Abstract
Nanodiscs are an example of discoidal nanoscale lipid/protein particles that have been extremely useful for the biochemical and biophysical characterization of membrane proteins. They are discoidal lipid bilayer fragments encircled and stabilized by two amphipathic helical proteins named membrane scaffolding protein (MSP), ~10 nm in size. Nanodiscs are homogeneous, easily prepared with reproducible success, amenable to preparations with a variety of lipids, and stable under a range of temperatures. Here we present solid-state NMR (SSNMR) studies on lyophilized, rehydrated POPC Nanodiscs prepared with uniformly 13C, 15N-labeled MSP1D1 (Δ1-11 truncated MSP). Under these conditions, by SSNMR we directly determine the gel-to-liquid crystal lipid phase transition to be at 3 ± 2 °C. Above this phase transition, the lipid 1H signals have slow transverse relaxation, enabling filtering experiments as previously demonstrated for lipid vesicles. We incorporate this approach into two- and three-dimensional heteronuclear SSNMR experiments to examine the MSP1D1 residues interfacing with the lipid bilayer. These 1H-13C and 1H-13C-13C correlation spectra are used to identify and quantify the number of lipid-correlated and solvent-exposed residues by amino acid type, which furthermore is compared with molecular dynamics studies of MSP1D1 in Nanodiscs. This study demonstrates the utility of SSNMR experiments with Nanodiscs for examining lipid-protein interfaces and has important applications for future structural studies of membrane proteins in physiologically relevant formulations.
The structure and function of membrane proteins is often significantly affected by the specific properties of the lipid bilayer in which the protein is embedded or tethered. A number of membrane bilayer properties such as cholesterol content, lipid headgroup charge, saturation of the lipid acyl chains, and length of the acyl chains can be crucial for proper folding and function of the membrane protein. Given that the native membrane itself can be closely tied to structural and functional properties of a membrane protein, comprehensive studies of membrane proteins should consider the structure and function of these proteins in the context of the native membrane environment and appropriate lipid components. Examples of membrane effects on protein structure and/or function can be found in proteins involved in the blood coagulation cascade (1, 2), drug metabolizing cytochrome P450s (3-5), G-protein coupled receptors (GPCRs) (6), ion channels (7, 8), and antimicrobial peptides (9-11). Constitution of lipid membranes can affect binding affinity between proteins, catalytic efficiency, and enzymatic activity of a membrane protein (3),(7),(12). The interaction between membrane proteins and the membrane bilayer implies that structural and functional studies of membrane proteins should take all necessary precautions in preparation of protein/bilayer mimetic assemblies, and assure that the chosen conditions do not alter protein’s native structure or function, which often represents a challenging prospect (13, 14).
In the last decade, a new membrane mimetic has been developed and well characterized for structural and functional studies of membrane proteins (15-23). This mimetic, called the Nanodisc (Figure 1a), consists of a 10 nm discoidal lipid bilayer encircled and stabilized by two molecules of amphipathic helical membrane scaffolding protein (MSP), derived from nascent high-density lipoproteins (HDLs) (16). Studies on Nanodiscs have been important in understanding many aspects of self-assembly and properties of HDLs (19, 20, 24, 25). More importantly, Nanodiscs have been instrumental in a number of functional and structural studies on various membrane proteins including rhodopsin and bacteriorhodopsin (21, 23), cytochrome P450s (17, 26-28), tissue factor (2), and others (29-31). To facilitate structural analysis of MSP, the protein component of Nanodiscs, we have developed methods for preparation of Nanodiscs for SSNMR studies (32). We have further shown that membrane proteins can be assembled in Nanodiscs and prepared in an active state suitable for SSNMR (33). Recently Nanodiscs have also been used to successfully study peptides and even integral membrane proteins using solution NMR (34-36). Dependant on the membrane protein of interest, Nanodiscs can offer a number of advantages over other traditionally used membrane mimetics. They can be assembled with a variety of lipids and lipid mixtures, allowing precise control of the lipid composition in the microenvironment of the embedded protein (16, 27, 37). This proved to be quite useful in the studies of blood clotting, where the use of Nanodiscs gave insights into the effects of increasing amount of charged phospholipids in the bilayer and that of bilayer completely made of charged phospholipids on the activity of the proteins involved in the initiation of the blood clotting cascade (38, 39). Nanodiscs also provide access to both sides of the embedded transmembrane proteins, and allow control of the oligomeric state of the incorporated protein (17, 21), which is quite difficult in detergent micelles or liposomes. This has previously allowed investigations into functional properties of a monomeric human drug-metabolizing protein cytochrome P450 3A4 (40, 41), whose function has been known to be affected by its aggregation state (42) impossible to control in other membrane mimetics. Nanodiscs have also allowed functional assessments and comparisons between monomeric and oligomeric protein assemblies in the case of seven transmembrane proteins (43, 44), bacterial chemoreceptor (37), and the peptide translocon complex (45, 46). In addition, Nanodiscs are amendable to incorporation of well-defined protein assemblies, and studies of a single P450 complexed with its redox partner in a Nanodisc have provided valuable insights (40, 47, 48). Reviews of Nanodiscs, as well numerous membrane proteins studies that have benefited from the use of Nanodiscs have recently been published (49, 50). Advantages provided by the Nanodisc system allow one to probe changes in protein conformation, protein/membrane interfaces and depth of membrane insertion as a function of lipid composition. The combination of these advantages and the stability of Nanodiscs under SSNMR experimental conditions make Nanodiscs a promising platform for structural studies of membrane proteins in their native membrane environment using SSNMR.
Figure 1. Gel to liquid crystalline transition of lipids in Nanodiscs.
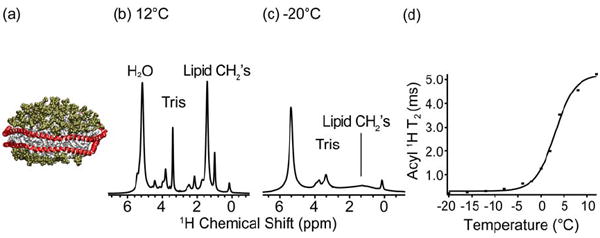
(a) A cartoon representation of a Nanodisc with MSP1D1 in red and POPC headgroups in green and tails in white. 1H 1D spectra of POPC Nanodiscs at (b) 12 °C and (c) -20 °C. Spectra were acquired on a 600 MHz (1H frequency) spectrometer. The acyl 1H linewidth and peak height indicates the phase transition. (d) Graph of T2 values as a function of temperature for the acyl methylene 1H signals (1.3 ppm). The lipid phase transition in POPC Nanodiscs is determined to be at +3 °C.
SSNMR has been extensively applied in the recent years to structural studies of membrane proteins (51-54), fibrils (55-57) and other supramolecular assemblies that remained inaccessible to other biophysical techniques. In particular, SSNMR is a powerful tool for obtaining detailed information about the membrane-protein interface and can yield information on the assembly of membrane proteins into functional oligomeric complexes, orientation (58),(59),(60), dynamics, location, and depth of insertion of proteins in the membrane (61),(62),(63),(64),(65-67), as recently reviewed (68).
In this study, we use two- and three-dimensional magic-angle spinning (MAS) SSNMR to investigate lipid-protein interfaces using the Nanodisc platform. The implemented experiments use proton spin diffusion, where magnetization from a spin source is selected based on mobility and transferred to the target protein. In this particular case, lipids are the spin source, since they are mobile and have relatively long T2 relaxation times (several ms or more, depending on temperature) above the lipid phase transition. Our studies are modeled after similar studies in which lipids within vesicles were used as the magnetization source (65), but complementary information with water as the magnetization source can be obtained with proteins embedded in bilayer below lipid phase transition temperature (69). In either case, a 1H T2 filter is used to select for the mobile 1H spins, and the 1H signals from the relatively rigid protein are suppressed by this filter. In the studies where lipid protons are the source of magnetization, the spin diffusion from lipid protons is fast for the transmembrane domains of a protein, while this is not the case for proteins with domains residing on the bilayer/water interface.
Here we use these proton spin-diffusion experiments to examine the lipid/protein interface of MSP in a Nanodisc. We describe a new preparation method of Nanodisc samples for SSNMR that is compatible with the more physiologically relevant POPC lipids. Studies presented here demonstrate that Nanodiscs retain the phase transition behavior of lipid bilayers in biological membranes under SSNMR sample preparation conditions, and that the lipid to protein ratio is sufficiently low (i.e sample sensitivity high) to obtain three-dimensional 1H-13C-13C spectra of MSP. The experimental spectra are compared to the molecular dynamic simulation results for MSP. The SSNMR spectra of MSP allow for amino acid type assignments, and the quantification of the 2D and 3D spectra by amino acid type are consistent with the results of molecular dynamics simulations. This work shows the potential of combining SSNMR with the Nanodisc platform for examining membrane protein topology and protein/membrane interfaces.
EXPERIMENTAL PROCEDURES
Sample Preparation
Nanodisc were made with uniformly 13C, 15N labeled MSP1D1 (a Δ1-11 truncation construct of MSP1, shown to form more stable Nanodiscs) (18) and 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC) lipids, and the SSNMR sample was lyophilized and rehydrated. Uniformly 13C, 15N-labeled MSP1D1 was expressed in Escherichia coli following a protocol described previously (32), and purified using Chelating Sepharose Fast Flow (Amersham Biosciences, Piscataway, NJ) charged with Ni2+ according to manufacturer’s instruction, and a 7-His-tag on the scaffolding protein. The expression protocol yields ~20 mg of isotopically labeled MSP1D1 per liter of growth media. The preparation of empty Nanodisc was performed as described previously (16), using POPC lipids (Avanti Polar Lipids, Alabaster, AL). In short, lipids from a chloroform stock were dried under a stream of nitrogen gas and left in vacuo overnight to remove residual chloroform. They were subsequently solubilized in a sodium cholate containing buffer (10 mM Tris/HCl, 0.1 M NaCl, pH 7.4) where the lipid:cholate ratio is 1:2, and the lipid concentration is approximately 50 mM. Incubation of the mixture in warm water and subsequent sonication result in complete solubilization of the lipids. MSP1D1 is then added to this mixture in a 1:65 ratio with POPC lipids (16). This mixture is incubated for approximately 1 hour at 4 °C. Finally, the self-assembly is initiated by removal of detergent during incubation with hydrophobic Amberlite XAD2 beads (Supelco, Bellefonte, PA) for 4 to 5 hours at 4 °C with moderate agitation. The final step of Nanodisc preparation is gel permeation chromatography, performed on a Superdex 200 HR 10/30 column (Amersham Biosciences) using Waters Millenium HPLC system with diode array detection system. The column was equilibrated in buffer (10 mM Tris/HCl, 100 mM NaCl pH 7.4). The mobile phase flow rate was 0.5 ml/min, with monitoring of protein absorbance at 280 nm, and one fraction collected per minute. The gel permeation chromatography can also be used to test the stability of the sample after experimental manipulations.
After purification Nanodiscs were exchanged into a 40 mM Tris/HCl, 10 mM NaCl (pH 7.4) buffer, and concentrated to approximately 300 μM Nanodisc concentration using an Amicon Ultra Centrifugal Filter unit with a 10 000 molecular weight cut-off (Millipore, Billerica, MA). This ensured that the NaCl concentration would remain low in the lyophilized pellet. Trehalose was added in 1:1 w:w ratio with the lipid as a lyoprotectant. The sample was then flash frozen in liquid nitrogen and lyophilized overnight. The stability of POPC Nanodiscs under this lyophilization procedure was tested by resuspension of samples in water and examination by size exclusion chromatography.
The uniformly 13C, 15N-labeled MSP1D1 POPC Nanodisc sample was transferred into a 3.2 mm thin wall rotor (Varian NMR, Palo Alto, CA) with a working volume of ~36 μL, and confined to active sample region of the rotor by Kel-F and rubber spacers as described elsewhere (70). The mass of MSP1D1 in the rotor was determined to be ~4 mg by comparing the intensity of 1D 13C spectra to standard proteins of known quantity. The mass of lipids, based upon stoichiometry of the Nanodisc, was estimated to be ~10 mg, and the mass of trehalose ~10 mg. This material was directly rehydrated in the SSNMR rotor with 10 μL water, which was determined to be sufficient to observe a bulk H2O signal at ~5 ppm in the 1H 1D spectrum.
SSNMR Spectroscopy
All SSNMR experiments were carried out on a 14.1 T (600 MHz 1H frequency) wide bore magnet with Varian InfinityPlus NMR spectrometer, equipped with a 3.2 mm HXY T3 probe tuned in 1H-13C-15N mode. Spectra were acquired at a magic-angle spinning (MAS) rate of 10 kHz. Pulse sequences were implemented with tangent ramped cross-polarization (CP) (71) with two pulse phase modulation (TPPM) (72) 1H decoupling at ~85 kHz. The typical π/2 pulse widths were 2.35 μs on 1H and 2.75 μs on 13C. Data were processed with NMRPipe (73) with back linear prediction, and zero filling and Lorentzian-to-Gaussian apodization were employed for each dimension before Fourier transformation. Additional acquisition and processing parameters for each spectrum are included in the figure captions. Chemical shifts were referenced externally with adamantane (74).
Molecular Dynamics Simulations of Nanodiscs
Each simulated Nanodisc was comprised of two MSP1D1 proteins and 160 DPPC lipids, and these Nanodiscs were constructed using the program Visual Molecular Dynamics (VMD) (75). The simulation parameters were described in detail elsewhere (24).
RESULTS AND DISCUSSION
Sample Preparation
We have shown previously that 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) Nanodiscs can be prepared for SSNMR by precipitation with PEG (32). This PEG precipitation procedure is not applicable to the POPC Nanodiscs, which disassemble under PEG precipitation. Development of new, robust SSNMR sample preparation protocols was necessary for wider application of Nanodiscs, in particular for POPC Nanodiscs due to higher physiological relevance of POPC lipids and their utility in studying membrane proteins. The lyophilization and rehydration procedure for preparation of Nanodiscs presented here is simple, highly reproducible, and allows ease of sample handling, which includes concentrating Nanodiscs into a very small sample volume without any loss of valuable sample that would occur with the use of common concentrating units. While POPC Nanodiscs can be readily lyophilized from 10 mM Tris/HCl, 100 mM NaCl in the presence of trehalose as a lyoprotectant, this results in a relatively large amount of mobile ions in the rehydrated SSNMR samples. This preparation results in deterioration of the 1H pulse widths and increased dielectric heating. Although we have previously developed probes that minimize these effects through improved resonator designs (76), most commercially available SSNMR probes (including those available for the instruments used in this study) are not designed to perform optimally with samples with >50 mM ionic strength. For SSNMR studies, we desired to decrease the ionic strength for optimal RF probe performance, and so we optimized the total ionic strength to address this concern. Therefore, we evaluated the lowest ionic strength that could be achieved while maintaining the integrity of POPC Nanodiscs upon lyophilization.
We found that POPC Nanodiscs are stable under lyophilization in the presence of trehalose if the starting Tris concentration is increased to 40 mM, and the NaCl concentration is decreased to 10 mM. After rehydration, these samples exhibit only a small fraction of the detrimental tuning effects of mobile ions in SSNMR, such that T1 relaxation times of the 1H signals determined the optimal repetition rate. A size exclusion chromatogram of the resuspended lyophilized Nanodiscs shows that the integrity of Nanodiscs is retained in the process of lyophilization (Supporting Information, Figure S1). The integrity of the sample is subsequently monitored by SSNMR spectra.
While Nanodisc samples were fully hydrated during SSNMR experiments, the bulk solution contained a high concentration of trehalose. It is known that sugars do not readily permeate the membrane (77), however it is likely that the OH groups of trehalose form hydrogen bonds with the polar headgroups of the lipid, and in this case with MSP as well, effectively replacing water molecules. This is known as the water replacement hypothesis (77-79). However, the results of size exclusion chromatography of lyophilized and rehydrated Nanodisc samples show no increase in size due to specific interactions of trehalose with the Nanodisc (either the bilayer or the MSP), and they indicate that the Nanodisc particles remain intact.
Lipid 1H 1D spectra and Bilayer Phase Transition
As noted in the introduction, the SSNMR lipid-protein correlation experiments rely on the ability to separate the magnetization from the mobile protons on the lipids or water, relative to protons from the protein. The lipid protons are highly mobile in the liquid crystalline phase. Therefore, it was a prerequisite to examine the behavior of the lipid bilayer in the Nanodisc, as prepared for SSNMR, and confirm that the gel to liquid crystalline phase transition was well defined, and in agreement with the transition found in lipid bilayers and vesicles. To this end, we performed a series of experiments measuring the 1H T2 values of lipids in directly detected 1H spectra from -20 °C to +15 °C. (The actual sample temperature for these experiments which, due to frictional and RF heating, varies from the reading on the thermocouple of the probe, was calibrated using ethylene glycol (80).)
The limiting cases of liquid crystalline phase (Figure 1b) and gel phase (Figure 1c) are easily distinguishable in the 1D spectra as monitored by the intensity of the lipid CH2 and methyl peaks. The transition temperature was fit as the inflection point of the T2 curve as a function of temperature and thereby determined to be 3 °C (Figure 1d). The relatively narrow (~0.2 ppm) lipid CH2 and methyl peaks observed above the lipid phase transition are indicative of high mobility of lipid acyl chains. Although these T2 values do not approach values observed for isotropic liquids, they are more than sufficient for the proposed 1H-13C 2D and 1H-13C-13C 3D lipid protein correlation experiments, where ~1 ms T2 filters are utilized.
Fully hydrated POPC liposomes undergo gel to liquid crystalline phase transition at ~ -3 °C, dehydrated POPC liposomes undergo the same transition at ~ 60 °C (79, 81), while dehydrated POPC liposomes lyophilized in the presence of trehalose undergo the transition at ~ -20 °C (79). The SSNMR measured phase transition temperature of the lipid bilayer in a Nanodisc of ~3 °C is certainly consistent with fully hydrated POPC bilayer in the Nanodisc. This temperature is indeed a few degrees higher than that observed in fully hydrated POPC liposomes, however, this is likely due to the properties of Nanodiscs, rather than due to effects of trehalose on the Nanodisc. In support of this, the same phenomenon has been observed in fully hydrated dipalmitoyl phosphatidylcholine (DPPC) and dimyristyol phosphatidylcholine (DMPC) Nanodiscs when compared to DPPC and DMPC liposomes. Previously, the phase transition behavior in Nanodiscs was measured by Differential Scanning Calorimetry (DSC), Small Angle X-ray Scattering (SAXS) and Laurdan fluorescence, and showed (for DMPC and DPPC) slightly higher values for the phase transition from that recorded in vesicles, as well as a relative broadening of this transition (19, 20). The increase in the melting temperature of the lipids in the Nanodiscs is a consequence of additional lateral pressure exerted by the MSP. Namely, the lipid phase transition from the gel to the liquid crystalline phase results in the thermal expansion of the lipid bilayer, which requires an increase in the surface area of the thermodynamically unfavorable lipid/MSP interface, as well as an increase in the MSP belt (82). The broadening of the phase transition in Nanodiscs is an established consequence of the size of the lipid bilayer in a Nanodisc, and the relatively small number of lipids participating in the cooperative phase transition. In addition, the boundary lipid associated with the MSP, which constitutes ~50% of the lipids in these Nanodiscs is thought to be excluded from the cooperative lipid domain (82, 83).
1H-13C-13C 3D Spin-Diffusion Pulse Sequence with a Double Quantum Filter
To examine the lipid-protein interface, we developed a 1H-13C-13C pulse sequence that takes advantage of the lipid phase transition behavior. Figure 2 shows the 3D 1H spin-diffusion pulse sequence with 13C detection, which we have adapted from earlier versions (65) by the addition of an (optional) indirect 13C dimension and double quantum mixing. The four initial 90° pulses on carbon destroy residual 13C magnetization. The 1H magnetization from the mobile protons (lipids or water) is selected by a T2 filter after the initial 90° pulse, and frequency labeled in the first dimension. The 1H signals from the comparatively rigid protein relax during this time due to strong 1H-1H dipolar couplings, while the lipids have a comparatively long T2 relaxation time. The 180° pulse in the middle of the filter refocuses the isotropic chemical shift evolution and B0 field inhomogeneity. The lipid magnetization is then allowed to evolve under the isotropic chemical shift interaction during t1. A subsequent 90° pulse stores the magnetization along the z-axis and allows magnetization to exchange from mobile protons to the (less mobile) protein protons by 1H-1H spin diffusion during a mixing time τmix. After this mixing time another 90° pulse flips the magnetization, which is then transferred to 13C-spins via tangent ramped CP, followed by a 13C evolution period. An SPC-5 mixing period in the 1H-13C-13C 3D is used to observe only one-bond 13C correlations (84). The sequence also includes an optional double quantum filter to attenuate the natural-abundance 13C background coming from the large number of natural abundance lipids in the samples. Finally, signals are detected on 13C under TPPM decoupling.
Figure 2. 1H-13C-13C 3D pulse sequence used to obtain lipid to protein correlations.
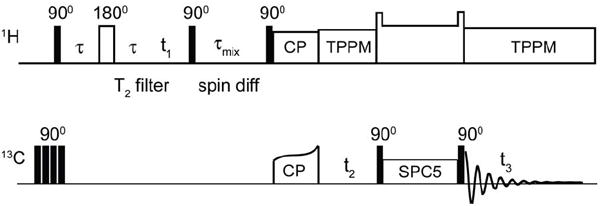
Following presaturation of 13C Boltzmann polarization, mobile 1H signals are selected by a T2 filter. 1H polarization is then transferred by spin diffusion to immobile protons, and to the attached 13C by cross polarization. Following evolution of the 13C chemical shifts, 13C-13C mixing is achieved by SPC-5 (84), and signals detected under TPPM decoupling (72).
1H-13C 2D Lipid-MSP Correlations in Nanodiscs
To obtain the 1H-13C 2D correlations of the MSP1D1 in a Nanodisc, we first identified and evaluated the strength the 13C resonances from the natural abundance lipids. To do this a 13C 1D spectrum was acquired with a 1 ms T2 filter, and without 1H-1H mixing (Figure 3a). Observed lipid signals are consistent with those previously published belonging to lipid carbons (9, 85). In the 13C 1D the strongest lipid signals at ~32 ppm belong to the CH2 carbons of the lipid acyl chains. The carbons at the end of the lipid chains (ω) are at ~18 ppm, followed by ω-1 carbons at ~23 ppm, and C3 (~27 ppm), carbons neighboring the oleate double bond carbons (~29 ppm), and subsequently ω-2 and C2 carbons (~33 and ~35 ppm, respectively). Headgroup carbons (α, β, γ, G1, and G3) are observed between 55 and 70 ppm, and the carbons C9 and C10 involved in the double bond are observed at ~130 ppm. Notably, the spectrum observed here arises exclusively from the natural abundance lipids. We also observe individually site-resolved 1H resonances in the 1H-13C 2D correlation spectrum, which are assigned (Figure 3b). The majority of lipid 1H polarization, from the long methylene chain, resides at ~1.3 ppm.
Figure 3. 13C 1D and 1H-13C 2D spectra of POPC bilayer in a Nanodisc.
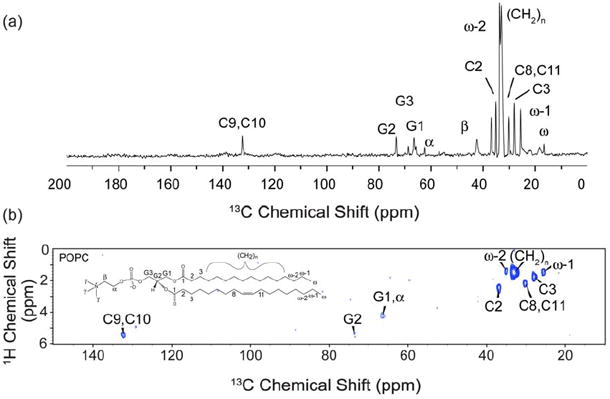
(a) 1D 13C spectrum of Nanodiscs with uniformly-13C,15N-labeled MSP1D1 and natural abundance POPC lipids at 8 °C on a 600 MHz (1H frequency) spectrometer. The lipid spectrum was acquired with a 1 ms T2 filter, no 1H-1H mixing and 0.4 ms 1H-13C contact time. The data was processed with 25 Hz Lorentzian-to-Gaussian line-broadening. (b) 1H-13C 2D spectrum of the same sample acquired under the same conditions for 6.5 hrs, with a schematic of a POPC lipid molecule. The spectrum was processed with 100 Hz of Lorentzian-to-Gaussian line broadening in each dimension.
Although the number of 13C sites in the protein is significantly greater than 13C sites in natural abundance lipids, the lipid signals present a potential complication due to the high number (~130 POPC lipids per Nanodisc) and very narrow linewidths (~0.2 ppm for 13C). We evaluated the relative intensities by incrementing the 1H-1H spin diffusion time in a series of 13C 1D spectra (Figure 4). The 13C 1D spectrum at very short 1H-1H mixing (effectively zero mixing, but with the identical phase cycle and number of pulses as subsequent spectra) corresponds to the assigned lipid spectrum in Figure 3a. With increasing 1H-1H mixing time, in addition to lipid signals, the protein signals are polarized on the timescale of ~30 ms. The protein signal intensity is derived both from the mobile lipid protons, as well as water, although the latter would have no appreciable contribution to the initial lipid spectrum. At the longer mixing time, the protein signals dominate the total intensity; for example, carbonyl (~175 ppm), Cα (55-65 ppm), aromatic (100-130 and ~160 ppm), Leu Cβ (~40 ppm) and various methyl and methylene 13C sites (15-35 ppm) become prominent in intensity at ~3 ms, and dominate the overall spectrum at 30 ms. Nevertheless, sharp lipid peaks are still observed superimposed upon the protein spectrum; this is most readily appreciated by the relative linewidths, since the lipids do not experience broadening due to one-bond (residual) 13C-13C dipolar and scalar couplings. Therefore the intensity of natural abundance lipids would present a challenge in the interpretation of the 1H-13C 2D spectrum of MSP1D1, if superimposed on some protein cross peaks of significantly lower intensity. Consequently, the 1H-13C 2D sequence implemented on MSP1D1 in Nanodiscs contained a double quantum filter to reduce the 13C background from natural abundance lipids by a factor of 100.
Figure 4. 13C 1D spectra of MSP1D1 in POPC Nanodiscs.
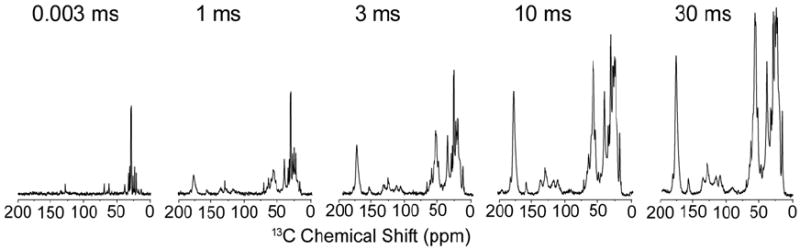
Spectra were acquired on U-13C,15N-labeled MSP1D1 at 8 °C. 13C 1D spectra were acquired with a 1 ms T2 filer, a 0.4 ms HC contact time, where 1H-1H mixing time was incremented and sequentially set to 3 μs, 1 ms, 3 ms, 10 ms and 30 ms. The data were processed with 30 Hz Lorentzian-to-Gaussian line broadening.
The 1H-13C 2D correlation spectrum of uniformly 13C, 15N-labeled MSP1D1 in a Nanodisc (Figure 5) contains only water and mobile lipid signals in the 1H dimension, due to the 1 ms T2 filter. With 30 ms 1H-1H mixing, a significant fraction of the polarization is transferred to the protein, as confirmed by the retention of signal following the double quantum filter. The number of protein resonances correlated to the lipids (~1.3 ppm) and the intensity of lipid/protein cross peaks is smaller than that of water/protein correlations (at ~5 ppm 1H frequency), which we attribute to the larger initial polarization and longer T2 for the water resonance. Furthermore, the exposed surface area on the outside of the Nanodisc (exposed to water) is greater than the internal surface area (exposed to lipid). This 1D 13C spectrum has minimal resolution, enabling only a small subset of peaks (e.g., Ala) to be tentatively assigned by amino acid type. Clearly a 3D is required to conduct more detailed analysis.
Figure 5. 1H-13C 2D spectra of MSP1D1 in POPC Nanodiscs.
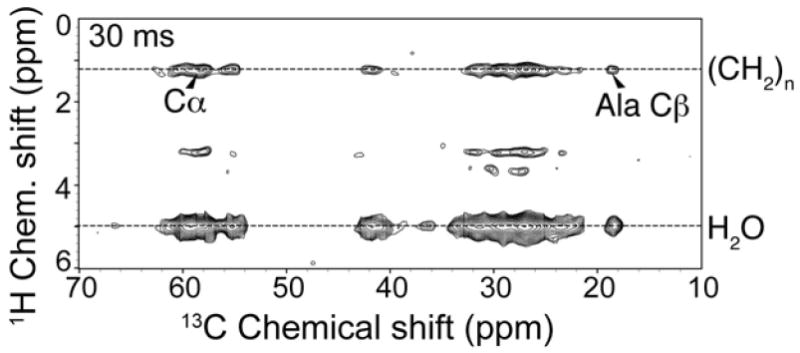
Experimental spectrum acquired on U-13C,15N-labeled MSP1D1 at 15 °C. Spectra were acquired with a 1 ms T2 filer, a 0.4 ms 1H-13C contact time, and a 30 ms 1H-1H mixing time. The spectrum was processed with back linear prediction, 100 Hz net line broadening in the direct, and 80 Hz in the indirect dimension (Lorentzian-to-Gaussian apodization), and zero filled to 4096 (ω2) × 16 384 (ω1) complex points before Fourier transformation. Data were acquired in ~7 hrs.
1H-13C-13C 3D for Identification of Lipid Interfaced MSP Residues by Amino Acid Type
MSP, like ApoA-I, has a repetitive amino acid sequence with several similar amphipathic alpha helices. The 13C chemical shifts are only partially resolved in two 13C dimensions; however, this same uniform secondary structure enables us to readily identify the chemical shifts of specific amino acids (Ala, Leu, Val, Pro, Lys) and classify the remaining signals into groups (e.g., Glu, Gln, Arg). In order to separate the lipid and water correlations, we retained the 1H dimension, while also digitizing the indirect 13C dimension, resulting in a 1H-13C-13C 3D experiment. In this case we chose SPC-5 13C-13C double quantum mixing (but without the coherence filter), since this was more sensitive under these experimental conditions. The short SPC-5 mixing time results in only one-bond 13C-13C cross peaks, simplifying the spectral analysis and enabling the identification of several amino acid types without cancellation of signals due to multi-bond transfers.
Figure 6 shows the 13C-13C planes of the 1H-13C-13C 3D at the water proton (a) (1H of 5 ppm) and lipid protons (b) (1H of 1.3 ppm) chemical shifts. Resolution in the proton dimension is sufficient to differentiate between water plane and lipid acyl chain plane (~1 ppm). Resolving the protons of lipid acyl chains from lipid terminal methyl group would be challenging with these experiments, although it is easily achieved in 1H-13C 2D experiments described earlier. Both strips show cross peaks that can be confidently assigned by amino acid type, allowing further analysis. Both the water and lipid planes show peaks corresponding to the uniquely resolved amino acid types Ala, Leu, Val, as well as intensity grouped among Glu, Gln and Arg; all of these shift patterns are consistent with helical secondary structure. In addition, the water plane (Figure 6a) shows correlations corresponding to Lys and Pro. The relative intensity of the cross peaks within each plane, as well as the absolute intensity of each plane in total, varies depending on the source of polarization. The peaks in both 13C-13C planes were picked in NMRPipe (73) and integrated using the nlinLS package. The intensities provide a semi-quantitative assessment of which amino acids are in proximity to water and/or lipid. We then compared these results with those from a 4.5 ns molecular dynamics (MD) simulation of MSP1D1 in Nanodiscs (24).
Figure 6. 1H-13C-13C 3D spectrum of MSP1D1 in Nanodiscs.
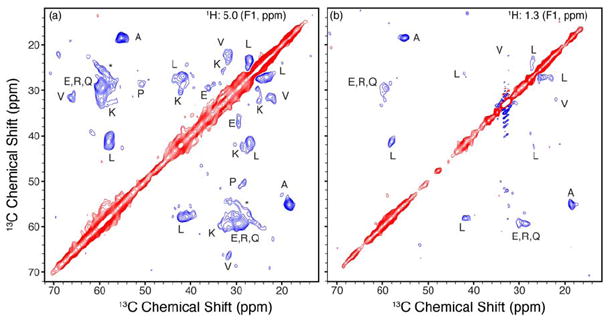
1H-13C-13C spectrum acquired at 600 MHz (1H frequency), 8 °C sample temp in ~115 hours. The T2 filter time was set to 1 ms to eliminate protein 1H polarization. A 1H-1H mixing time of 30 ms was used followed by 1H-13C cross polariation. 13C-13C mixing with 12 ms of SPC-5 was used without the double quantum filiter. Water (a) and lipid (b) 13C-13C planes are show with peaks labeled by amino acid type (88). Positive contours shown in red, negative in blue. The seeming artifact in the lipid plane is due to a strong signal originating from lipids correlated to themselves.
It should be noted that the Nanodiscs used in the MD simulations were composed of DPPC lipids in a lipid gel phase, but given the timescales simulated (<ten nanoseconds), lipid diffusion is not actually observed within the simulations. The simulations do however provide sufficient time for the lipids to relax around the amphipathic helices of MSP, allowing for determination of which amino acid residues are in contact with the lipids. As the comparisons with the NMR measurements are only looking at the matching of hydrophobic amino acids with lipid acyl chains, the difference in lipid type in the experimental data and the MD simulations are expected to affect the resulting analysis.
We analyzed the MD trajectories according to the following strategy. The majority of cross peaks in the 2D 13C-13C planes are CA-CB or CB-CG, consistent with the expectation that polarization is transferred from lipid or water to the amino acid sidechain. We averaged the distance (over the final 0.75 ns of the MD trajectory) from each CB to the closest lipid acyl chain or water molecule. Since the absolute intensity of each plane depends upon a number of factors that cannot be predicted from theory (absolute polarization, range of T2 values, diffusion rates, etc.), we normalized the data to common ratio of intensities. For the MD simulations, this was the ratio W:L, where W is the number of residues within 4.0 Å of water, and L is the number of residues within 4.0 Å of lipids. We then calculated the observed ratio of NMR signal intensity for W and L, corresponding to the 13C-13C cross peaks occurring in the respective planes (~5.0 ppm for W and 1.3 ppm for L). These results are summarized in Table 1 and Figure 7. Figure 7 shows a graph of the W:L ratios in the NMR and MD experiments, along with a line of best fit. Error bars for the NMR data were calculated using the signal to noise of the peaks in the same manner as Mani et al. (64), in the case of Leu, averaging the multiple observed cross peaks. As expected, the data show a higher W:L ratio for charged and polar residues (Glu, Gln, Arg and Lys) than hydrophobic residues (Ala, Leu, Val) in both cases. Indeed, for Lys and Glu, the lipid correlations were weak, but still observable. This is consistent with previously published results from modeling (86) and published x-ray structure of truncated apo A-I (87) where a number of charged residues can be found associated with the lipid interface.
Table 1.
Intensity Ratio for Water-Protein and Lipid-Protein Correlations.
| Amino Acid | MD Ratioa | NMR Ratiob | NMR Confidencec |
|---|---|---|---|
| A | 3.7 | 2.4 | 0.3 |
| EQR | 8.3 | 3.7 | 0.6 |
| K | 6.8 | 3.7 | 0.8 |
| L | 3.0 | 3.1 | 0.4 |
| V | 1.2 | 2.4 | 0.5 |
Calculated as the ratio of CB carbons within 4.0 Å of water or lipid over the last 0.75 ns of a 4.5 ns MD simulation of MSP Nanodiscs
Calculated as the ratio of 13C-13C crosspeaks to lipid and water in the 1H-13C-13C 3D
Calculated as Δ = ( (1 / Iwater)2 + (1 / Ilipid)2 )1/2 where Iwater and Ilipid where the signal to noise ratio of the water and lipid peaks used in the calculation
Figure 7. Plot of Water to Lipid ratios determined by NMR and Molecular Dynamics.
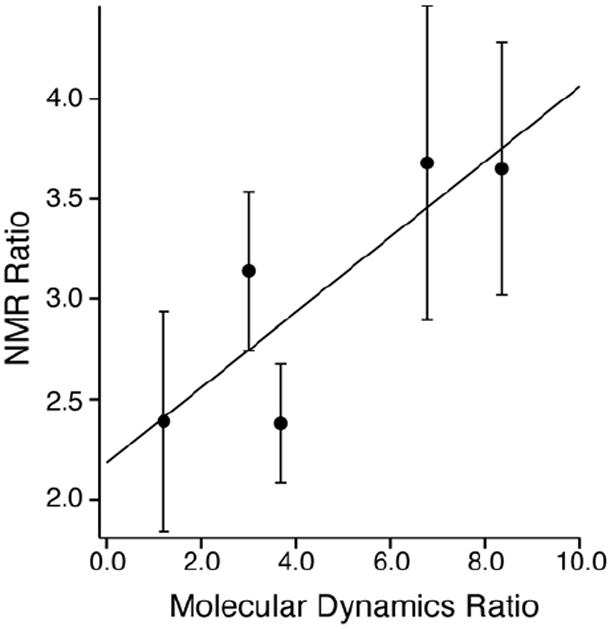
Ratio of CB carbons in MSP within 4.0 Å of a water molecule to the ratio of CB carbons within 4.0 Å of lipid plotted versus ratio of cross peak intensities for the same residues at the water and lipid 1H frequencies in the 1H-13C-13C 3D spectrum. MD ratios are the average number of CB atoms for a given residue within 4.0 Å of lipid or water averaged over two MSP molecules for the last 0.75 ns of a 4.5 ns molecular dynamics simulation. NMR ratios were calculated using integrated peak intensities for 13C-13C cross peaks corresponding to the same residues appearing at both the lipid and water 1H frequency in the 1H-13C-13C 3D spectrum. Error bars for the NMR ratio were calculated using the signal to noise of the peaks in the 3D using the method of Mani et al. (64). The linear best fit line is plotted. The NMR ratio is scaled due to different 1H T2 as well as differences in total initial polarization.
These results on MSP1D1 in Nanodiscs demonstrate the potential of the 1H-13C-13C pulse sequence for obtaining information on the membrane protein topology and lipid/protein interfaces. The information that can be obtained in the case of MSP is limited by the large portion of the protein interacting with the lipid bilayer (approximately a third of 211 residues), and the degeneracy in the chemical shift distribution due to all amino acids being present in a helical secondary structure. More qualitative information can certainly be obtained from experiments like these in combination with amino acid type-specific isotope labeling schemes of the protein of interest, and more quantitative information if site-specific assignments for the protein are available.
CONCLUSIONS
In the work presented here, we have demonstrated a new sample preparation method for POPC Nanodiscs and applied SSNMR to examine membrane/protein interfaces. SSNMR was used to assess the phase transition behavior of the lipid bilayer in a Nanodisc and confirm that under the sample preparation conditions POPC Nanodiscs exhibited the expected gel to liquid crystalline phase transition characteristic of POPC lipid bilayers. Using MSP1D1 Nanodiscs, it was demonstrated that the 1H-13C-13C 3D pulse sequence developed provides qualitative information on the protein residues at the lipid/protein boundary. The experimental information was compared to results of molecular dynamic simulation of Nanodiscs, and it was found that the experimental results are consistent with the predicted lipid/protein interface from the MD simulations in the amino acid types and the number of residues in the interface. The study demonstrates the potential for use of Nanodiscs platform in combination with presented proton spin diffusion experiments for examining lipid/protein interfaces and membrane topology for membrane proteins embedded in Nanodiscs.
Supplementary Material
Acknowledgments
The authors thank Dr. Ying Li for her involvement in early stages of the SSNMR experiment development and application, and John M. Boettcher, Rebecca L. Davis-Harrison, and Yelena V. Grinkova for sample preparation discussions.
This work was supported by the National Institutes of Health grants GM33775 and GM31756 (to S. G. Sligar), GM79530 (to C.M. Rienstra and M. A. Schuler), R01-GM067887 and P41-RR05969 (to K. Schulten), as well as grant MCB02-34938 (to K. Schulten) from National Science Foundation. Amy Y. Shih acknowledges support from a Beckman Postdoctoral Fellowship.
Abbreviations
- CP
cross-polarization
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DPPC
1,2-dipalmitoyl-sn-glycero-3-phosphocholine
- DSC
Differential Scanning Calorimetry
- MAS
magic-angle spinning
- MD
molecular dynamics
- MSP
membrane scaffolding protein
- MSP1D1
Δ(1-11) truncated membrane scaffolding protein
- PEG
polyethylene glycol
- POPC
1-palmitoyl-2-oleoyl-phosphatidylcholine
- SPC5
Supercycled POST-C5
- SSNMR
solid-state nuclear magnetic resonance
- TPPM
two-pulse phase modulation
Footnotes
Supporting Information Available The normalized gel filtration chromatograms of POPC Nanodiscs before and after lyophilization and rehydration are presented in Figure S1. Table S1 provides the ratio of intensities for the water-protein and lipid-protein correlations in the water plane of the 3D experiment. Supporting information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zwaal RF, Comfurius P, Bevers EM. Lipid-protein interactions in blood coagulation. Biochim Biophys Acta. 1998;1376:433–453. doi: 10.1016/s0304-4157(98)00018-5. [DOI] [PubMed] [Google Scholar]
- 2.Shaw AW, Pureza VS, Sligar SG, Morrissey JH. The local phospholipid environment modulates the activation of blood clotting. J Biol Chem. 2007;282:6556–6563. doi: 10.1074/jbc.M607973200. [DOI] [PubMed] [Google Scholar]
- 3.Yun CH, Ahn T, Guengerich FP. Conformational change and activation of cytochrome P450 2B1 induced by salt and phospholipid. Arch Biochem Biophys. 1998;356:229–238. doi: 10.1006/abbi.1998.0759. [DOI] [PubMed] [Google Scholar]
- 4.Kim KH, Ahn T, Yun CH. Membrane properties induced by anionic phospholipids and phosphatidylethanolamine are critical for the membrane binding and catalytic activity of human cytochrome P450 3A4. Biochemistry. 2003;42:15377–15387. doi: 10.1021/bi035280k. [DOI] [PubMed] [Google Scholar]
- 5.Ahn T, Guengerich FP, Yun CH. Membrane insertion of cytochrome P450 1A2 promoted by anionic phospholipids. Biochemistry. 1998;37:12860–12866. doi: 10.1021/bi980804f. [DOI] [PubMed] [Google Scholar]
- 6.Chini B, Parenti M. G-protein coupled receptors, cholesterol and palmitoylation: facts about fats. J Mol Endocrinol. 2009;42:9. doi: 10.1677/JME-08-0114. [DOI] [PubMed] [Google Scholar]
- 7.Bernier LP, Ase AR, Chevallier S, Blais D, Zhao Q, Boue-Grabot E, Logothetis D, Seguela P. Phosphoinositides regulate P2X4 ATP-gated channels through direct interactions. J Neurosci. 2008;28:12938–12945. doi: 10.1523/JNEUROSCI.3038-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas AM, Tinker A. Determination of phosphoinositide binding to K(+) channel subunits using a protein-lipid overlay assay. Methods Mol Biol. 2008;491:103–111. doi: 10.1007/978-1-59745-526-8_8. [DOI] [PubMed] [Google Scholar]
- 9.Mani R, Buffy JJ, Waring AJ, Lehrer RI, Hong M. Solid-state NMR investigation of the selective disruption of lipid membranes by protegrin-1. Biochemistry. 2004;43:13839–13848. doi: 10.1021/bi048650t. [DOI] [PubMed] [Google Scholar]
- 10.Matsuzaki K, Murase O, Miyajima K. Kinetics of pore formation by an antimicrobial peptide, magainin 2, in phospholipid bilayers. Biochemistry. 1995;34:12553–12559. doi: 10.1021/bi00039a009. [DOI] [PubMed] [Google Scholar]
- 11.Prenner EJ, Lewis RN, McElhaney RN. The interaction of the antimicrobial peptide gramicidin S with lipid bilayer model and biological membranes. Biochim Biophys Acta. 1999;1462:201–221. doi: 10.1016/s0005-2736(99)00207-2. [DOI] [PubMed] [Google Scholar]
- 12.Morrissey JH, Pureza V, Davis-Harrison RL, Sligar SG, Ohkubo YZ, Tajkhorshid E. Blood clotting reactions on nanoscale phospholipid bilayers. Thromb Res. 2008;122(Suppl 1):S23–26. doi: 10.1016/S0049-3848(08)70014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinogradova O, Sonnichsen F, Sanders CR. On choosing a detergent for solution NMR studies of membrane proteins. J Biomol NMR. 1998;11:381–386. doi: 10.1023/a:1008289624496. [DOI] [PubMed] [Google Scholar]
- 14.Sorgen PL, Cahill SM, Krueger-Koplin RD, Krueger-Koplin ST, Schenck CC, Girvin ME. Structure of the Rhodobacter sphaeroides light-harvesting 1 beta subunit in detergent micelles. Biochemistry. 2002;41:31–41. doi: 10.1021/bi011576j. [DOI] [PubMed] [Google Scholar]
- 15.Bayburt TH, Carlson JW, Sligar SG. Reconstitution and imaging of a membrane protein in a nanometer-size phospholipid bilayer. J Struct Biol. 1998;123:37–44. doi: 10.1006/jsbi.1998.4007. [DOI] [PubMed] [Google Scholar]
- 16.Bayburt TH, Grinkova YV, Sligar SG. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2002;2:853–856. [Google Scholar]
- 17.Baas BJ, Denisov IG, Sligar SG. Homotropic cooperativity of monomeric cytochrome P450 3A4 in a nanoscale native bilayer environment. Arch Biochem Biophys. 2004;430:218–228. doi: 10.1016/j.abb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 19.Shaw AW, McLean MA, Sligar SG. Phospholipid phase transitions in homogeneous nanometer scale bilayer discs. Febs Letters. 2004;556:260–264. doi: 10.1016/s0014-5793(03)01400-5. [DOI] [PubMed] [Google Scholar]
- 20.Denisov IG, McLean MA, Shaw AW, Grinkova YV, Sligar SG. Thermotropic phase transition in soluble nanoscale lipid bilayers. J Phys Chem B. 2005;109:15580–15588. doi: 10.1021/jp051385g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayburt TH, Grinkova YV, Sligar SG. Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch Biochem Biophys. 2006;450:215–222. doi: 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Nath A, Atkins WM, Sligar SG. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 23.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 24.Shih AY, Denisov IG, Phillips JC, Sligar SG, Schulten K. Molecular dynamics simulations of discoidal bilayers assembled from truncated human lipoproteins. Biophys J. 2005;88:548–556. doi: 10.1529/biophysj.104.046896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shih AY, Freddolino PL, Sligar SG, Schulten K. Disassembly of nanodiscs with cholate. Nano Lett. 2007;7:1692–1696. doi: 10.1021/nl0706906. [DOI] [PubMed] [Google Scholar]
- 26.Denisov IG, Baas BJ, Grinkova YV, Sligar SG. Cooperativity in cytochrome P450 3A4: linkages in substrate binding, spin state, uncoupling, and product formation. J Biol Chem. 2007;282:7066–7076. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- 27.Civjan NR, Bayburt TH, Schuler MA, Sligar SG. Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. Biotechniques. 2003;35:556–560. 562–553. doi: 10.2144/03353rr02. [DOI] [PubMed] [Google Scholar]
- 28.Nath A, Grinkova YV, Sligar SG, Atkins WM. Ligand binding to cytochrome P450 3A4 in phospholipid bilayer nanodiscs: the effect of model membranes. J Biol Chem. 2007;282:28309–28320. doi: 10.1074/jbc.M703568200. [DOI] [PubMed] [Google Scholar]
- 29.Alami M, Dalal K, Lelj-Garolla B, Sligar SG, Duong F. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 2007;26:1995–2004. doi: 10.1038/sj.emboj.7601661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leitz AJ, Bayburt TH, Barnakov AN, Springer BA, Sligar SG. Functional reconstitution of Beta2-adrenergic receptors utilizing self-assembling Nanodisc technology. Biotechniques. 2006;40:601–612. doi: 10.2144/000112169. [DOI] [PubMed] [Google Scholar]
- 31.Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci U S A. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Kijac AZ, Sligar SG, Rienstra CM. Structural analysis of nanoscale self-assembled discoidal lipid bilayers by solid-state NMR spectroscopy. Biophys J. 2006;91:3819–3828. doi: 10.1529/biophysj.106.087072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kijac AZ, Li Y, Sligar SG, Rienstra CM. Magic-angle spinning solid-state NMR spectroscopy of nanodisc-embedded human CYP3A4. Biochemistry. 2007;46:13696–13703. doi: 10.1021/bi701411g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyukmanova EN, Shenkarev ZO, Paramonov AS, Sobol AG, Ovchinnikova TV, Chupin VV, Kirpichnikov MP, Blommers MJJ, Arseniev AS. Lipid-protein nanoscale bilayers: A versatile medium for NMR investigations of membrane proteins and membrane-active peptides. J Am Chem Soc. 2008;130:2140. doi: 10.1021/ja0777988. [DOI] [PubMed] [Google Scholar]
- 35.Lyukmanova EN, Shenkarev ZO, Ovchinnikova TV, Chupin VV, Blommers MJJ, Arseniev AS. Reconstituted high density lipoprotein particles: a promising medium for high-resolution NMR investigations of membrane proteins and membrane-active peptides. FEBS J. 2008;275:171–171. [Google Scholar]
- 36.Gluck JM, Wittlich M, Feuerstein S, Hoffmann S, Willbold D, Koenig BW. Integral Membrane Proteins in Nanodiscs Can Be Studied by Solution NMR Spectroscopy. J Am Chem Soc. 2009;131:12060. doi: 10.1021/ja904897p. [DOI] [PubMed] [Google Scholar]
- 37.Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci U S A. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaw AW, Pureza VS, Sligar SG, Morrissey JH. The local phospholipid environment modulates the activation of blood clotting. J Biol Chem. 2007;282:6556–6563. doi: 10.1074/jbc.M607973200. [DOI] [PubMed] [Google Scholar]
- 39.Morrissey JH, Pureza V, Davis-Harrison RL, Sligar SG, Ohkubo YZ, Tajkhorshid E. Blood clotting reactions on nanoscale phospholipid bilayers. Thromb Res. 2008;122(Suppl 1):S23–26. doi: 10.1016/S0049-3848(08)70014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baas BJ, Denisov IG, Sligar SG. Homotropic cooperativity of monomeric cytochrome P450 3A4 in a nanoscale native bilayer environment. Arch Biochem Biophys. 2004;430:218–228. doi: 10.1016/j.abb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Denisov IG, Grinkova YV, Baas BJ, Sligar SG. The ferrous-dioxygen intermediate in human cytochrome P450 3A4. Substrate dependence of formation and decay kinetics. J Biol Chem. 2006;281:23313–23318. doi: 10.1074/jbc.M605511200. [DOI] [PubMed] [Google Scholar]
- 42.Davydov DR, Fernando H, Baas BJ, Sligar SG, Halpert JR. Kinetics of dithionite-dependent reduction of cytochrome P450 3A4: heterogeneity of the enzyme caused by its oligomerization. Biochemistry. 2005;44:13902–13913. doi: 10.1021/bi0509346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bayburt TH, Grinkova YV, Sligar SG. Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch Biochem Biophys. 2006;450:215–222. doi: 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 44.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 45.Alami M, Dalal K, Lelj-Garolla B, Sligar SG, Duong F. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 2007;26:1995–2004. doi: 10.1038/sj.emboj.7601661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dalal K, Nguyen N, Alami M, Tan J, Moraes TF, Lee WC, Maurus R, Sligar SS, Brayer GD, Duong F. Structure, Binding, and Activity of Syd, a SecY-interacting Protein. J Biol Chem. 2009;284:7897–7902. doi: 10.1074/jbc.M808305200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duan H, Civjan NR, Sligar SG, Schuler MA. Co-incorporation of heterologously expressed Arabidopsis cytochrome P450 and P450 reductase into soluble nanoscale lipid bilayers. Arch Biochem Biophys. 2004;424:141–153. doi: 10.1016/j.abb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 48.Denisov IG, Baas BJ, Grinkova YV, Sligar SG. Cooperativity in cytochrome P450 3A4: linkages in substrate binding, spin state, uncoupling, and product formation. J Biol Chem. 2007;282:7066–7076. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- 49.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borch J, Hamann T. The nanodisc: a novel tool for membrane protein studies. Biol Chem. 2009;390:805–814. doi: 10.1515/BC.2009.091. [DOI] [PubMed] [Google Scholar]
- 51.Li Y, Berthold DA, Gennis RB, Rienstra CM. Chemical shift assignment of the transmembrane helices of DsbB, a 20-kDa integral membrane enzyme, by 3D magic-angle spinning NMR spectroscopy. Protein Sci. 2008;17:199–204. doi: 10.1110/ps.073225008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frericks HL, Zhou DH, Yap LL, Gennis RB, Rienstra CM. Magic-angle spinning solid-state NMR of a 144 kDa membrane protein complex: E. coli cytochrome bo3 oxidase. J Biomol NMR. 2006;36:55–71. doi: 10.1007/s10858-006-9070-5. [DOI] [PubMed] [Google Scholar]
- 53.Shi L, Ahmed MA, Zhang W, Whited G, Brown LS, Ladizhansky V. Three-dimensional solid-state NMR study of a seven-helical integral membrane proton pump--structural insights. J Mol Biol. 2009;386:1078–1093. doi: 10.1016/j.jmb.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 54.Etzkorn M, Martell S, Andronesi OC, Seidel K, Engelhard M, Baldus M. Secondary structure, dynamics, and topology of a seven-helix receptor in native membranes, studied by solid-state NMR spectroscopy. Angew Chem Int Ed Engl. 2007;46:459–462. doi: 10.1002/anie.200602139. [DOI] [PubMed] [Google Scholar]
- 55.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 56.Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer’s beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 57.Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Structure of a protein determined by solid-state magic-angle- spinning NMR spectroscopy. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- 58.Park SH, Mrse AA, Nevzorov AA, Mesleh MF, Oblatt-Montal M, Montal M, Opella SJ. Three-dimensional structure of the channel-forming trans-membrane domain of virus protein “u” Vpu from HIV-1. J Mol Biol. 2003;333:409–424. doi: 10.1016/j.jmb.2003.08.048. [DOI] [PubMed] [Google Scholar]
- 59.Kamihira M, Vosegaard T, Mason AJ, Straus SK, Nielsen NC, Watts A. Structural and orientational constraints of bacteriorhodopsin in purple membranes determined by oriented-sample solid-state NMR spectroscopy. J Struct Biol. 2005;149:7–16. doi: 10.1016/j.jsb.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 60.Park SH, Prytulla S, De Angelis AA, Brown JM, Kiefer H, Opella SJ. High-resolution NMR spectroscopy of a GPCR in aligned bicelles. J Am Chem Soc. 2006;128:7402–7403. doi: 10.1021/ja0606632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu W, Crocker E, Constantinescu SN, Smith SO. Helix packing and orientation in the transmembrane dimer of gp55-P of the spleen focus forming virus. Biophys J. 2005;89:1194–1202. doi: 10.1529/biophysj.104.057844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu W, Crocker E, Siminovitch DJ, Smith SO. Role of side-chain conformational entropy in transmembrane helix dimerization of glycophorin A. Biophys J. 2003;84:1263–1271. doi: 10.1016/S0006-3495(03)74941-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toke O, O’Connor RD, Weldeghiorghis TK, Maloy WL, Glaser RW, Ulrich AS, Schaefer J. Structure of (KIAGKIA)3 aggregates in phospholipid bilayers by solid-state NMR. Biophys J. 2004;87:675–687. doi: 10.1529/biophysj.103.032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mani R, Cady SD, Tang M, Waring AJ, Lehrer RI, Hong M. Membrane-dependent oligomeric structure and pore formation of a beta-hairpin antimicrobial peptide in lipid bilayers from solid-state NMR. Proc Natl Acad Sci U S A. 2006;103:16242–16247. doi: 10.1073/pnas.0605079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huster D, Yao X, Hong M. Membrane protein topology probed by (1)H spin diffusion from lipids using solid-state NMR spectroscopy. J Am Chem Soc. 2002;124:874–883. doi: 10.1021/ja017001r. [DOI] [PubMed] [Google Scholar]
- 66.Buffy JJ, Hong T, Yamaguchi S, Waring AJ, Lehrer RI, Hong M. Solid-state NMR investigation of the depth of insertion of protegrin-1 in lipid bilayers using paramagnetic Mn2+ Biophys J. 2003;85:2363–2373. doi: 10.1016/s0006-3495(03)74660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prosser RS, Luchette PA, Westerman PW. Using O2 to probe membrane immersion depth by 19F NMR. Proc Natl Acad Sci U S A. 2000;97:9967–9971. doi: 10.1073/pnas.170295297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong M. Oligomeric structure, dynamics, and orientation of membrane proteins from solid-state NMR. Structure. 2006;14:1731–1740. doi: 10.1016/j.str.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 69.Kumashiro KK, Schmidt-Rohr K, Murphy OJ, Ouellette KL, Cramer WA, Thompson LK. A novel tool for probing membrane protein structure: Solid-state NMR with proton spin diffusion and X-nucleus detection. J Am Chem Soc. 1998;120:5043–5051. [Google Scholar]
- 70.Franks WT, Zhou DH, Wylie BJ, Money BG, Graesser DT, Frericks HL, Sahota G, Rienstra CM. Magic-angle spinning solid-state NMR spectroscopy of the beta1 immunoglobulin binding domain of protein G (GB1): 15N and 13C chemical shift assignments and conformational analysis. J Am Chem Soc. 2005;127:12291–12305. doi: 10.1021/ja044497e. [DOI] [PubMed] [Google Scholar]
- 71.Hediger S, Meier BH, Kurur ND, Bodenhausen G, Ernst RR. NMR cross-polarization by adiabatic passage through the Hartmann-Hahn condition (APHH) Chem Phys Lett. 1994;223:283–288. [Google Scholar]
- 72.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- 73.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe - a multidimensional spectral processing system based on Unix pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 74.Morcombe CR, Zilm KW. Chemical shift referencing in MAS solid state NMR. J Magn Reson. 2003;162:479–486. doi: 10.1016/s1090-7807(03)00082-x. [DOI] [PubMed] [Google Scholar]
- 75.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 76.Stringer JA, Bronnimann CE, Mullen CG, Zhou DH, Stellfox SA, Li Y, Williams EH, Rienstra CM. Reduction of RF-induced sample heating with a scroll coil resonator structure for solid-state NMR probes. J Magn Reson. 2005;173:40–48. doi: 10.1016/j.jmr.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 77.Golovina EA, Golovin AV, Hoekstra FA, Faller R. Water Replacement Hypothesis in Atomic Detail-Factors Determining the Structure of Dehydrated Bilayer Stacks. Biophys J. 2009;97:490–499. doi: 10.1016/j.bpj.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsvetkova NM, Phillips BL, Crowe LM, Crowe JH, Risbud SH. Effect of sugars on headgroup mobility in freeze-dried dipalmitoylphosphatidylcholine bilayers: solid-state 31P NMR and FTIR studies. Biophys J. 1998;75:2947–2955. doi: 10.1016/S0006-3495(98)77736-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Crowe LM, Crowe JH. Dry Dipalmitoylphosphatidylcholine and Trehalose Revisited. Biophys J. 1988;53:A127–A127. doi: 10.1016/0005-2736(88)90392-6. [DOI] [PubMed] [Google Scholar]
- 80.Ammann C, Meier P, Merbach AE. A Simple Multi-Nuclear Nmr Thermometer. J Magn Reson. 1982;46:319–321. [Google Scholar]
- 81.Koster KL, Webb MS, Bryant G, Lynch DV. Interactions between Soluble Sugars and Popc (1-Palmitoyl-2-Oleoylphosphatidylcholine) during Dehydration - Vitrification of Sugars Alters the Phase-Behavior of the Phospholipid. Biochim Biophys Acta, Biomembr. 1994;1193:143–150. doi: 10.1016/0005-2736(94)90343-3. [DOI] [PubMed] [Google Scholar]
- 82.Denisov IG, McLean MA, Shaw AW, Grinkova YV, Sligar SG. Thermotropic phase transition in soluble nanoscale lipid bilayers. J Phys Chem B. 2005;109:15580–15588. doi: 10.1021/jp051385g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shaw AW, McLean MA, Sligar SG. Phospholipid phase transitions in homogeneous nanometer scale bilayer discs. FEBS Lett. 2004;556:260–264. doi: 10.1016/s0014-5793(03)01400-5. [DOI] [PubMed] [Google Scholar]
- 84.Hohwy M, Rienstra CM, Jaroniec CP, Griffin RG. Fivefold symmetric homonuclear dipolar recoupling in rotating solids: Application to double quantum spectroscopy. J Chem Phys. 1999;110:7983–7992. [Google Scholar]
- 85.Forbes J, Husted C, Oldfield E. High-Field, High-Resolution Proton Magic-Angle Sample-Spinning Nuclear Magnetic-Resonance Spectroscopic Studies of Gel and Liquid-Crystalline Lipid Bilayers and the Effects of Cholesterol. J Am Chem Soc. 1988;110:1059–1065. [Google Scholar]
- 86.Segrest JP, Jones MK, Klon AE, Sheldahl CJ, Hellinger M, De Loof H, Harvey SC. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J Biol Chem. 1999;274:31755–31758. doi: 10.1074/jbc.274.45.31755. [DOI] [PubMed] [Google Scholar]
- 87.Borhani DW, Rogers DP, Engler JA, Brouillette CG. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc Natl Acad Sci U S A. 1997;94:12291–12296. doi: 10.1073/pnas.94.23.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang H, Neal S, Wishart DS. RefDB: a database of uniformly referenced protein chemical shifts. J Biomol NMR. 2003;25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


