Abstract
Background
Neuroblastoma (NB) is an important childhood cancer with a strong genetic component related to disease susceptibility. Approximately 1% of NB cases have a positive family history. Following a genome-wide linkage analysis and sequencing of candidate genes in the critical region, we identified ALK as the major familial NB gene. Dominant mutations in ALK are found in more than 50% of familial NB cases. However, in the families used for the linkage study, only about 50% of carriers of ALK mutations are affected by NB.
Methods
To test whether genetic variation may explain the reduced penetrance of the disease phenotype, we analyzed genome-wide genotype data in ALK mutation-positive families using a model-based linkage approach with different liability classes for carriers and non-carriers of ALK mutations.
Results
The region with the highest LOD score was located at chromosome 2p23–p24 and included the ALK locus under models of dominant and recessive inheritance.
Conclusions
This finding suggests that variants in the non-mutated ALK gene or another gene linked to it may affect penetrance of the ALK mutations and risk of developing NB in familial cases.
Key Words: Neuroblastoma, Penetrance, Linkage analysis, Genetic modifier
Introduction
Neuroblastoma (NB) is a childhood cancer affecting approximately 1 in 10,000 children, with one of the highest mortality rates of all pediatric malignancies [1]. Most NBs arise sporadically, but approximately 1% of all cases have a positive family history [1]. Following a whole-genome linkage analysis and sequencing of candidate genes in the critical region thus identified, we showed that dominant mutations in ALK on chromosome 2p23 are responsible for NB in approximately 50% of familial cases [2]. However, genotyping of relatives of affected individuals also demonstrated that only about 50% of all heterozygous carriers of ALK mutations are affected by NB in the families used for the linkage study. Variation at the major gene as well as elsewhere in the genome, or modifier genes, may explain reduced penetrance in Mendelian disorders [3]. In order to identify potential genetic modifiers of the disease penetrance, we re-evaluated the genome-wide data limiting the analysis to the families with ALK mutations using a model-based linkage approach. Our data suggest that variants in the non-mutated ALK gene or another gene linked to it on chromosome 2p23–p24 may influence the probability that a child develops NB in the presence of ALK mutations.
Subjects and Methods
NB Families
All NB families included in the present study were part of the linkage study described in Mossé et al. [2]. Only families with confirmed ALK mutations were included in the present study. These comprise all the families in figure 1 in Mossé et al. [2] with the exception of family 6 and family 12 that are not informative for linkage analysis, leaving a total of 6 pedigrees including 69 individuals with available genome-wide genotype data. A total of 21 individuals in these families were affected by NB, and of these, 20 were confirmed to be carriers of a single ALK mutation using methods already described [2]. One affected child who was not tested for the presence of ALK mutations was the affected male in family 56. He was the brother and the son of two ALK-positive individuals, and was therefore assumed to be a carrier of the same mutation present in his relatives. In addition, 22 unaffected individuals were identified as non-penetrant carriers of the same ALK mutations present in their affected relatives. The remaining 26 individuals were unaffected and did not carry an ALK mutation.
Fig. 1.
Chromosome 2 LOD scores (continuous line) and HLOD scores (broken line) under 4 models of inheritance. In all inheritance models, penetrance was fixed at 50%.
Linkage Analysis
Genome-wide genotype data used for this study was obtained using the Illumina Linkage IVb SNP panel, and was the same we utilized in the original linkage study which identified ALK as the major NB gene in these families [2]. For the purpose of this study, linkage analysis was performed with Merlin 1.1.2 [4] using maximum likelihood estimates of SNP allele frequencies based on the NB family data. In order to be able to include information in the analysis about all carriers of ALK mutations, whether affected or unaffected, we used a model-based approach, as model-free analysis is typically ‘affected-only’. Two liability classes were defined conditional on the presence of ALK mutations. In the first class we included all unaffected individuals without ALK mutations and assumed 0 penetrance for the disease. In the other liability class we included all individuals, affected and unaffected, who were carriers of an ALK mutation and assumed an incomplete penetrance of the disease equal to 0.5. Under this model, regions of the genome that are shared by ALK-positive affected individuals in the same family but not by their ALK-positive unaffected relatives should provide the strongest evidence for linkage; unaffected individuals who do not carry ALK mutations do not contribute any information. Analyses were performed using two different disease gene frequencies q, one rare (q = 0.001) and one common (q = 0.1), under both a dominant and a recessive mode of inheritance, for a total of 4 models. Linkage in the presence of heterogeneity was assessed using heterogeneity HLOD scores and their accompanying estimates of the proportion of linked families α.
Data for chromosome 2 were also analyzed assuming 3 liability classes, one for unaffected individuals without ALK mutations (probability of being affected equal 0 for all genotypes); one for all affected carriers of ALK mutations (probability of being affected equal 0 for the normal, and 0.5 for the disease-risk genotypes), and one for all unaffected carriers of ALK mutations (probability of being affected equal 0 for the normal, and 0.1 for the disease-risk genotypes). Finally, for chromosome 2 only, we used LAMP [5] to calculate a LOD score maximized over all possible model parameters (MOD score). Based on the family genotype data and a prevalence of NB equal to 1/10,000, LAMP estimates disease allele frequency and penetrance for the 3 disease genotypes (homozygous normal, +/+; heterozygous, +/–; and homozygous affected, –/–). All affected and unaffected individuals, both ALK positive and negative, with genotype data were included in this analysis with their observed phenotype.
Genetic map positions in centiMorgan for all SNPs were approximated dividing the physical map positions in base pairs, according to the NCBI Build 36, by 106. All reported genomic positions are according to the NCBI Build 36.
Results
Genome-wide LOD scores for all chromosomes under the 4 models (rare dominant; common dominant; rare recessive; common recessive), including heterogeneity LOD scores, are reported in online supplementary figures 1–4 (www.karger.com/doi/10.1159/000324843). The only region of the genome with LOD scores greater than 3 was on chromosome 2p (fig. 1, table 1). The highest LOD score for the dominant model was 3.50 for q = 0.001 at SNP rs1997325 located at 22,677 kb, and for the recessive model it was 3.58 for q = 0.1 at SNP rs520354 located at 21,113 kb. In both cases, the LOD and the HLOD scores were identical, and the estimate of the proportion of linked families α was equal to 1. Reduction of penetrance for unaffected carriers of ALK mutations from 0.5 to 0.1 did not modify the overall chromosome 2 results significantly, with a maximum LOD and HLOD of 4.62 for q = 0.001 at SNP rs1997325 under the dominant model, and a maximum LOD and HLOD of 3.14 for q = 0.1 at SNP rs520354 under the recessive model.
Table 1.
Maximum LOD scores in chromosome 2p
| Location, bp | SNP | LOD | HLOD | Alpha | Inheritance model | Gene frequency |
|---|---|---|---|---|---|---|
| 22,677,255 | rs1997325 | 3.50 | 3.50 | 1 | dominant | 0.001 |
| 22,677,255 | rs1997325 | 1.69 | 1.69 | 1 | dominant | 0.1 |
| 21,113,117 | rs520354 | 2.25 | 2.96 | 0.82 | recessive | 0.001 |
| 21,113,117 | rs520354 | 3.58 | 3.58 | 1 | recessive | 0.1 |
In all inheritance models, penetrance was fixed at 50%.
Alpha = Estimate of proportion of linked families.
Only two other genomic regions showed LOD and/ or HLOD scores greater than 2 (table 2, online suppl. fig. 1–4). SNP rs1396208 on chromosome 12 had LOD and HLOD of 2.11 and 2.14, respectively, under the common recessive model. A previous linkage analysis of one of these families, FNB52, using microsatellites and a model-based affected-only approach, supported linkage to a region on chromosome 12p containing SNP rs1396208 with a maximum LOD score of 3.01 [6], in addition to the 2p region where ALK is located. SNP rs756658 on chromosome 22 had HLOD scores of 2.50, 2.74, and 2.32 under the rare dominant, rare recessive, and common recessive models, respectively.
Table 2.
LOD scores ≥2 at locations other than chromosome 2p
| Chromosome | Location, bp | SNP | LOD | HLOD | Alpha | Inheritance model | Gene frequency |
|---|---|---|---|---|---|---|---|
| 12 | 24,449,587 | rs1396208 | 2.11 | 2.14 | 0.92 | recessive | 0.1 |
| 22 | 17,835,836 | rs756658 | 0.21 | 2.50 | 0.80 | dominant | 0.001 |
| 22 | 17,835,836 | rs756658 | −1.80 | 2.74 | 0.80 | recessive | 0.001 |
| 22 | 17,835,836 | rs756658 | 1.61 | 2.32 | 0.81 | recessive | 0.1 |
In all inheritance models, penetrance was fixed at 50%.
Alpha = Estimate of proportion of linked families.
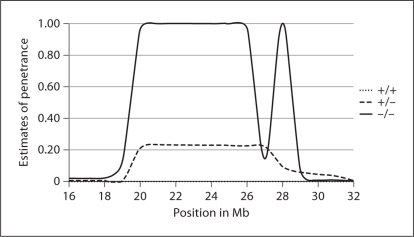
To estimate the penetrance of the 3 disease genotypes in the chromosome 2p region, we ran a MOD score analysis using the LAMP software [5] and data on all available individuals, affected and unaffected, including those who did not carry an ALK mutation. The maximum LOD score estimated by LAMP was 4.85 near rs2001795 at 23 Mb. Estimates of penetrance in the chromosome 2p region from 16 to 32 Mb around the location of the maximum LOD scores are reported in figure 2. The homozygous –/– genotype was estimated to have a penetrance equal to 1 at most positions between 20 and 29 Mb, whereas in the same region, the penetrance was 0.23 for the heterozygous +/– and close to 0 for the homozygous +/+ genotype. The dip in the homozygous –/– penetrance around 27 Mb may be explained by limited marker information at this position. In fact there are a total of 29 SNPs in the 16 Mb region of figure 2, but only one is located between 25 and 29 Mb.
Fig. 2.
Estimates of penetrance for the 3 disease locus genotypes on chromosome 2p region between 16 and 32 Mb.
Discussion
All together, our results suggest that genetic variants located in the 2p23–p24 region on the chromosome containing the non-mutated ALK allele affect the probability of an individual developing NB in the presence of an ALK mutation on the homologous chromosome. Given that ALK itself is located on 2p23, sharing among affected individuals of that region on the chromosome carrying the ALK mutation can explain the positive linkage signal observed under the dominant model; however, the positive LOD scores obtained under the recessive model requires the additional co-segregation with penetrance of the disease of the chromosome 2p region transmitted by the non-carrier parent. Accordingly, the probability of being affected is estimated at only 0.23 for heterozygous carriers of a putative disease variant in the same chromosome 2p region, but at 1 for homozygous carriers. ALK is located between 29,296 and 29,998 kb on chromosome 2p, and therefore approximately 7 Mb and 8 Mb away from the location of the maximum LOD scores under the dominant and recessive models, respectively. Only one intragenic ALK SNP, rs1358514, is included in our data; its LOD (and HLOD) scores, under the two models achieving maximum LOD scores >3, were 2.83 for the dominant, and 2.48 for the recessive model, respectively. Given the limited resolution of linkage analysis, it is possible that the positive linkage signal under the recessive model is due to segregation of ALK variants on the chromosome transmitted by the non-carrier parent that modify penetrance of the ALK mutations on the homologous chromosome. In other words, presence of otherwise silent ALK variants in trans to a pathogenic mutation would result in NB.
According to dbSNP build 132, there are 8,850 SNPs in the ALK gene region, 94 of which are reported as coding SNPs. It is possible to hypothesize that some of these variants may affect gene expression and thus influence penetrance of the mutations located on the homologous allele. A similar mechanism was suggested for autosomal dominant retinitis pigmentosa (RP) linked to the RP11 locus on chromosome 19q, on the basis of linkage data that showed that RP penetrance co-segregated with the transmission of wild-type alleles from the non-carrier parents [7]. In erythropoietic protoporphyria, another autosomal dominant disease with reduced penetrance due to mutations of the FECH gene, the presence of a FECH common single-nucleotide variant in trans to the mutated allele is necessary for expression of the disease [8]. In hereditary elliptocytosis caused by dominant mutations in the gene coding for the alpha subunit of spectrin, penetrance of mutations is affected by a high-frequency, otherwise-silent polymorphism at the same locus that determines the relative level of expression of alpha-spectrin alleles [9]. The limited data currently available to us on variation within the ALK gene itself in these families (only one intragenic SNP is included in the linkage panel) prevents us at this time to directly test this hypothesis. Higher density SNP genotyping or sequencing data at the ALK locus on all carriers, affected and unaffected, would be necessary for this purpose.
We cannot exclude that other loci in the proximity of the ALK locus, rather than ALK itself, have an effect on penetrance of its mutations. The 1-LOD support interval for the common recessive model spans approximately 14 Mb from 18,050 to 32,350 kb and includes more than 120 RefSeq genes. Given the rarity of familial NB, it is unlikely that many additional families will become available to allow a finer mapping of this region by linkage analysis. Other approaches such as association analysis following high-density SNP genotyping or targeted sequencing are more likely to be successful in providing a definitive answer as to the presence of modifier variants of the NB penetrance in this genomic region.
Supplementary Material
Supplemental Figures
Acknowledgements
This work was support in part by NIH grants R01-CA78454 (to J.M.) and R01-CA140198 (to Y.M.). L.L. is supported by the Italian Neuroblastoma Foundation.
References
- 1.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mossé YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, Laquaglia MJ, Sennett R, Lynch JE, Perri P, Laureys G, Speleman F, Kim C, Hou C, Hakonarson H, Torkamani A, Schork NJ, Brodeur GM, Tonini GP, Rappaport E, Devoto M, Maris JM. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nadeau JH. Modifier genes and protective alleles in humans and mice. Curr Opin Genet Dev. 2003;13:290–295. doi: 10.1016/s0959-437x(03)00061-3. [DOI] [PubMed] [Google Scholar]
- 4.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin – rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 5.Li M, Boehnke M, Abecasis GR. Joint modeling of linkage and association: identifying SNPs responsible for a linkage signal. Am J Hum Genet. 2005;76:934–949. doi: 10.1086/430277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Longo L, Panza E, Schena F, Seri M, Devoto M, Romeo G, Bini C, Pappalardo G, Tonini GP, Perri P. Genetic predisposition to familial neuroblastoma: identification of two novel genomic regions at 2p and 12p. Hum Hered. 2007;63:205–211. doi: 10.1159/000099997. [DOI] [PubMed] [Google Scholar]
- 7.McGee TL, Devoto M, Ott J, Berson EL, Dryja TP. Evidence that the penetrance of mutations at the RP11 locus causing dominant retinitis pigmentosa is influenced by a gene linked to the homologous RP11 allele. Am J Hum Genet. 1997;61:1059–1066. doi: 10.1086/301614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gouya L, Martin-Schmitt C, Robreau AM, Austerlitz F, Da Silva V, Brun P, Simonin S, Lyoumi S, Grandchamp B, Beaumont C, Puy H, Deybach JC. Contribution of a common single-nucleotide polymorphism to the genetic predisposition for erythropoietic protoporphyria. Am J Hum Genet. 2006;78:2–14. doi: 10.1086/498620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Randon J, Boulanger L, Marechal J, Garbarz M, Vallier A, Ribeiro L, Tamagnini G, Dhermy D, Delaunay J. A variant of spectrin low-expression allele alpha LELY carrying a hereditary elliptocytosis mutation in codon 28. Br J Haematol. 1994;88:534–540. doi: 10.1111/j.1365-2141.1994.tb05070.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figures