such as periostin and EMILIN-1. Correlation of secretome profiles with relative levels of invasiveness using Pavlidis template matching further indicated potential roles for these proteins in U87 glioblastoma invasion. Antibody inhibition of CH3L1 reduced U87 cell invasiveness by 30%.
Keywords: Glioblastoma, secretome, invasion, chitinase-3-like protein 1, YKL-40, osteopontin, neuropilin-1, semaphorin-7A, suprabasin, cathepsin
Introduction
Glioblastoma multiforme (GBM) is the most common type of tumor that arises in the brain and is also the most deadly, with a five-year survival rate of only 3%1. Contributing to the incurable nature of these tumors are their propensity towards extensive invasion into surrounding healthy tissue and resistance to both chemo- and radiation therapy. Unfortunately, little progress has been made in the treatment of malignant gliomas over the last twenty years, partly due to our poor understanding of their behavior and the lack of effective therapeutic targets.
The process of tumor cell invasion is a combination of complex cellular and molecular events involving the breakdown of extracellular matrix (ECM) components, cell detachment, and migration through the basement membrane and stroma. It is well known that ECM degradation is dictated by the overexpression of key proteases whose activity is normally kept in check in healthy tissue by endogenous inhibitors (reviewed by Rao2 and Levičar et al3). The main regulators of this process include urokinase-type plasminogen activator (uPA) and its receptor (uPAR), the families of matrix metalloproteases (MMP), a disintegrin and metalloproteinase domain-containing proteins (ADAM) and the cathepsins. Most of these are secreted as pro-proteases which are then activated through a complex cascade of cross- and self- activation leading to the degradation of all components of the ECM including collagen, fibronectin and laminin.
While the overexpression of these proteases and their involvement in glioma cell invasion is well documented at the cellular level, it is ultimately the balance between proteases, their inhibitors, various adhesion molecules and other not yet defined molecules that dictates the invasive and aggressive behavior of a tumor cell. Thus, comprehensive analysis of secreted proteins, or secretome profiling, is a valuable approach to further our understanding of GBM biology and to define the molecular signature of the invasive glioblastoma secretome.
In this study, we sought to characterize the secretome profiles of four malignant glioblastoma cell lines (LN18, T98, U118 and U87) using a stable isotope labeling by amino acids in cell culture (SILAC) strategy4,5,6 and then correlate the secretome profiles with their invasive behavior. We found that the balance between ECM proteases and their inhibitors alone may not determine the overall invasiveness of a tumor cell, but that this process may be enhanced by additional, non-proteolytic factors. In this study we have identified several new candidate proteins that may actively promote tumor cell invasiveness.
Methods
Cell lines and metabolic labeling
Malignant astrocytoma cell lines LN18, T98, U118 and U87 were purchased from American Type Culture Collection (Manassas, VA). Cell lines were grown to confluency and subsequently washed six times with serum-free medium, followed by a 24 hour incubation in serum-free medium containing either “heavy” or “light” amino acids. Since T98 cell lines exhibited the lowest invasion characteristics these cells were cultured in “heavy” (or “labeled”) Dulbecco's modified Eagle's medium (DMEM) (Atlanta Biologicals, Lawrenceville, GA) where Arg and Lys were replaced by 13C6-Arg and 13C6, 15N2-Lys (Cambridge Isotopes, Andover, MA), while the comparative astrocytoma cell lines U87, LN18 and U118 were grown in “light” (or “unlabeled”) DMEM (Atlanta Biologicals, Lawrenceville, GA) containing 12C6-Arg and 12C6, 14N2-Lys (Sigma-Aldrich, St. Louis, MO). A comparison of labeled T98 with unlabeled T98 was included as a control. Incorporation of the heavy isotopes was verified by mass spectrometry to be > 98% after at least five cell doublings (data not shown). Conditioned media from labeled and unlabeled cells were collected and cellular debris removed by centrifugation at 1,000 × g for 10 minutes at 4°C. Protein concentration of each sample was determined using the Bradford assay (Pierce Protein Research Products, Rockford, IL). Conditioned media for western blot analysis were collected in the same manner, with all cells grown in unlabeled DMEM.
Sample preparation
Conditioned medium (50 μg total secreted protein) from each of one unlabeled cell line (T98, U118, LN18 or U87) was mixed with an equal amount of conditioned medium (50 μg of total secreted protein) from the SILAC labeled T98 cell line. To validate the data for the most significant sample pair (e.g. unlabeled U87 versus labeled T98) we performed a reverse SILAC experiment in which we compared unlabeled T98 to labeled U87 conditioned media. Paired samples were concentrated to dryness using Amicon Ultra-4 Ultracel 3K centrifugal filter units (Millipore Corporation, Billerica, MA) followed by vacuum centrifugation. Resulting residues were then dissolved in Laemmli buffer containing 50 mM DTT, denatured at 95°C for five minutes and alkylated with iodoacetamide (final concentration of 100 mM). Proteins were then separated by SDS-PAGE on Criterion 8–16% Tris-HCl midi-format gels (Bio-Rad, Hercules, CA) at 200 volts for one hour. Gels were stained with Bio-Safe Coomassie blue (Bio-Rad, Hercules, CA) and each gel lane sliced into approximately 40 bands followed by digestion with trypsin (Promega, Madison, WI) using an established protocol7. Resultant peptides from each band were dried by vacuum centrifugation and resuspended in 6 μL of 0.1% TFA for analysis by mass spectrometry.
Mass spectrometry analysis
NanoLC MS and MS/MS were performed using the Eksigent nanoLC 2D HPLC system (Eksigent Technologies, Inc., Dublin, CA) coupled to the LTQ-Orbitrap XL hybrid mass spectrometer (Thermo Electron Corporation, San Jose, CA). Samples were desalted via autosampler loading onto a C18 trap column (300 μm i.d. × 1 mm, LC Packings, Sunnyvale, CA) for 10 minutes at a flow rate of 5 μL/min then loaded onto the PepMap 100 C18 (3.5 μm, 100 Å) reverse-phase nanocolumn (75 μm i.d. × 15 cm) (LC Packings, Sunnyvale, CA) at a flow rate of 250 nL/min. The mobile phases comprised ultrapure Optima LC/MS water (Thermo Fisher Scientific, Inc, Pittsburgh, PA) with 0.1% formic acid (A) and 90% acetonitrile with 0.1% formic acid (B). Peptides were eluted over a 45-min linear gradient increasing from 5% to 95% B. Peptides were introduced into the mass spectrometer via a 10-μm silica tip (New Objective Inc., Ringoes, NJ) adapted to a nano-electrospray source (Thermo Electron Corp, San Jose, CA). The spray voltage was set at 1.4 kV with the heated capillary at 200°C. The LTQ-Orbitrap XL was operated using Xcalibur 2.0.7 (Thermo Fisher Scientific, Inc.) in data-dependent mode in which one cycle of experiments consisted of one full-MS survey using the high resolution Orbitrap mass analyzer (300–2000 m/z, R=30,000) and subsequently five sequential MS/MS events of the most intense peaks using collision-induced dissociation with helium in the LTQ.
Protein identification and quantitation
Protein identification was performed using BioWorks 3.3.1 software (Thermo Electron Corp, San Jose, CA) in which raw data files were searched against the 22,222 human proteins extracted from the SwissProt database (release 55.0) using the Sequest search engine with a peptide tolerance of 50 ppm, fragment ions tolerance of 1 AMU and allowing up to two missed cleavages. Parameters for modifications included a static cysteine modification of +57.02146 Da and the following variable protein modifications: +15.99492 Da shift for oxidized Met; and +6.0204 and +8.0142 Da shifts for stable isotope labeled Arg and Lys respectively. Acceptable peptide identification scores were set as follows: DeltaCn (ΔCn) > 0.1; variable threshold of Xcorr vs. charge state (Xcorr = 1.9 for z = 1, Xcorr = 2.5 for z = 2, and Xcorr = 3.5 for z ≥ 3); and peptide probability based score with p value < 0.001. A false discovery rate of 1.69 % was determined by using the same filtration criteria and searching the raw data files against an indexed reversed human SwissProt database (data not shown). Quantitative analysis was performed using the Census algorithm8 and the following filters: determination factor = 0.5, outlier threshold p-value = 0.1, peptides with negative R = removed. To correct for any inaccuracies in equal mixing of conditioned media from heavy and light labeled samples, we established a histogram of log-transformed Census ratios and used the off-set of the histogram center from zero as the correction factor. Only proteins identified with two or more peptides of unique sequence were retained for quantitative analysis. Known contaminant proteins from processing or serum (e.g. keratin, albumin, hemoglobin) were not included in the analysis.
Cell death examination
To assess the relative levels of cell death in our experiments, confluent cells were washed six times in serum free medium, then incubated for 24 hours in serum free medium. Total cells including floating cells were collected and counted using a hemacytometer with trypan blue exclusion to distinguish living cells from dead cells. Percent cell death was calculated as the number of dead cells over total cell count and tested for significance using Kruskal-Wallis.
Tumor cell invasion assay
Cell suspensions containing 100,000 live cells from each cell line were seeded into the upper region of a Boyden chamber comprising a Fluoroblok membrane with 8 μm pores coated with Matrigel (BD Biosciences, San Jose, CA). Cells were allowed to attach to the Matrigel-coated membrane, then washed six times with serum free medium and synchronized for 24 hours in the same medium. DMEM containing 10% FBS was then placed in the lower chamber to act as a chemottractant. Cells were allowed to invade through the Matrigel-coated membrane for 12 hours, after which they were incubated with a 1 μM calcein AM/5 μM Hoechst 33342 solution for 30 minutes. Fluorescent cells in ten fields per well were imaged on the Zeiss Apotome (Munich, Germany) at 470/508 nm (Calcein AM) and 359/461 nm (Hoescht 33342). Both invasive cells on the bottom of the membrane and non-invasive cells on the top of the membrane were counted using ImageJ software and averaged for each cell line. Percent invasion was calculated as the average number of invasive cells per microscope field that passed through the Matrigel into the lower chamber divided by the average number of total cells (average invasive cells per field + average non-invasive cells per field). This controlled for any differences in cell number that may have arisen due to differing levels of apoptosis, proliferation or seeding density. ANOVA followed by Student-Newman-Keuls was used to test for significant differences in levels of invasiveness among the four cell lines. For antibody inhibition of CH3L1 during invasion, synchronized U87 cells were seeded in serum-free DMEM containing a final concentration of 5 μg/mL of α-CH3L1 antibody (R&D Systems, Minneapolis, MN, #AF2599), 5 μg/mL normal goat IgG (R&D Systems, Minneapolis, MN, #AB-108-C) or no antibody and allowed to invade for 12 hours.
Correlation of invasion assay data with secretome data
Spectral counts were extracted after Sequest processing and normalized against a reference sample to remove technical bias. Pavlidis template matching, also known as Feature Selection9, was used to correlate the spectral count for each protein detected with the relative levels of invasiveness for each cell line as determined by Matrigel invasion assay. This correlative approach, executed in MultiExperiment Viewer (Dana-Farber Cancer Institute, Boston, MA, USA; www.tm4.org/mev), has been successfully implemented with microarray expression data as a means to identify candidate genes associated with invasion and foci formation10,11. Secretome-invasion associations were based on Pearson Correlation and significance was defined at p < 0.05. Only proteins with known or predicted localization to the extracellular, cell membrane, endosomal or lysosomal compartment were included in the analysis.
Western blot analysis
Conditioned media (30 μg) from each cell line were concentrated using Amicon Ultra-4 Ultracel 3K centrifugal filter units (Millipore Corporation, Billerica, MA) followed by vacuum centrifugation. Resulting residues were dissolved in NuPage LDS Sample Buffer containing NuPage Reducing Agent (Invitrogen Corp, Carlsbad, CA) and denatured at 95°C for five minutes. Proteins were resolved by SDS-PAGE on a NuPage Novex 10% Bis-Tris midi format gel (Invitrogen Corp, Carlsbad, CA) at 200 volts for one hour. Proteins were transferred to an Amersham Hybond ECL 0.45μm nitrocellulose membrane (GE Healthcare, Piscataway, NJ) at 100 v for 1.5 hours. The membrane was probed with 1:500 α-CH3L1 antibody (R&D Systems, Minneapolis, MN, #AF2599) followed by 1:2000 donkey α-goat-HRP (Jackson ImmunoResearch Laboratories, West Grove, PA, #705-035-147) or with 1:1000 α-vinculin antibody (Sigma-Aldrich, St. Louis, MO, #V9131) followed by 1:2000 goat α-mouse-HRP (Bio-Rad, Hercules, CA, #170-6516). Bands were visualized by enhanced chemiluminescence using Amersham ECL (GE Healthcare, Piscataway, NJ).
Results
Relative invasiveness of astrocytoma cell lines
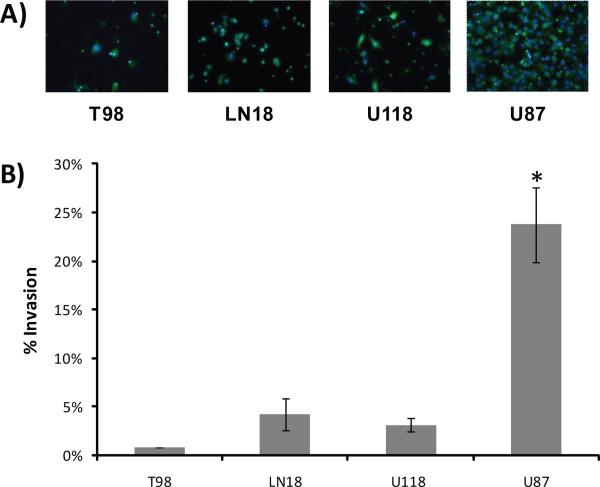
We first characterized the invasive phenotypes of the four astrocytoma cell lines LN18, T98, U118 and U87 using a Matrigel-coated Boyden chamber. Figure 1 shows the invasive behavior of each cell line based on the infiltration of the cells through the Matrigel after 12 hours incubation. While T98, LN18 and U118 cells exhibited low to moderate invasive properties, U87 cells were significantly more invasive, with 6 to 29 fold higher levels of invasiveness compared to the other cell lines.
Figure 1. Matrigel invasion assay of glioblastoma cell lines.
A) Fluorescent images (20×) of cells that invaded through the Matrigel-coated Boyden chamber after twelve hours of incubation. Cytosol = green/calcein AM; Nucleus = blue/Hoescht 33342. B) Percent invasion of cells (average invasive cells per field normalized to average number of total invasive plus noninvasive cells per field). Data are reported as mean values (± SD) of at least three replicates for each cell line. * Student-Newman-Keuls p<0.05.
Secretome profiling of glioblastoma cell lines
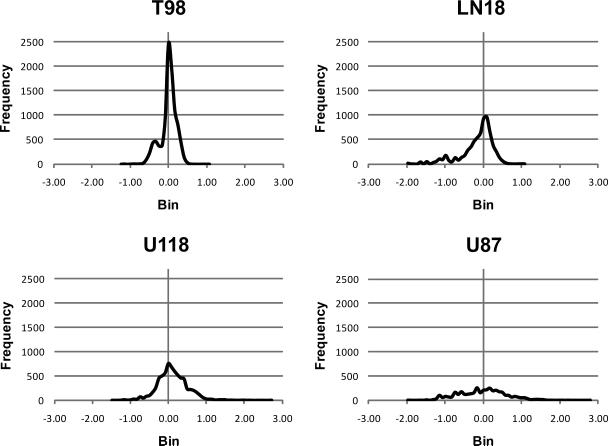
To determine which secreted proteins may contribute to the highly invasive nature of the U87 cell line, we quantified the relative expression levels of all secreted proteins in the four cell lines using a differential SILAC strategy12,13. Since this quantitation method is most easily performed between pairs of samples, and since T98 cells exhibited the lowest invasive behavior compared to the other cell lines, we chose to label T98 cells with heavy isotope bearing amino acids (13C6-Arg and 13C6, 15N2-Lys) and use them as a reference sample by which to define the relative secretome profiles of unlabeled U118, LN18 and U87 cells. As a control, we also compared the secretome of labeled T98 cells to unlabeled T98 cells. In this study we quantified 1140 proteins in the conditioned media of all four cell lines combined (Supplemental Tables 1 and 2a–e, Supplemental Information and PRoteomics IDEntifications database14 (PRIDE) at: http://www.ebi.ac.uk/pride/). Figure 2 shows the ratio distribution profiles of labeled and unlabeled tryptic peptide pairs from each comparison. The peptide ratio distributions of our control (T98 unlabeled vs. T98 labeled) shows a narrow distribution with an average ratio of 1.01 ± 0.46 reflecting the expected similarity between the two samples. Peptide ratio distribution derived from LN18/T98 and U118/T98 pairs provide broader distributions with average ratios of 0.87 ± 0.65 and 2.15 ± 8.79 respectively, though their similarity to each other could reflect the similarity between their secretome profiles. The U87/T98 peptide ratio distribution had a much broader distribution, with no real definable peak and an average peptide ratio of 2.49 ± 11.31, suggesting highly different secretome profiles between U87 and T98 cell lines.
Figure 2. Distribution of SILAC peptide ratios between glioblastoma cell lines.
Peptide ratios obtained from T98 labeled samples paired with U87, U118, LN18 or T98 unlabeled control samples were obtained using the Census algorithm. Ratios were log transformed and plotted into bins of 0.08 against frequency (number of peptides in each bin).
To ensure that this differential protein secretion is not a result of differential cell death we used trypan blue exclusion and confirmed that the percentage of cell death across the four cell lines did not differ significantly after 24 hours serum deprivation (data not shown).
Quantitative secretome analysis
Although there was slight variation between SILAC ratios in the unlabeled T98 versus labeled T98 secretomes (ratios averaging 1.01 ± 0.46), this same set of samples showed little to no variation in the SILAC ratios of the cytosolic fraction (ratios averaging 1.04 ± 0.20, see Supplemental Figure 1, Supplemental Information). This suggests that the secretome is more susceptible to biological and/or technical variation than the intracellular compartment. Thus, we used a stringent criterium to determine which proteins were differentially secreted by U118, LN18 and U87 cells as compared to the T98 control. We calculated the average, standard deviation and 99% confidence interval in the control pair (unlabeled T98 and SILAC labeled T98) and considered ratios that fell outside this confidence interval to be significant. This method of calculating significant expression levels from SILAC data is commonly used in SILAC experiments, though normally only a 95% confidence interval is used15,16,17. To further validate the data obtained for the U87 versus T98 sample pair, we also performed a reverse SILAC experiment in which T98 cells were unlabeled and U87 cells were labeled.
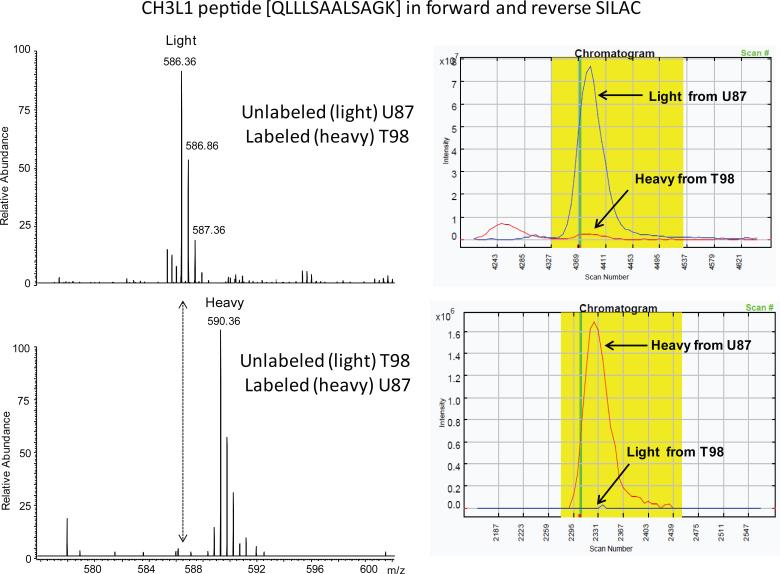
Proteins that were detected in one member of a SILAC pair but not the other were also considered significant and these spectra were validated in both the reverse and forward SILAC experiments using Census peak ratio viewers. See the example in Figure 3 for a peptide corresponding to chitinase-3-like protein 1 (CH3L1) that was exclusively secreted by U87 cells and not by T98 cells. This same peptide was validated in both forward and reverse SILAC and clearly indicates that T98 cells express undetectable amounts of CH3L1.
Figure 3. Manual validation of uniquely expressed proteins.
Peptides that were detected in one member of a SILAC pair but not the other were manually validated for absence of the corresponding peptide. A) Sample spectra show peaks mapping to the QLLLSAALSAGK peptide of chitinase-3-like protein 1 in both forward and reverse SILAC experiments, indicating the absence of the peptide in the T98 conditioned media. B) Census quantitation of the same peptide with forward and reverse labeling.
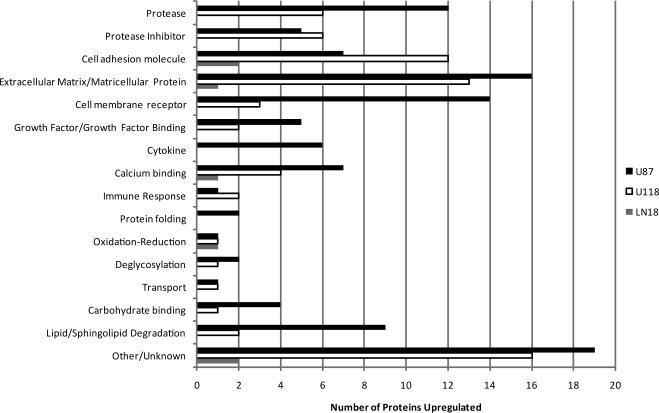
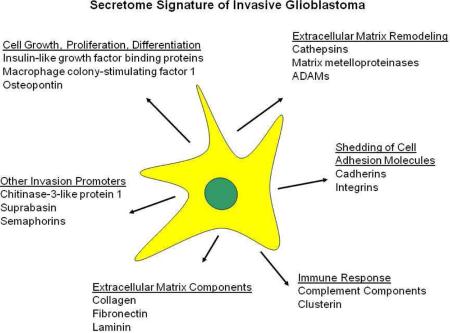
Using these stringent criteria we found 209 proteins differentially expressed among the three cell lines in comparison to the T98 control (see Supplemental Table 3, Supplemental Information). These differentially secreted proteins were classified by functional category using the UniProtKB database (www.uniprot.org) and the results are summarized in Figure 4. The number of proteases significantly upregulated in the U87 conditioned medium was 2 to 12- fold greater than that in the LN18 and U118 media. These proteases include cathepsins, matrix metalloproteinases and members of the ADAM family and in general showed the highest levels of expression when compared to the other cell lines. Although the U118 secretome comprised a similar number and category of proteases, the majority of expression levels were 2 to16- fold lower than in the conditioned medium of the U87 cells. U87 cells also had a greater number of upregulated cell membrane receptors, growth factors/growth factor binding proteins, ceramide/sphingolipid degrading enzymes and cytokines as compared to the other cell lines (see Figure 4 and Supplemental Table 3, Supplemental Information).
Figure 4. Functional annotation of proteins upregulated in the glioblastoma secretome.
Proteins differentially secreted by the glioblastoma cell lines U87, U118 and LN18 relative to T98 were classified based on their overall functional category. The total number of overexpressed proteins per cell line is shown in parentheses. Only proteins with known extracellular, cell membrane and lysosomal/endosomal localizations are included.
Concomitantly, the U87 cell line had decreased or no secretion of some extracellular cell-adhesion promoting proteins (e.g. periostin, EMILIN-1, vascular adhesion protein 1 and thrombospondin-1) and increased shedding of cadherin 2 (N-cadherin) and cadherin 13 (T-cadherin; H-cadherin) (Supplemental Table 3, Supplemental Information).
Correlation of secretome profiles with invasive phenotypes
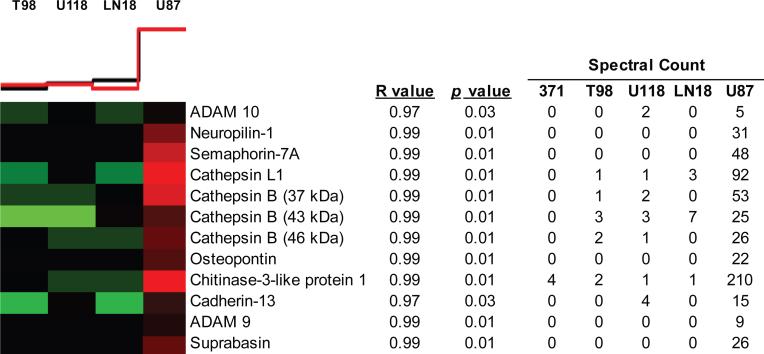
To define additional invasion promoting proteins in the glioblastoma conditioned media, we used Pavlidis template matching. The template matching algorithm matches cell invasion values measured by Matrigel assay to protein spectral counts across the four astrocytoma cell lines. More specifically, we used invasion values of the four cell lines as a template and correlated the pattern of expression levels of each protein across the four cell line to this template. Protein expression patterns were considered to correlate with the invasion pattern if they had a coefficient correlation (R value) > 0.95 and p-value < 0.05. We chose to use spectral count over SILAC ratios for template matching because the former reflects the abundance of proteins in the conditioned media and allowed us to include analysis of proteins which were exclusively expressed by one cell line. We verified correlation between spectral count and SILAC ratios for these differentially secreted proteins. With this method, we identified additional candidate invasion promoting proteins in the U87 conditioned medium as well as a few proteins that may have an inhibitory effect on tumor cell invasion (see Supplemental Table 4, Supplemental Information). This list includes ADAM9, ADAM10, cathepsin B, cathepsin L1, osteopontin, neuropilin-1, semaphorin-7A, suprabasin and CH3L1 which were all highly or exclusively secreted by U87 cells compared to the other three cell lines and healthy primary astrocytes, and have potential roles in tumor cell invasion (Figure 5).
Figure 5. Correlation of glioblastoma secretome profiles with invasive phenotype.
Levels of invasiveness were correlated with spectral counts of secreted, cell membrane and lysosomal protein expression by each cell line using Pavlidis template matching. Black template line above heat map represents relative invasion potential of each cell line, and red line indicates mean relative expression of all correlated proteins within each column. Red heat map blocks reflect relative high protein expression and green represent lower expression. Spectral counts from healthy primary astrocytes (371) were not included in the analysis, but are shown as a reference. Select proteins positively correlating with U87 invasiveness are shown, for a complete list see Supplemental Table 4, Supplemental Information.
U87 cells secrete a high level of CH3L1
Of the proteins that positively correlated with U87 cell invasiveness, CH3L1 exhibited the highest level of expression based on the number of peptide spectral counts detected in the conditioned medium (Figure 5). We validated CH3L1 expression levels by western blot analysis of conditioned media collected from all four cell lines. CH3L1 was detected at its expected molecular mass (~ 40 kDa) and was highly abundant in the U87 conditioned medium but almost undetectable in the conditioned media of the three other cell lines as well as healthy primary astrocytes (Figure 6).
Figure 6. Invasive U87 cells express high levels of CH3L1.
Conditioned media from each cell line were collected after 24-hours incubation in serum-free medium and 30 μg analyzed by western blot against CH3L1 protein. The top panel shows relative expression levels of CH3L1 in the tumor cell lines and healthy human astrocytes (ACBRI 371 obtained from Applied Cell Biology Research Institute, Kirkland, WA). Vinculin was used as a loading control (bottom panel).
U87 cell invasion is reduced in the presence of CH3L1-specific antibody
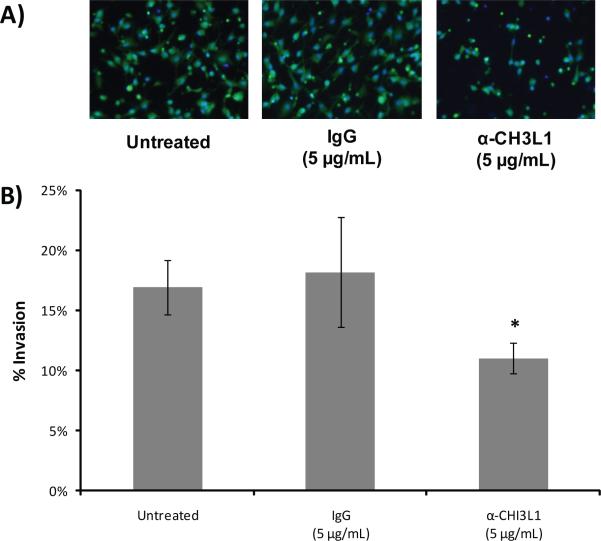
To investigate a possible role for CH3L1 in the invasiveness of U87 cells we inhibited its activity via antibody during the Matrigel invasion assay. Addition of 5 μg/mL of CH3L1-specific antibody to the U87 culture medium reduced the invasive behavior of these cells by more than 30% as compared to cells left untreated or those treated with a non-specific IgG (Figure 7).
Figure 7. Antibody inhibition of CH3L1 during invasion.
A) Fluorescent images (20×) of U87 cells that invaded through the Matrigel-coated Boyden chamber after treatment with α-CH3L1 antibody, IgG or no antibody. Cytosol = green/calcein AM; Nucleus = blue/Hoescht 33342. B) Percent invasion of cells. Data are reported as mean values (± SD) of four replicates. *Wilcoxon Rank Sum p<0.05.
Discussion
Comparison of the secretome profiles of the highly invasive U87 glioblastoma cell line with the less invasive LN18, T98 and U118 cell lines correlate well with our current understanding of tumor cell invasiveness and further validate the model used for this study. As expected, invasive U87 cells secreted significant levels of enzymes known for their roles in tumor cell invasion, including key members of the ADAM, cathepsin and MMP families2,3 while the majority of complementary protease inhibitors detected exhibited low levels of expression. At the same time the U87 cell conditioned media contained increased levels of cadherins 2 and 13, which may be indicative of a cadherin shedding mechanism utilized to decrease cellular attachment18,19,20, and decreased expression of some extracellular adhesion promoting proteins such as periostin, EMILIN-1 and thrombospondin-1. Overall, this secretome signature appears to favor invasive behavior.
Though for the most part the expression levels of well-known invasive proteases were lower in the U118 cell conditioned medium than those of U87 cells, several MMPs (MMP1, MMP2 and MMP14) may still be considered highly expressed by the U118 cells compared to LN18 and T98 cells, yet the U118 cells exhibited levels of invasiveness comparable to the LN18 and T98 cells, which exhibited little to no protease expression. Additionally, although compared to U87 cells, the U118 cell line secreted 2.6 fold higher levels of the ECM protein tenascin, which has been shown to enhance glioblastoma cell invasion21,22, U118 cells had an eight-fold lower level of invasiveness than U87 cells, suggesting the contribution of additional invasion enhancing proteins in the U87 cell secretome.
The most interesting proteins identified via correlation analysis as potential invasion promoters are perhaps those which exhibit almost exclusive expression by U87 cells. These proteins include ADAM9, ADAM10, cathepsin B, cathepsin L1, osteopontin, semaphorin-7A, neuropilin-1, suprabasin and CH3L1. These proteins also exhibited little to no secretion by healthy primary astrocytes, further suggesting potential contributions of these proteins to the invasive nature of U87 cells.
U87 cells had higher expression levels of both ADAM9 and ADAM10 relative to the other cell lines and was implicated in U87 cell invasiveness through our statistical analysis. ADAM proteins are cell surface proteins comprising adhesion and metalloprotease domains which are capable of regulating cell-cell and cell-matrix interactions via integrin binding and are known for cleaving a number of membrane-bound substrates to produce biologically active proteins, such as growth factors and cytokines23. A role for ADAM9 in U87 cell invasiveness is supported by reports of its soluble isoform promoting invasion in liver and lung carcinoma cell lines through β-1 integrin signaling24,25. A recent study demonstrated that ADAM10 is necessary for the shedding of cadherin 2 from the surface of glioblastoma cells to promote migration in vitro26. Indeed, we saw a two-fold or more increase in the level of cadherin 2 in the conditioned media of U87 cells compared to the other cell lines. Cadherin 13 also correlated with U87 invasiveness in our study. Also known as T-cadherin or H-cadherin, cadherin 13 is an atypical member of the cadherin family which lacks a transmembrane domain and is instead associated with the surface of the plasma membrane via glycosylphosphatidylinositol (GPI) linkage. The downregulation of cadherin 13 has been associated with poor prognosis in a variety of carcinomas including lung, ovarian, and prostate cancer27 and its expression has been linked to increased cell attachment and decreased cell motility in the C6 glioma cell line28. However, whether shedding of cadherin 13 is another mechanism by which cancer cells alter surface protein expression to increase their invasive behavior and whether ADAM10 contributes to its release remains to be investigated.
Cathepsins are well known to be overexpressed by astrocytoma cells compared to healthy astrocytes and have been shown to play a role in tumor cell invasion as well as proliferation and angiogenesis29. The four glioblastoma cell lines used in this study secreted cathepsins B, D, F, L1 and Z, with cathepsins B and L1 levels significantly higher in the conditioned medium of the highly invasive U87 cell line compared to the less invasive T98 cell line. Furthermore, these cathepsins were not detected in the conditioned media of healthy primary astrocytes. While our data suggest that the cathepsin B and L1 isoforms detected in this study correspond to the proenzymes only it is possible that these highly expressed pro-enzymes are partially responsible for the degradation of extracellular proteins necessary to facilitate U87 cell invasion since a number of in vitro studies have demonstrated the ability of the cathepsin B and L pro-enzymes to degrade components of the extracellular matrix, including laminin and fibronectin30,31,32. Collectively these data suggest a contribution of cathepsins B and L1 to U87 cell invasiveness. Indeed, selective inhibition of cathepsins B and L1 by gene silencing successfully reduced glioblastoma invasion up to 70%33,34.
Osteopontin was exclusively secreted by the U87 cell line but not by the other, less invasive glioblastoma cell lines. Interestingly, this cytokine was found elevated in glioblastoma tissue as well as the serum and cerebrospinal fluid of patients with glioblastoma tumors35,36. Its role in tumor cell invasion may derive from an autocrine function that promotes cellular motility and migration, as silencing of osteopontin in U87 cells by siRNA significantly reduced cell migration while induction of its expression was shown to enhance cell motility37,38.
Neuropilin-1 (NRP1) was present at significant levels in the U87 conditioned medium (i.e. 31 spectral counts, 12 unique peptides) and absent in the conditioned media of the other three glioma cell lines. NRP1 exists mainly as a transmembrane receptor for the axonal chemorepellant semaphorin 3A (Sema3A)39,40 on the surface of neuronal cells as well as for VEGF16541, which has been shown to stimulate chemotaxis of endothelial cells. Overexpression of NRP1 has been demonstrated to promote glioma growth and correlates with tumor progression in high versus low grade astrocytomas42,43. A soluble isoform (sNRP1) has also been identified in the PC3 prostate cancer cell line and appears to be a VEGF165 antagonist with anti-tumor properties44. Close examination of the peptides detected for NRP1 revealed that the U87 cells expressed this protein in three distinct forms, with the majority of peptides (19 spectral counts) identified at the high molecular mass range of approximately 200–250 kDa, a small number (3 spectral counts) located near 130 kDa and a third form identified with nine spectral counts migrating near 56–87 kDa. These three NRP1 species likely correspond to a high-molecular weight chondroitin-sulphate modified NRP1 (NRP1-CS), unmodified NRP1 and sNRP1, respectively. Recent studies suggest that the glycan modified NRP1-CS has a dampening effect on tumor cell invasiveness since S612A site-directed mutagenesis of the glycosylation site resulted in a 50% increase in U87 cell invasiveness45. The release of this chondroitin-sulphate modified isoform into the conditioned media of U87 cells may be a regulatory mechanism by which these cells counteract any anti-invasive properties that NRP1-CS would invoke at the cell surface. The functional relevance of the sNRP1 isoform to the highly invasive properties of the U87 cell line remains to be elucidated.
Semaphorin-7A (Sema7A) was also exclusively expressed by the highly invasive U87 cell line (48 spectral counts; 14 unique peptides). Semaphorins are a family of membrane bound and soluble proteins that play a major role in axonal guidance and outgrowth. While Sema7A is a GPI-anchored membrane protein, the existence of a soluble isoform released from the cell membrane via proteolysis has been demonstrated46. Sema7A has been shown to promote the migration and chemotaxis of both monocytes and osteoblasts47,48, but its influence on cell adhesion appears to be receptor and cell-type specific. Binding of Sema7A to beta 1-integrins on the monocyte surface has been shown to promote cell attachment through MAPK signaling while cell adhesion is decreased through Sema7A interaction with plexin C147. Conversely, it has been suggested that adhesion is disrupted through beta-integrin signaling in osteoclasts48. To our knowledge, Sema7A has not been previously implicated in glioblastoma, however, studies have shown a loss of plexin C1 expression with metastatic melanoma progression as well as an inverse correlation with melanoma cell invasiveness49,50. Indeed, mass spectrometric analysis of U87 cell membrane proteins did not detect the presence of plexin C1, but rather the presence of integrin beta-1 (19 spectral counts, 12 unique peptides, data not shown) suggesting that perhaps Sema7A contributes to the increased motility and decreased adhesion necessary for U87 cell invasion via integrin signaling and the downregulation of plexin C1.
The absence of periostin and EMILIN-1 in the conditioned medium of the U87 cell line could represent an additional factor contributing to the differences in levels of invasiveness between U87 cells and the U118, LN18 and T98 cell lines. Periostin and EMILIN-1 function as cell adhesion proteins51,52 while periostin also acts to strengthen the architecture of the extracellular matrix by aiding in the deposition of matricellular proteins and regulating the fibrillar formation of collagens53,54. Both proteins were highly expressed by the less invasive cell lines LN18 and U118 but were absent in the U87 conditioned medium. Neither did the T98 cells express these proteins, however, these cells may reduce their levels of invasion in part through the high levels of expression of other adhesion-promoting factors such as thrombospondin-1. The role of EMILIN-1 in glioblastoma or tumor cell invasion in general has not been described. The effect of periostin expression on tumor cell invasion may be dependent on cellular context, since some studies have reported that increased periostin expression correlates with invasion and metastasis of some cancer types, including breast55,56, colon57, pancreatic58 and esophageal squamous cell cancer59, while there was no correlation with lung cancer metastasis56 and periostin was found to suppress invasion and metastasis in bladder cancer60. It has also been suggested that cellular localization may determine whether periostin promotes or inhibits tumor cell invasion, such that secreted periostin may act to promote cellular adhesion while a cytosolic isoform may facilitate invasion via organization of the cytoskeleton60. Thus, our data implicate periostin as an invasion suppressing protein in the extracellular environment of glioblastoma cells.
We also detected suprabasin in the conditioned medium of U87 but not in the conditioned medium of the other glioblastoma cell lines. Suprabasin transcripts were first identified in differentiating keratinocytes and have since been detected in muscle tissue, but are absent in mouse embryonic and adult brain61,62,63,64. Suprabasin is thus far believed to be involved in skin development, but its exact function is not well known at present and to our knowledge this is the first time that this protein has been shown to be secreted by invasive glioblastoma cells.
Perhaps the most interesting protein that might contribute to U87 cell invasiveness is CH3L1. This protein was highly secreted by the U87 cell line (210 spectral counts) compared to the other cell lines (≤ 4 spectral counts). CH3L1 is an extracellular carbohydrate-binding lectin devoid of chitinase activity and is not believed to possess proteolytic abilities65. However, its expression level has been shown to increase with tumor grade in many cancer types66, including glioblastoma67,68,69,35 and has been suggested as a possible biomarker for glioma subtypes68,70,71. A role for CH3L1 in tumor cell invasion is strongly suggested in our study by the decrease in U87 cell invasion observed upon addition of CH3L1 neutralizing antibodies to the cell culture medium. This finding is supported by two recent studies in which shRNA and siRNA inhibition of CH3L1 expression also blunted levels of U87 cell invasion72,69 as well as studies showing an increase in tumor cell and astrocyte invasiveness upon CH3L1 overexpression68,72. The mechanism by which CH3L1 may enhance the invasive properties of tumor cells has yet to be fully understood. Collagens type I, II and III are thus far the only known binding partners for this protein and it has been shown that bovine cartilage-derived CH3L1 inhibits the fibrillar assembly of type I collagen. It has been proposed that the retention of collagen solubility upon CH3L1 binding may render collagen more susceptible to proteolytic degradation, thereby enhancing the invasion process73. Another study indicates that CH3L1 may affect tumor cell invasiveness via multiple mechanisms including regulation of MMP2 expression and cell adhesion and by inducing changes in the actin cytoskeleton that result in the morphological organization necessary for cell mobility72. Although binding partners have not been identified for these processes, cell surface proteins are likely targets given the heparin-binding properties of CH3L174.
Conclusions
In this study we have presented the secretome profile of four glioblastoma cell lines and correlated their unique signatures with tumor cell invasiveness. While the complex interactions of proteases and their inhibitors are fairly well-characterized with respect to tumor cell invasiveness, this study highlights the need to further investigate the functions of proteins not directly involved in extracellular degradation, cell adhesion or motility that may also play important roles in tumor cell invasion. It will be interesting to further investigate the mechanisms by which proteins such as CH3L1 might enhance or promote this phenomenon. It is anticipated that new therapeutic targets may be discovered among this overlooked protein population and that their inhibition may be successful in blocking the invasion of tumor cells into surrounding healthy tissues.
Supplementary Material
Synopsis.
We performed a comprehensive examination of the secretome of four glioblastoma cell lines and correlated protein expression with invasive phenotype. We identified a number of proteins highly or exclusively expressed by the highly invasive U87 cells as compared to the less invasive cell lines, including ADAM9, ADAM10, cathepsin B, cathepsin L1, osteopontin, neuropilin-1, semaphorin-7A, suprabasin and chitinase-3-like protein 1 (CH3L1). Antibody inhibition of CH3L1 reduced U87 cell invasiveness by 30%.
Acknowledgements
This research was supported by grants from the Intellectual and Developmental Disabilities Research Center (NIH IDDRC P30HD40677), Children's National Medical Center (FY09 RAC Award 2967), the National Cancer Institute (Grant CA120316) and the Cosmos Club Foundation (2009 Cosmos Scholars Grant).
Abbreviations
- ADAM
a disintegrin and metalloproteinase domain-containing protein
- Arg
arginine
- CH3L1
chitinase-3-like protein 1
- DMEM
Dulbecco's modified Eagle's medium
- ECM
extracellular matrix
- LC
liquid chromatography
- Lys
lysine
- MMP
matrix metalloproteinase
- MS
mass spectrometry
- NRP1
neuropilin-1
- Sema3A
semaphorin-3A
- Sema7A
semaphorin-7A
- SILAC
stable isotope labeling by amino acids in cell culture
Footnotes
All authors read and approved the final manuscript.
Competing Interests The authors declare that they have no competing interests.
Supporting Information Supplemental Table 1 lists the SILAC ratios for all proteins quantified in the T98, U118, LN18 and U87 conditioned media. Supplemental Table 2a–e summarizes the peptides quantified in the T98, U118, LN18 and U87 conditioned media. Supplemental Table 3 summarizes the proteins considered to be differentially expressed in the conditioned media of the glioblastoma cell lines. Supplemental Table 4 is a complete list of proteins expressed by U87 cells that correlate with their highly invasive phenotype. Supplemental Figure 1 compares the distribution of SILAC peptide ratios between cytosolic and secreted proteins quantified in the control T98 unlabeled:T98 labeled sample. This material is available free of charge via the internet at http://pubs.acs.org.
Raw data have been submitted to the Proteomics IDEntifications database (PRIDE) at: http://www.ebi.ac.uk/pride/
References
- 1.Ries LAG, Young JL, Keel GE, Eisner MP, Lin YD, Horner MJ, editors. SEER Survival Monograph: Cancer Survival Among Adults: U.S. SEER Program, 1988–2001, Patient and Tumor Characteristics, National Cancer Institute, SEER Program, NIH Pub. No. 07-6215. Bethesda, MD: 2007. [Google Scholar]
- 2.Rao JS. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev Cancer. 2003;3:489–501. doi: 10.1038/nrc1121. [DOI] [PubMed] [Google Scholar]
- 3.Levičar N, Nutall RK, Lah TT. Proteases in brain tumor progression. Acta Neurochir. 2003;145:825–838. doi: 10.1007/s00701-003-0097-z. [DOI] [PubMed] [Google Scholar]
- 4.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 5.Ong SE, Kratchmarova I, Mann M. Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC) J Proteome Res. 2003;2:173–181. doi: 10.1021/pr0255708. [DOI] [PubMed] [Google Scholar]
- 6.Gehrmann ML, Hathout Y, Fenselau C. Evaluation of metabolic labeling for comparative proteomics in breast cancer cells. J Proteome Res. 2004;3:1063–1068. doi: 10.1021/pr049906k. [DOI] [PubMed] [Google Scholar]
- 7.Jensen ON, Wilm M, Shevchenko A, Mann M. Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. Methods Mol Biol. 1999;112:513–530. doi: 10.1385/1-59259-584-7:513. [DOI] [PubMed] [Google Scholar]
- 8.Park SK, Venable JD, Zu T, Yates JR. A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods. 2008;5:319–322. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavlidis P, Noble WS. Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-10-research0042. RESEARCH0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teramoto H, Castallone MD, Malek RL, Letwin N, Frank B, Gutkind JS, Lee NH. Autocrine activation of an osteopontin-CD44-Rac pathway enhances invasion and transformation by H-RasV12. Oncogene. 2005;13:489–501. doi: 10.1038/sj.onc.1208209. [DOI] [PubMed] [Google Scholar]
- 11.Irby RB, Malek RL, Bloom G, Tsai J, Letwin N, Frank BC, Verratii K, Yeatman TJ, Lee NH. Iterative microarray and RNA interference-based interrogation of the SRC-induced invasive phenotype. Cancer Res. 2005;65:1814–1821. doi: 10.1158/0008-5472.CAN-04-3609. [DOI] [PubMed] [Google Scholar]
- 12.An E, Lu X, Flippin J, Devaney JM, Halligan B, Hoffman E, Csaky K, Hathout Y. Secreted proteome profiling of human RPE cell cultures derived from donors with age related macular degeneration and age matched healthy donors. J Proteome Res. 2006;5:2599–2610. doi: 10.1021/pr060121j. [DOI] [PubMed] [Google Scholar]
- 13.Grønborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, Sato N, Molina H, Jensen ON, Hruban RH, Goggins MG, Maitra A, Pandey A. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics. 2006;5:157–171. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Vizcaíno JA, Côté R, Reisinger F, Foster JM, Mueller M, Rameseder J, Hermjakob H, Martens L. A guide to the Proteomics Identifications Database proteomics data repository. Proteomics. 2009;9:4276–83. doi: 10.1002/pmic.200900402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seyfried NT, Gozal YM, Dammer EB, Xia Q, Duong DM, Cheng D, Lah JJ, Levey AI, Peng J. Multiplex SILAC analysis of a cellular TDP-43 proteinopathy model reveals protein inclusions associated with SUMOylation and diverse polyubiquitin chains. Mol Cell Proteomics. 2010;9:705–718. doi: 10.1074/mcp.M800390-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rinschen MM, Yu MJ, Wang G, Boja ES, Hoffert JD, Pisitkun T, Knepper MA. Quantitative phosphoproteomic analysis reveals vasopressin V2-receptor-dependent signaling pathways in renal collecting duct cells. Proc Natl Acad Sci USA. 2010;107:3882–3887. doi: 10.1073/pnas.0910646107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soulard A, Cremonesi A, Moes S, Schütz F, Jenö The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol Biol Cell. 2010;21:3475–3486. doi: 10.1091/mbc.E10-03-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paradies NE, Grunwald GB. Purification and characterization of NCAD90, a soluble endogenous form of N-cadherin, which is generated by proteolysis during retinal development and retains adhesive and neurite-promoting function. J Neurosci Res. 1993;36:33–45. doi: 10.1002/jnr.490360105. [DOI] [PubMed] [Google Scholar]
- 19.Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, Saftig P. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and β-catenin nuclear signaling. EMBO J. 2005;24:742–752. doi: 10.1038/sj.emboj.7600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paradis S, Harrar DB, Lin Y, Koon AC, Hauser JL, Griffith EC, Zhu L, Brass LF, Chen C, Greenberg ME. An RNAi-based approach identifies molecules required for glutamatergic and GABAergic synapse development. Neuron. 2007;53:217–232. doi: 10.1016/j.neuron.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarkar S, Nuttall RK, Liu S, Edwards DR, Yong VW. Tenascin-C stimulates glioma cell invasion through matrix metalloproteinase-12. Cancer Res. 2006;66:11771–11780. doi: 10.1158/0008-5472.CAN-05-0470. [DOI] [PubMed] [Google Scholar]
- 22.Hirata E, Arakawa Y, Shirahata M, Yamaguchi M, Kishi Y, Okada T, Takahashi JA, Matsuda M, Hashimoto N. Endogenous tenascin-C enhances glioblastoma invasion with reactive change of surrounding brain tissue. Cancer Sci. 2009;100:1451–1459. doi: 10.1111/j.1349-7006.2009.01189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein T, Bishoff R. Active Metalloproteases of the a disintegrin and metalloprotease (ADAM) family: biological -function and structure. J Proteome Res. 2011;10:17–33. doi: 10.1021/pr100556z. [DOI] [PubMed] [Google Scholar]
- 24.Mongaret C, Alexandre J, Thomas-Schoemann AT, Bermudez E, Chéreau C, Nicco C, Goldwasser F, Weill B, Batteux F, Lemare F. Tumor invasion induced by oxidative stress is dependent on membrane ADAM 9 protein and its secreted form. Int J Cancer. 2011;129 doi: 10.1002/ijc.25746. n/a doi: 10.1002/ijc.25746. [DOI] [PubMed] [Google Scholar]
- 25.Mazzocca A, Coppari R, De Franco R, Cho JY, Libermann TA, Pinzani M, Toker A. A secreted form of ADAM9 promotes carcinoma invasion through tumor-stromal interactions. Cancer Res. 2005;65:4728–4738. doi: 10.1158/0008-5472.CAN-04-4449. [DOI] [PubMed] [Google Scholar]
- 26.Kohutek ZA, diPierro CG, Redpath GT, Hussaini IM. ADAM-10-mediated N-cadherin cleavage is protein kinase C-alpha dependent and promotes glioblastoma cell migration. J Neurosci. 2009;29:4605–4615. doi: 10.1523/JNEUROSCI.5126-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreeva AV, Kutuzov MA. Cadherin 13 in cancer. Genes Chromosomes Cancer. 2010;49:775–790. doi: 10.1002/gcc.20787. [DOI] [PubMed] [Google Scholar]
- 28.Huang ZY, Wu Y, Hedrick N, Gutmann DH. T-cadherin-mediated cell growth regulation involves G2 phase arrest and requires p21CIP1/WAF1 expression. Mol Cell Biol. 2003;23:566–578. doi: 10.1128/MCB.23.2.566-578.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levičar N, Strojnik T, Kos J, Dewey RA, Pilkington GJ, Lah TT. Lysosomal enzymes, cathepsins in brain tumor invasion. J Neurooncol. 2002;58:21–32. doi: 10.1023/a:1015892911420. [DOI] [PubMed] [Google Scholar]
- 30.Mason RW, Gal S, Gottesman MM. The identification of the major excreted protein (MEP) from a transformed mouse fibroblast cell line as a catalytically active precursor form of cathepsin L. Biochem J. 1987;248:449–454. doi: 10.1042/bj2480449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishido K, Kominami E. Procathepsin L degrades extracellular matrix proteins in the presence of glycosaminoglycans in vitro. Biochem Biophys Res Commun. 1995;217:624–631. doi: 10.1006/bbrc.1995.2820. [DOI] [PubMed] [Google Scholar]
- 32.Klose A, Zigrino P, Dennhöfer R, Mauch C, Hunzelmann N. Identification and discrimination of extracellularly active cathepsins B and L in high-invasive melanoma cells. Anal Biochem. 2006;353:57–62. doi: 10.1016/j.ab.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 33.Levičar N, Dewey RA, Daley E, Bates TE, Davies D, Kos J, Pilkington GJ, Lah TT. Selective suppression of cathepsin L by antisense cDNA impairs human brain tumor cell invasion in vitro and promotes apoptosis. Cancer Gene Ther. 2003;10:141–151. doi: 10.1038/sj.cgt.7700546. [DOI] [PubMed] [Google Scholar]
- 34.Lakka SS, Gondi CS, Yanamandra N, Olivero WC, Dinh DH, Gujrati M, Rao JS. Inhibition of cathepsin B and MMP-9 gene expression in glioblastoma cell line via RNA interference reduces tumor cell invasion, tumor growth and angiogenesis. Oncogene. 2004;23:4681–4689. doi: 10.1038/sj.onc.1207616. [DOI] [PubMed] [Google Scholar]
- 35.Skreekanthreddy P, Srinivasan H, Kuman DM, Nijaguna MB, Sridevi S, Vrinda M, Arivazhagan A, Balasubramaniam A, Hegde AS, Chandramouli BA, Santosh V, Rao MR, Kondaiah P, Somasundaram K. Identification of potential serum biomarkers of glioblastoma: serum osteopontin levels correlate with poor prognosis. Cancer Epidemiol Biomarkers Prev. 2010;19:1409–1422. doi: 10.1158/1055-9965.EPI-09-1077. [DOI] [PubMed] [Google Scholar]
- 36.Schuhmann MW, Zucht HD, Nassimi R, Heine G, Schneekloth CG, Steurenburg HJ, Selle H. Peptide screening of cerebrospinal fluid in patients with glioblastmoa multiforme. Eur J Surg Oncol. 2010;36:201–207. doi: 10.1016/j.ejso.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 37.Lamour V, Le Mercier M, Lefranc F, Hagedorn M, Javerzat S, Bikfalvi A, Kiss R, Castronovo V, Bellahcène A. Selective osteopontin knockdown exerts anti-tumoral activity in a human glioblastoma model. Int J Cancer. 2010;126:1797–1805. doi: 10.1002/ijc.24751. [DOI] [PubMed] [Google Scholar]
- 38.Kim MS, Park MJ, Moon EJ, Kim SJ, Lee CH, Yoo H, Shin SH, Song ES, Lee SH. Hyaluronic acid induces osteopontin via the phosphatidylinositol 3-kinase/Akt pathway to enhance the motility of human glioma cells. Cancer Res. 2005;65:686–691. [PubMed] [Google Scholar]
- 39.He Z, Tessier-Lavigne M. Neuropilin is a receptor for the axonal chemorepellent semaphorin III. Cell. 1997;90:739–751. doi: 10.1016/s0092-8674(00)80534-6. [DOI] [PubMed] [Google Scholar]
- 40.Kolodkin AL, Levengood DV, Rowe EG, Tai YT, Giger RJ, Ginty DD. Neuropilin is a semaphorin III receptor. Cell. 1997;90:753–762. doi: 10.1016/s0092-8674(00)80535-8. [DOI] [PubMed] [Google Scholar]
- 41.Soker S, Takashima S, Miao HQ, Neufeld G, Klagsbrun M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell. 1998;92:735–745. doi: 10.1016/s0092-8674(00)81402-6. [DOI] [PubMed] [Google Scholar]
- 42.Broholm H, Laursen H. Vascular endothelial growth factor (VEGF) receptor neuropilin-1's distribution in astrocytic tumors. AMPIS. 2004;112:257–263. doi: 10.1111/j.1600-0463.2004.apm11204-0505.x. [DOI] [PubMed] [Google Scholar]
- 43.Nassare C, Roth M, Jacob L, Roth L, Koncina E, Thien A, Labourdette G, Poulet P, Hubert P, Crémel G, Roussel G, Aunis D, Bagnard D. Peptide-based interference of the transmembrane domain of neuropilin-1 inhibits glioma growth in vivo. Oncogene. 2010;29:2381–2392. doi: 10.1038/onc.2010.9. [DOI] [PubMed] [Google Scholar]
- 44.Gagnon ML, Bielenberg Gechtman Z, DR., Miao HQ, Takashima S, Soker S, Klagsbrun M. Identification of a natural soluble neuropilin-1 that binds vascular endothelial growth factor: In vivo expression and antitumor activity. Proc Natl Acad Sci USA. 2000;97:2573–2578. doi: 10.1073/pnas.040337597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frankel P, Pellet-Many C, Lehtolainen P, D'Abaco GM, Tickner ML, Cheng L, Zachary IC. Chondroitin sulphate-modified neuropilin-1 is expressed in human tumour cells and modulates 3D invasion in the U87MG human glioblastoma cell line through a p130Cas-mediated pathway. EMBO Rep. 2008;9:983–989. doi: 10.1038/embor.2008.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Angelisová P, Drbal K, Cerný J, Hilgert I, Horejsí V. Characterization of the human leukocyte GPI-anchored glycoprotein CDw108 and its relation to other similar molecules. Immunobiology. 1999;200:234–245. doi: 10.1016/s0171-2985(99)80073-4. [DOI] [PubMed] [Google Scholar]
- 47.Scott GA, McClelland LA, Fricke AF. Semaphorin 7a promotes spreading and dendricity in human melanocytes through beta 1-integrins. J Invest Dermatol. 2008;128:151–161. doi: 10.1038/sj.jid.5700974. [DOI] [PubMed] [Google Scholar]
- 48.Delorme G, Saltel F, Bonnelye E, Jurdic P, Machuca-Gayet I. Expression and function of semaphorin 7A in bone cells. Biol Cell. 2005;97:589–597. doi: 10.1042/BC20040103. [DOI] [PubMed] [Google Scholar]
- 49.Lazova R, Gould Rothberg BE, Rimm D, Scott G. The semaphorin 7A receptor plexin C1 is lost during melanoma metastasis. Am J Dermatopathol. 2009;31:177–181. doi: 10.1097/DAD.0b013e318196672d. [DOI] [PubMed] [Google Scholar]
- 50.Scott GA, McClelland LA, Fricke AF, Fender A. Plexin C1, a receptor for semaphorin 7A, inactivates cofilin and is a potential tumor suppressor for melanoma progression. J Invest Dermatol. 2009;129:954–963. doi: 10.1038/jid.2008.329. [DOI] [PubMed] [Google Scholar]
- 51.Takeshita S, Kikuno R, Tezuka K, Amann E. Osteoblast-specific factor 2: cloning of a putative bone adhesion protein with homology with the insect protein fasciclin I. Biochem J. 1993;294:271–278. doi: 10.1042/bj2940271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spessotto P, Cervi M, Mucignat MT, Mungiguerra G, Sartoretto I, Doliana R, Colombatti A. beta 1 integrin-dependent cell adhesion to EMILIN-1 is mediated by the gC1q domain. J Biol Chem. 2003;278:6160–6167. doi: 10.1074/jbc.M208322200. [DOI] [PubMed] [Google Scholar]
- 53.Norris RA, Damon B, Mironov V, Kasyanov V, Ramamurthi A, Moreno-Redriguez R, Trusk T, Potts JD, Goodwin RL, Davis J, Hoffman S, Wen X, Sugi Y, Kern CB, Mjaatvedt CH, Turner DK, Oka T, Conway SJ, Molkentin JD, Forgacs G, Markwald RR. Periostin regulates collagen fibrillogenesis and the biomechanical properties of connective tissues. J Cell Biochem. 2007;101:695–711. doi: 10.1002/jcb.21224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kii I, Nishiyama T, Li M, Matsumoto K, Saito M, Amizuka N, Kudo A. Incorporation of tenascin-C into the extracellular matrix by periostin underlies an extracellular meshwork architecture. J Biol Chem. 2010;285:2028–39. doi: 10.1074/jbc.M109.051961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shao R, Bao S, Bai X, Blanchette C, Anderson RM, Dang T, Gishizky ML, Marks JR, Wang XF. Acquired expression f periostin by human breast cancers promotes tumor angiogenesis through up-regultaion of vascular endothelial growth factor receptor 2 expression. Mol Cell Biol. 2004;24:3992–4003. doi: 10.1128/MCB.24.9.3992-4003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sasaki H, Yu CY, Dai M, Tam C, Loda M, Auclair D, Chen LB, Elias A. Elevated serum periostin levels in patients with bone metastases from breast but not lung cancer. Breast Cancer Res Treat. 2003;77:245–252. doi: 10.1023/a:1021899904332. [DOI] [PubMed] [Google Scholar]
- 57.Bao S, Ouyang G, Bai X, Huang Z, Ma C, Liu M, Shao R, Anderson RM, Rich JN, Wang XF. Periostin potently promotes metastatic growth of colon cancer by augmenting cell survival via the Akt/PKB pathway. Cancer Cell. 2004;5:329–339. doi: 10.1016/s1535-6108(04)00081-9. [DOI] [PubMed] [Google Scholar]
- 58.Ben QW, Jin XL, Liu J, Cai X, Yuan F, Yuan YZ. Periostin, a matrix specific protein, is associated with proliferation and invasion of pancreatic cancer. Oncol Rep. 2011;25:709–716. doi: 10.3892/or.2011.1140. [DOI] [PubMed] [Google Scholar]
- 59.Michaylira CZ, Wong GS, Miller CG, Gutierrez CM, Nakagawa H, Hammond R, Klein-Szanto AJ, Lee JS, Kim SB, Herlyn M, Diehl JA, Gimotty P, Rustgi AK. Periostin, a cell adhesion molecule, facilitates invasion in the tumor microenvironment and annotates a novel tumor-invasive signature in esophageal cancer. Cancer Res. 2010;70:5281–5292. doi: 10.1158/0008-5472.CAN-10-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim CJ, Yoshioka N, Tambe Y, Kushima R, Okada Y, Inoue H. Periostin is down-regulated in high grade human bladder cancers and suppresses in vitro cell invasiveness and in vivo metastasis of cancer cells. Int J Cancer. 2005;117:51–58. doi: 10.1002/ijc.21120. [DOI] [PubMed] [Google Scholar]
- 61.Park GT, Lim SE, Jang SI, Morasso MI. Suprabasin, a novel epidermal differentiation marker and potential cornified envelope precursor. J Biol Chem. 2002;47:45195–45202. doi: 10.1074/jbc.M205380200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bazzi H, Fantauzzo KA, Richardson GD, Jahoda CAB, Christiano AM. Transcriptional profiling of developing mouse epidermis reveals novel patterns of coordinated gene expression. Dev Dyn. 2007;236:961–970. doi: 10.1002/dvdy.21099. [DOI] [PubMed] [Google Scholar]
- 63.Brass EP, Peters MA, Hinchcliff KW, He YD, Ulrich RG. Temporal pattern of skeletal muscle gene expression following endurance exerc ise in Alaskan sled dogs. J Appl Physiol. 2009;107:605–612. doi: 10.1152/japplphysiol.91347.2008. [DOI] [PubMed] [Google Scholar]
- 64.Moffatt P, Salois P, St-Amant N, Gaumond MH, Lanctôt C. Identification of a conserved cluster of skin-specific genes encoding secreted proteins. Gene. 2004;334:123–131. doi: 10.1016/j.gene.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 65.Hakala BE, White C, Recklies AD. Human Cartilage gp-39, a major secretory product of articular chondrocytes and synovial cells, is a mammalian member of a chitnase protein family. J Biol Chem. 1993;268:25803–25810. [PubMed] [Google Scholar]
- 66.Coffman FD. Chitinase 3-like-1 (CHI3L1): a putative disease marker at the interface of proteomics and glycomics. Crit Rev Clin Lab Sci. 2008;45:531–562. doi: 10.1080/10408360802334743. [DOI] [PubMed] [Google Scholar]
- 67.Nigro JM, Misra A, Zhang L, Smirnov I, Colman H, Griffin C, Ozburn N, Chen M, Pan E, Koul D, Yung WKA, Feurerstein BG, Aldape KD. Integrated array-comparative genomic hybridization and expression array profiles identify clinically relevant molecular subtypes of glioblastoma. Cancer Res. 2005;65:1678–1686. doi: 10.1158/0008-5472.CAN-04-2921. [DOI] [PubMed] [Google Scholar]
- 68.Hormigo A, Gu B, Karimi S, Reidel E, Panageas KS, Edgar MA, Tanwar MK, Rao JS, Fleisher M, DeAngelis LM, Holland EC. YKL-40 and matrix metalloproteinase-9 as potential serum biomarkers for patients with high-grade-gliomas. Clin Cancer Res. 2006;12:5698–5704. doi: 10.1158/1078-0432.CCR-06-0181. [DOI] [PubMed] [Google Scholar]
- 69.Zhang W, Murao K, Zhang X, Matsumoto K, Diah S, Okada M, Miyake K, Kawai N, Fei Z, Tamiya T. Resveratrol represses YKL-40 expression in human glioma U87 cells. BMC Cancer. 2010;10:593. doi: 10.1186/1471-2407-10-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rousseau A, Nutt CL, Betensky RA, Iafrate AJ, Han M, Ligon KL, Rowtich DH, Louis DN. Expression of oligodendroglial and astrocytic lineage markers in diffuse gliomas: use of YKL-40, ApoE, ASCL1, and NKX2-2. J Neuropathol Exp Neurol. 2006;65:1149–1156. doi: 10.1097/01.jnen.0000248543.90304.2b. [DOI] [PubMed] [Google Scholar]
- 71.Nutt CL, Betensky RA, Brower MA, Batchelor TT, Louis DN, Stemmer-Rachamimov AO. YKL-40 is a differential diagnostic marker for histologic subtypes of high-grade-gliomas. Clin Cancer Res. 2005;11:2258–2264. doi: 10.1158/1078-0432.CCR-04-1601. [DOI] [PubMed] [Google Scholar]
- 72.Ku BM, Lee YK, Ryu J, Jeong JY, Choi J, Eun KM, Shin HY, Kim DG, Hwang EM, Yoo JC, Park JY, Roh GS, Kim HJ, Cho GJ, Choi WS, Paek SH, Kang SS. CHI3L1 (YKL-40), is expressed in human gliomas and regulates the invasion, growth and survival of glioma cells. Int J Cancer. 2010;128:1316–1326. doi: 10.1002/ijc.25466. [DOI] [PubMed] [Google Scholar]
- 73.Bigg HF, Wait R, Rowan AD, Cawston TE. The mammalian chitinase-like lectin, YKL-40, binds specifically to type I collagen and modulates the rate of type I collagen fibril formation. J Biol Chem. 2006;281:21082–21095. doi: 10.1074/jbc.M601153200. [DOI] [PubMed] [Google Scholar]
- 74.Nyirkos P, Golds EE. Human synovial cells secrete a 39 kDa protein similar to a bovine mammary protein expressed during the non-lactating period. Biochem J. 1990;269:265–268. doi: 10.1042/bj2690265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.