Abstract
γδ T cells function in innate and adaptive immunity and are primed for secondary responses by procyanidin components of unripe apple peel (APP). Here we investigate the effects of APP and purified procyanidins on γ δ T cell gene expression. A microarray analysis was performed on bovine γ δ T cells treated with APP; increases in transcripts encoding GM-CSF, IL-8, and IL-17, but not markers of TCR stimulation such as IFNγ , were observed. Key responses were confirmed in human, mouse, and bovine cells by RT-PCR and/or ELISA, indicating a conserved response to procyanidins. In vivo relevance of the cytokine response was shown in mice following intraperitoneal injection of APP, which induced production of CXCL1/KC and resulted in neutrophil influx to the blood and peritoneum. In the human γ δ T cell-line, MOLT-14, GM-CSF and IL-8 transcripts were increased and stabilized in cells treated with crude APP or purified procyanidins. The ERK1/2 MAPK pathway was activated in APP-treated cells, and necessary for transcript stabilization. Our data describe a unique γ δ T cell inflammatory response during procyanidin treatment and suggest that transcript stability mechanisms could account, at least in part, for the priming phenotype.
Keywords: γ δ T cells, cytokines, gene regulation, procyanidin, ERK1/2, transcript stability
Introduction
γδ T cells are a class of lymphocytes involved in both the innate and adaptive arms of the immune system and thus possess numerous immune functions.1 They are a unique T cell whose TCR (T-cell receptor) can recognize unprocessed, non-peptide antigens2,3 and do not require antigen presentation by classical MHC molecules4. γ δ T cells can also process and present antigen as classic antigen presenting cells (APCs)5,6. They are found at many portals of entry into the body and thus are well-positioned to respond immediately to infection and/or insult. They are a major source of immune cytokines early in many infections and can also perform direct cytotoxic functions. Our laboratory has recently described the ability of γ δ T cells from bovine and human to respond to a procyanidin component (specifically oligomeric procyanidins) from unripe apple peel (APP) in a manner called priming7–10. A primed γ δT cell expresses surface IL-2Rα and CD69 in addition to producing select inflammatory cytokines, but not the prototypic inflammatory cytokine, IFNγ 7. Furthermore, in the absence of a secondary stimulus, a primed γ δT cell demonstrates limited proliferation 8–10. This observation is particularly interesting with respect to the innate immune capacity of these cells, as this phenotype suggests that select dietary procyanidins prime γ δ T cells to be in a heightened state of responsiveness without fully activating them. In support of this, feeding mice APP in their drinking water leads to expansion of the γ δ T cell pool within the intestinal mucosa11. The mechanism by which procyanidin-induced priming of γ δ T cells and eventual expansion in vivo occurs has not been defined, but elaborating upon our understanding of these responses is important in understanding how immune responses could potentially be fine-tuned to benefit host immunity.
Procyanidins are members of the polyphenol group of metabolites unique to plants. They can be assembled into oligomers to result in a very diverse body of procyanidin species that function on both plant and mammalian systems, including antioxidant, anti-pathogen, and anti-cancer activities, as well as immunostimulatory effects10. We have previously shown that oligomeric procyanidins from APP are active on both primary human γ δ T cell populations, Vδ2 and Vδ18. Additionally, a large subset of human NK cells, and a small subset of αβ T cells and B cells also respond to these agonists. Therefore, it is likely that procyanidins do not signal through the TCR but rather through a mechanism common to these cell types8. Larger oligomeric species, including trimer molecules, exhibit greater specificity towards γ δ T cells than other cell types. Of the commercially-available purified procyanidins, procyanidin C1 (PC1), a trimer isolated from grape seed, had the most robust priming activity on human PBMCs whereas smaller oligomers were less potent10.
In bovine γ δ T cells, APP-derived procyanidins induce a priming phenomenon very similar to that observed by PAMPs. Thesepriming responses are characterized by the rapid production of a select set of immune cytokines, such as GM-CSF and myeloid cell chemokines (IL-8), but not the prototypic γ δ T cell cytokine IFNγ 7,12, is observed. IL-8 is a chemokine whose primary role is to recruit neutrophils to sites of inflammation and/or infection13, whereas GM-CSF is a growth factor that acts on hematopoietic stem cells to generate granulocytes (including neutrophils, eosinophils, and basophils) and monocytes. GM-CSF and IL-8 both contain similar adenosine/uridine-rich elements (AURE*) 14,15 found in the 3’UTR of many transcripts whose expression must be tightly regulated due to the inflammatory or oncogenic nature of the encoded protein16.
AUREs are targeted by proteins aptly named AURE-binding proteins (AURE-BP), and interaction of the element with an AURE-BP can either increase or decrease the stability of the transcript17. AUREs are classified into three distinct groups that differ both in their sequence as well as in the mechanism of transcript degradation 18. GM-CSF and IL-8, as well as many other immune-associated genes, contain class II AUREs, which are defined by their multiple overlapping copies of the AUUUA motif14,15. Gene expression of IL-8 and GM-CSF can also be controlled similarly, as transcription of both genes is regulated by the NFκB and MAPK pathways19–21. Due to such similarities, we hypothesized that the molecular details of the unique inflammatory profile in cells treated with APP procyanidins could rely on these post-transcriptional mechanisms.
In this study we took advantage of three different model systems (mice, cattle, and humans) to define conserved responses of γ δ T cells towards plant-derived procyanidins. Microarray analysis of sorted bovine γ δ T cells showed that a number of myeloid cell cytokines, including IL-8 and GM-CSF, were up-regulated after APP treatment, confirming a PAMP-like γ δT cell priming response during procyanidin culture. Furthermore, mice injected intraperitoneally with APP displayed neutrophil influx and elevated CXCL1/KC (mouse equivalent of IL-8) levels in both the peritoneum and the blood, affirming the relevance of the expression of these cytokines in vivo. To test the impact of procyanidins on the stability of these AURE-containing transcripts, we used the human γ δ T cell line, MOLT-14. Both IL-8 and GM-CSF transcripts were stabilized in cells treated with either APP or purified procyanidin (PC1), and this event likely occurred via the ERK1/2 MAPK pathway. Together, our data suggest that a transcript stabilization mechanism is activated in APP-treated cells and could account, at least in part, for theγ δT cell priming response since transcript stabilization could enable the cell to rapidly produce large amounts of cytokines in preparation for secondary stimulation or insult.
Materials and Methods
Cell culture
MOLT-14 cells (DSMZ, Brunswick, Germany) were maintained at a density of approximately 1.5 × 106 cells/mL in complete RPMI (Cellgro, Manassas, VA, USA) containing 10% fetal bovine serum22 (Hyclone, Waltham, MA, USA) at 37°C and 10% CO2. Twenty-four hours prior to any experiment, cells were collected by brief centrifugation at 500xg, then suspended at 1.5 × 106 cells/mL in serum-free medium, X-VIVO-15 (X-VIVO; BioWhittaker, Walkersville, MD, USA), in six- or twelve- well tissue culture plates (BD Falcon; Becton Dickinson, Franklin Lakes, NJ), depending on the experiment. Primary bovine PBLs were obtained as previously described8. Briefly, freshly collected PBMCs were prepared by Histopaque 1077 (Sigma-Aldrich, St. Louis, MO, USA) density gradient and monocytes were removed using anti-CD14 microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). Monocyte depletion was confirmed by FACS (FACSCalibur, BD Biosciences). For transcript stabilization studies, cells in X-VIVO were treated with APP (Apple PolyR, Littleton, CO, USA) at 54μg/mL, recombinant human TNFα (PeproTech, Rocky Hill, NJ, USA) at 50ng/mL, recombinant bovine TNFα (Thermo Scientific, Waltham, MA), PC1 (Phytolab, Vestenbergsgreuth, Germany) at 50μg/mL, or PBS for 4h, then with actinomycin D (Sigma-Aldrich) at 5μg/mL. At the time-points indicated in the figures, cells were harvested from the wells and transferred to centrifuge tubes, then collected by centrifugation for 5m at 500xg. Supernatant fluids were removed and discarded, and then RNA was extracted from the cell pellets using the Qiagen RNeasy kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. For studies using the U0126 inhibitor, cells in X-VIVO were treated with PBS or APP at 54μg/mL for 2h, then U0126 (Cell Signaling, Danvers, MA, USA) at 20μM was added and cells were incubated for an additional 2h, followed by treatment with actinomycin D as described above. For splenocyte culture, spleens were removed from WT (C57BL/6) or γ δ T cell-deficient animals and were disrupted with ten strokes of a Dounce homogenizer in 10mL X-VIVO medium. Suspensions were poured through a 70μm filter to remove clumped cells and were plated in twelve-well plates at a density of 2 × 106 cells/mL in X-VIVO.
Mouse splenocyte priming assays
Carboxyfluorescein succinimidyl ester (CFSE)-labeled BALB/c splenocytes were prepared as above and cultured in X-VIVO medium with PBS or PC1 (15μg/mL) for 24h. Culture supernatant fluid was then replaced with fresh medium containing 1ng/mL rhIL-2 and cultured for an additional 72h. Cells were then stained for γ δTCR (GL3) and analyzed by FACS.
Microarrays
Microarray experiments were performed as previously described23,24. Bovine PBMCs were collected from two different calves and γ δ T cells were stained with GD3.8 (pan γ δ T cell mAb) and sorted using a FACS Vantage (Becton Dickson, Franklin Lakes, NJ, USA) to purities greater than 97%. Sorted cells were rested overnight in X-VIVO, and then stimulated with either PBS or APP (70μg/mL) for 4h. RNA was extracted and analyzed on an Agilent Bioanalyzer (Agilent Technologies, Wilmington, DE, USA) to assess integrity. RNA was then amplified and used to probe GenechipR Bovine Genome Arrays (Affymetrix, Santa Clara, CA, USA). Identical experiments were performed using cells from two different calves (four total arrays). The intensity of each gene on the APP array was normalized to its corresponding spot on the PBS array for each specific calf. Only genes determined to be present in all four arrays using the Gene Chip Operating Software program (Affymetrix) were used for further analysis. The fold-change in gene expression over PBS from each calf was then averaged, and those with a 3-fold or greater increase after treatment with APP were included in the analysis in Table 1. Microarray data was submitted to GEO and is accessible using the series number GSE13321.
Table I. Immune relevant genes with greater than 3-fold induction after treatment of bovine γ δ T cells with APP.
Complete microarray data are accessible under GEO series number GSE13321.
| APP fold change over PBS | Gene Description |
|---|---|
| 85.2 | GM-CSF (colony stimulating factor 2) |
| 21.3 | Vascular endothelial junction-associated molecule (VE-JAM) |
| 19.5 | IL-2 |
| 19.1 | Killer cell lectin-like receptor F1 |
| 11.8 | Chemokine (C-C motif) ligand 3-like 1 |
| 9.2 | MIP-1β |
| 8.8 | IL-4R |
| 8.8 | CD83 (B-cell activation protein) |
| 7.3 | VEGF (vascular endothelial growth factor) |
| 7.2 | TGFβ1 (transforming growth factor beta 1) |
| 7.0 | IL-13 |
| 6.9 | CD69 (early T-cell activation antigen) |
| 6.7 | CCR5 (chemokine receptor 5) |
| 6.1 | Chemokine (C-X-C motif) ligand 2 |
| 6.0 | IL-1Rα |
| 6.0 | IL-2Rα |
| 5.8 | TNFα (tumor necrosis factor, alpha) |
| 5.8 | Chemokine (C-X-C motif) ligand 16 |
| 5.7 | Chemokine (C-C motif) ligand 20 |
| 5.4 | MHC class I heavy chain |
| 5.2 | Chemokine (C-X-C motif) ligand 17 |
| 4.1 | CD82 antigen |
| 4.1 | Chemokine (C motif) ligand 1 |
| 3.9 | IL-17 |
| 3.7 | ODC 1 (ornithine decarboxylase 1) |
| 3.1 | IL-8 |
Real-time RT-PCR
RNA was used in reverse transcription reactions using random hexamer primers and SuperScript III enzyme (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The resulting cDNA was used in quantitative PCR experiments using SYBR green reagent (Applied Biosystems, Foster City, CA, USA) and primers specific for 18S (Forward: 5’-TCGAGGCCCTGTAATTGGAAT-3’ and Reverse: 5’-CCCAAGATCCAACTACGAGCTT-3’), GM-CSF (Forward: 5’-GCAGCCTCACCAAGCTCAAG-3’ and Reverse: 5’-TCTTTGCACAGGAAGTTT-3’), IL-8 (Forward: 5’-TTCAGAGACAGCAGAGCACACA-3’ and Reverse: 5 ’-GGCCAGCTTGGAAGTCATGT-3’), beta-actin (Forward: 5’-AAGGATTCCTATGTGGGCGA-3’ and Reverse: 5’-TCCATGTCGTCCCAGTTGGT-3’), and c-jun (Forward: 5’-CAGAGTCCCGGAGCGAACT-3 ’ and Reverse: 5’-CCTTCTTCTCTTGCGTGGCT-3’). For mouse experiments the primers used were specific for beta-actin (Forward: 5’-CCTAAGGCCAACCGTGAAAA-3 ’ and Reverse: 5 ’-GAGGCATACAGGGACAGCACA-3’), GM-CSF (Forward: 5 ’-GGTTCAGGGCTTCTTTGATGG-3’ and Reverse: 5’-CCTGGGCATTGTGGTCTACAG-3’), and for CXCL1/KC (Forward: 5’-TGCTAGTAGAAGGGTGTTGTGCGA-3’ and Reverse: 5’-TCCCACACATGTCCTCACCCTAAT-3’). Reactions were performed on a 7500 Fast Real-time PCR System (Applied Biosystems) in triplicate, and data were normalized to 18S or beta-actin to obtain relative transcript levels. Each individual experiment was performed a minimum of three times, and data are presented as average ± standard deviation.
ELISA analyses
CXCL1/KC protein was detected in the blood and IP fluid of mice immunized with APP using the Quantikine KC Immunoassay kit (R & D Systems, Minneapolis, MN, USA). IL-8 was detected in supernatant fluids from MOLT-14 cells treated with APP using the Quantikine IL-8 Immunoassay kit (R & D Systems). GM-CSF was detected in supernatant fluids of MOLT-14 cells treated with APP using the GM-CSF ELISAMax kit (BioLegend, San Diego, CA, USA). All experiments were performed in triplicate according to the respective manufacturer’s instructions. Data are presented as average ± standard deviation.
Peritonitis studies
BALB/c or CXCR2 KO (gift of A. Harmsen, Montana State University) mice were injected in the peritoneum with APP (1mg in PBS, optimal dose and timing were empirically determined) or different doses of purified procyanidins, and after 3h blood was collected into blood collection MicrovetteR 200 tubes (Sarstedt, Newton, NC, USA) with sera separated by centrifugation at 12 000 x g for 10m. Red blood cells were lysed in ACK buffer (0.15M NH4Cl, 1mM KHCO3, 0.1mM EDTA), washed in Hank’s Buffered Saline Solution (HBSS, Cellgro), and leukocytes were analyzed by flow cytometry as described below. After euthanasia, peritoneal cavities were washed with 300μL HBSS (injected and retrieved) followed by a 5mL wash with HBSS. Retrieved HBSS from the 300μL wash was used for CXCL1/KC ELISAs. Cells from the 5mL wash were collected by centrifugation at 500xg for 5m and similarly subjected to flow cytometric analysis. Blood and peritoneal cells were stained with directly conjugated RB6-8C5-FITC (anti-granulocyte receptor-1; Bright on all neutrophils), anti-CD45.2-APC (all leukocytes; disregards RBCs) and anti-CD11b-PE (bright on neutrophils) and read on a FACS-Calibur equipped with an HTS loader (BD Biosciences, San Jose, CA, USA). Viable leukocytes were gated based on FSC/SSC and positive CD45 staining. Of the viable leukocytes, the percentage of neutrophils (RB6-8C5bright/CD11bbright and distinctive light scatter profile) was determined. All animal procedures were performed in accordance with National Institutes of Health guidelines and approved by the Institutional Animal Care and Use Committee of Montana State University (Bozeman, MT).
Western Immunoblots
MOLT-14 cells were treated with PBS or APP for 30m, 60m, and 4h at the concentrations described above. At the indicated time-points, cells were transferred to tubes and collected by centrifugation for 5m at 500xg. Supernatant fluids were removed and discarded and pellets were suspended in buffer containing 0.15M NaCl, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, and 10mM Tris-HCl pH 7.2, and were incubated on ice for 30m. Samples were boiled for 5m prior to resolution on a 10% SDS-PAGE gel. Proteins in the gels were transferred to nitrocellulose then blocked in 10% Blotto (10% w/v nonfat dry milk in PBS) for 1h at room temperature. Blots were incubated with the following antibodies diluted 1:1 000 in 0.5% Blotto at room temperature overnight: anti-phospho ERK 1/2 MAPK (Thr202/Tyr204)(Cell Signaling, Danvers, MA, USA), anti-phospho p38 MAPK (Thr180/Tyr182) (Cell Signaling), anti-phospho JNK1/JNK2 MAPK (Thr183/Tyr185) (Abcam, Cambridge, MA, USA), anti-ERK1/2 MAPK (Cell Signaling), anti-p38 MAPK (Cell Signaling), or anti-JNK1/JNK2 MAPK (also known as SAPK/JNK) (Cell Signaling). Blots were washed three times with 0.5% Blotto and incubated with HRP-goat anti-rabbit secondary antibody at 1:5 000 in 0.5% blotto for 2h at room temperature. Blots were washed and developed with ECL reagent (GE Healthcare, Piscataway, NJ, USA) and exposed to film for autoradiography.
Results
APP induces robust changes in global transcript levels in primary bovine γ δ T cells
To broaden our understanding of the transcriptome profile induced in γ δ T cells treated with APP beyond a few select cytokines, a microarray experiment was performed using primary sorted bovine γ δ T cells. We used the advantage of the bovine system for these experiments as large numbers of γ δ T cells could be purified to near homogeneity from neonatal animals25. Importantly, the APP preparation used in these studies has been documented to contain 95% polyphenolic compounds, the majority of which are procyanidins, with undetectable or very little endotoxin8. Cells were treated with buffer carrier or APP for 4h and then RNA was extracted and used to probe an Affymetrix Bovine Genechip array as described23. APP treatment of γ δ T cells significantly increased transcript levels for approximately 2 000 genes at least two-fold while there were few genes significantly down-regulated in the APP- versus PBS-treated γ δT cells (see GEO series number GSE13321). The most robust response was observed for the GM-CSF transcript, which was induced approximately 85-fold over PBS-treated cells (Table I). Transcripts for activation markers such as CD69 and IL-2Rα were also induced over PBS-treated control cells. Other transcripts with increased expression included VEGF, an endothelial growth factor, and NK cell molecules including the killer cell lectin-like receptor 1, but the significance of their expression was not further analyzed. Interestingly, overt inflammation markers such as IFNγ were not detected in these experiments. However, transcripts for other inflammatory molecules such as tumor-necrosis factor-α (TNFα) and IL-8 were induced 5.8-fold and 3.1-fold, respectively. This pattern of gene expression was consistent with γ δT cell priming8,9 and other work in our laboratory7 where procyanidins induce a unique inflammatory profile in γ δ T cells that does not include expression of IFNγ.
APP procyanidins prime mouse γ δT cells in vitro and alter the mouse cytokine profile in vivo, resulting in neutrophil influx to the blood and peritoneum
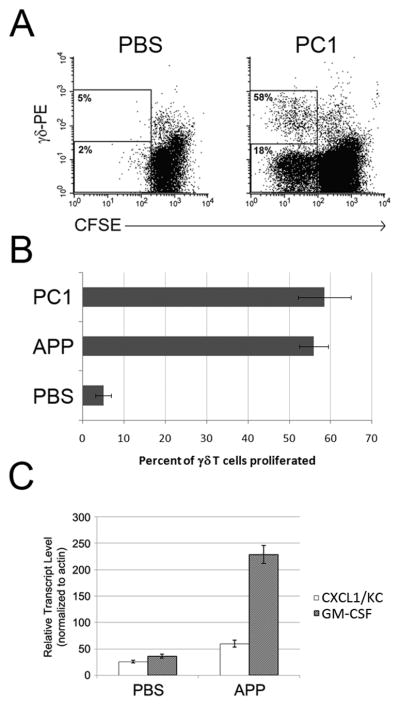
An increasedγ δT cell population in the intestinal mucosa of mice 11 and rats 26 is observed after consumption of APP, suggesting that a murine response occurs similar to that described for bovine and human γ δT cells7–10. As such, we wanted to determine if mouse γ δT cells similarly respond to APP in vitro. To test for a mouse response to oligomeric procyanidins, we used a commercially-available, highly-purified oligomeric trimer (PC1). Priming was quantified by the number of γ δT cells that proliferated following APP and PC1 treatment, compared to the un-primed (buffer control) culture. Procyanidin treatment induced γ δT cell proliferation in response to IL-2 (Figure 1A[BALB/C] and 1B [C57Bl/6]), indicating APP and purified procyanidins acted as γ δT cell priming agents in two common inbred mouse strains. Similar to previous experiments with bovine and human cells, a small percentage of non-γ δT cell lymphocytes proliferated. In human cultures, the non-γ δT cells responding to procyanidin are predominantly NK cells, however this procyanidin-responsive population was not characterized in mice 8.
Figure 1. APP and/or purified procyanidin (PC1) prime mouse γ δT cells to proliferate in response to IL-2 and induce CXCL1/KC and GM-CSF transcript expression in murine splenocytes.
A) CFSE-labeled BALB/c splenocytes were cultured in X-VIVO-15 medium with PBS or PC1 (15μg/mL) for 24h. Culture supernatant fluid was then replaced with fresh medium containing 1ng/mL rhIL-2 and cultured for an additional 72h. Cells were then stained for γ δTCR (GL3) and analyzed by flow cytometry. A representative example of a two-color FACS plot from one of three experiment repeats is shown. Values represent the percent proliferated of either the γ δT cell (top region) or non-γ δT cell (bottom region) populations. Percent proliferation was measured by dividing the number CFSE-dim cells (gated) from the number of total cells in the respective population. B) Experiments performed as described in (A) were also completed using spleen cells from C57Bl/6 mice, and responses to APP and PC1 compared. Data represent the average +/− SD from 3 experiments (P<0.001 for both APP and PC1 vs. buffer control). C) Spleen cells isolated from BALB/c mice were treated with APP or PBS in X-VIVO for four hours. RNA was extracted from each sample and subjected to quantitative RT-PCR for actin, GM-CSF, and CXCL1/KC. Results represent mean +/− SD from triplicate values.
Next we tested for the ability of mouse cells to increase expression of the prototypical priming transcripts encoding GM-CSF and IL-8. Spleen cell preparations from BALB/c mice were treated with buffer/carrier or APP for 4h, and CXCL1/KC (IL-8 homolog) as well as GM-CSF transcripts were measured by quantitative RT-PCR. As shown in Figure 1C, both cytokine transcripts were readily detected following APP treatment10. Since IL-8 and GM-CSF transcripts are expressed in procyanidin-treated cells from human (unpublished data) and bovine (Table 1), and were confirmed in mouse, we chose to focus the remainder of our studies on these two cytokines.
We next performed in vivo experiments to assess a functional role for procyanidin-induced CXCL1/KC and GM-CSF. Since GM-CSF and CXCL1/KC induce the development of a peritonitis response27,28, we chose this model to measure APP responses in vivo. Neutrophil influx was measured as the ratio of neutrophils (RB6-8C5bright, CD11bbright) to total leukocytes (CD45.2+). Substantial neutrophil influxes were observed in both the blood (neutrophilia) and the peritoneum (recruitment) of mice injected with APP (Figure 2A). Administration of purified procyanidins likewise induced neutrophilia; as little as 50μg PC1 induced significant neutrophil accumulation (data not shown). As shown in Figure 2B, approximately 785 pg/mL CXCL1/KC was detected in the IP fluid wash, and 1,400 pg/mL was detected in the blood of mice injected with APP, confirming that the neutrophil response observed in APP-injected mice correlates with the expression of CXCL1/KC. Likewise, neutrophil recruitment to the peritoneum was observed when CXCR2−/ − mice, which lack the major CXCL1/KC receptor, were used in the same procedure, thereby confirming a role for CXCL1/KC in this process (Figure 2C). GM-CSF was not detected by ELISA in the same sample fluids at the 4h time point (unpublished data). It is possible that GM-CSF could be detected at time points different than those tested, but it is also possible that the ELISA method used here was not sensitive enough for this model. These gene and protein expression data and in vivo responses suggested that GM-CSF and CXCL1/KC play a key role in affecting the procyanidin response in vivo, and thus these transcripts were further analyzed.
Figure 2. APP induces CXCL1/KC in the peritoneum and blood of mice resulting in neutrophilia.
A) BALB/c mice were injected IP with 1 mg of APP prepared in PBS, or with PBS alone. After four hours, blood and IP fluid were harvested from each animal and the cells in each sample were subjected to FACS analysis. Data are presented as the mean percentage (+/−SD from 3 mice) of neutrophils out of the total number of viable leukocytes in the peritoneum and are representative of a minimum of three independent experiments. B) BALB/c mice were injected IP with 1 mg of APP prepared in PBS, or with PBS alone. After four hours, blood and IP fluid were harvested from each animal and were subjected to ELISA for CXCL1/KC. Data are the mean (+/− SD) concentration (pg/mL) of CXCL1/KC in each sample. C) Experiments were done as described for (A) and comparisons in the APP response in the peritoneum of BALB/c mice and mice lacking CXCR2 were made. p-value was determined using Student’s unpaired T test.
GM-CSF and IL-8 transcripts are stabilized in human γ δT cells during treatment with APP
After validating that procyanidin effects in the mouse were similar to previous reports for other species8 and demonstrating an in vivo relevance for procyanidins, we next sought to ascertain the mechanism for the rapid response to secondary stimuli after priming with APP8. In an effort to control for variability in primary cell cultures and the limited number of γ δT cells available in such cultures, we selected a cell line. Several cell lines were tested for procyanidin responsiveness including: THP1, Monomac-1, Jurkat, Lucy, PEER, and MOLT-14 cells (data not shown). Of those tested, only MOLT-14 cells, a γ δTCR+ human leukemia cell line, up-regulated GM-CSF and IL-8 transcripts, as well as, generated GM-CSF in response to procyanidin culture (Supplemental Figure 1 and data not shown). Thus, using the MOLT-14 cell line, we designed experiments to begin to define the mechanism by which GM-CSF and IL-8 transcript and protein are induced in cells treated with APP.
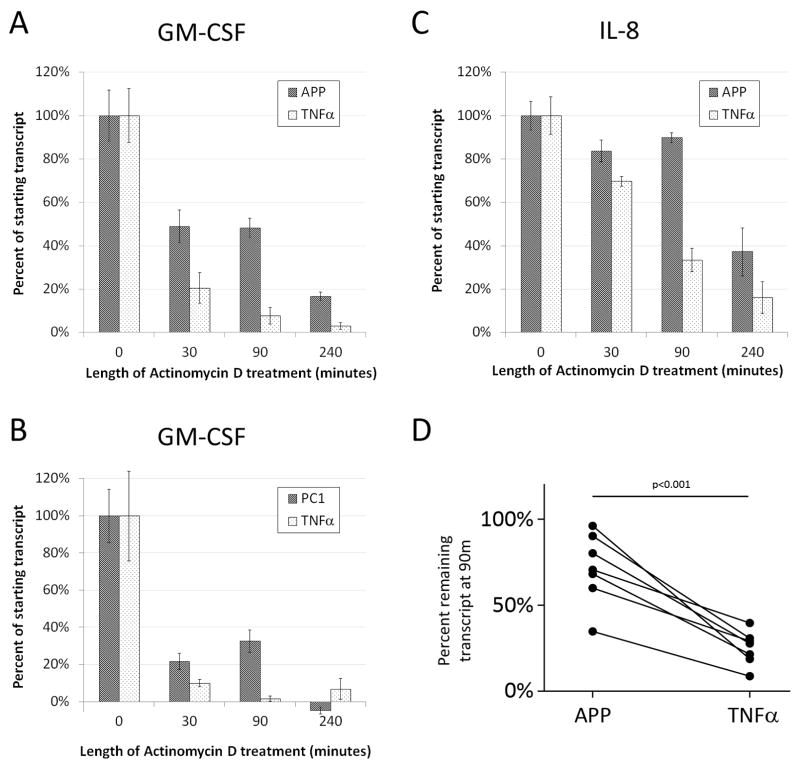
Based on the rapid responses we previously observed in primed γ δT cells, we hypothesized that these transcripts contain structural features enabling this rapid induction8. Upon analyzing the transcripts for similar structures, we observed that GM-CSF and IL-8 contain similar AU-rich elements (AUREs) that function to modulate transcript degradation rates16. Therefore, the stability of these transcripts was analyzed following treatment with APP. MOLT-14 cells were treated with APP or TNFα for 4h, and then with actinomycin D to prevent de novo transcription. RNA was harvested at time 0 (collected immediately before actinomycin-D treatment), 30m, 90m, and 4h post-actinomycin D treatment, and samples were analyzed by quantitative RT-PCR using 18S to determine relative expression. Since MOLT-14 cells constitutively express very low GM-CSF and IL-8 transcripts (data not shown), treatment with TNFα was used as a negative control as it induces expression of GM-CSF transcript but does not stabilize it29. GM-CSF transcript at the time of collection (t=0) in TNFα-treated cells was approximately 11.8-fold less than in APP-treated cells, and 2.1-fold greater than in PC1-treated cells. Initial IL-8 transcript was similar in APP- and TNFα-treated cells (1.4-fold greater than TNFα-induced transcript). In APP-treated cells, the GM-CSF transcript decreased to 49% of the initial amount thirty minutes after actinomycin D-induced transcription blockade, and remained at 48% and 17% of the initial amount after 90m and 4h, respectively (Figure 3A). By comparison, the transcript was degraded quickly in TNFα-treated cells; GM-CSF transcripts decreased to 20% of the initial amount at 30m, and were nearly equivalent to PBS-treated cells at both 90m and 4h following TNFα-induced induction. These results indicate that APP- but not TNFα-induced GM-CSF transcripts were likely stabilized in MOLT-14 cells.
Figure 3. APP stabilizes GM-CSF and IL-8 transcript in MOLT-14 cells.
A–C) MOLT-14 cells were treated with PBS, APP, TNFα, or PC1 in cRPMI for four hours and then collected (t=0) or treated with actinomycin D for the length of time indicated in the figure. RNA was extracted from each sample and subjected to quantitative RT-PCR for 18S, GM-CSF (panels A and B), or IL-8 (panel C). Data represent the percent remaining transcript as measured by the mean relative expression of triplicate values minus background (PBS-treated cells 4h after actinomycin D treatment) +/− SD. Data are representative of three independent experiments. PC1 experiments (panel B) were performed as in A except separately and with different TNFα-and PBS-treated controls. D) Primary bovine PBLs from calves were cultured with APP (50μg/mL) or TNFα (50ng/mL) for 4h. The cultures were then treated with actinomycin D (ActD) by spiking directly into the culture (n=4) or by replacing the media with fresh medium containing actinomycin D (n=3). Cultures were harvested immediately after actinomycin D treatment and again 90m later. RT-PCR was performed to determine GM-CSF transcript relative to 18s. The relative percentage of transcript remaining for TNFα and APP at 90m vs. 0m is shown. Error bars represent SD. The p-value was determined using a one-tailed Wilcoxon’s signed rank test. Data from APP-treated PBMCs with medium replacement were also used as part of the data set in Supplemental Figure 2B. Without these data, the p-value is significant to p<0.01.
We also tested whether the purified PC1 trimer also influenced the stability of the GM-CSF transcript in MOLT-14 cells. As shown in Figure 3B, PC1 both induced and stabilized the GM-CSF transcript. Though the induced expression of GM-CSF by PC1 was not as robust as that induced by APP (t=0 was 22-fold less than with APP), treatment with PC1 resulted in a stability pattern similar to that for APP. These data confirm that defined procyanidin species, such as the PC1 trimer, induce transcript stability in γ δ T cells.
We then performed similar experiments using the IL-8 transcript. As shown in Figure 3C, the IL-8 transcript was similarly regulated by APP and TNFα, although induced IL-8 levels were less when compared to GM-CSF (4-fold increase versus background vs. 919.7-fold increase for GM-CSF). TNFα-induced IL-8 transcript expression was not stabilized and was equivalent to cells treated with PBS by 90m. In comparison, treatment with APP resulted in IL-8 stabilization, which remained significantly greater than background (PBS), even 4h later.
Previous reports demonstrate predicted half-lives for GM-CSF and IL-8 to be ~30m30 and ~20m31 respectively. Therefore, we predicted un-stabilized transcripts would be greatly diminished at 90m (3 half-lives for GM-CSF and 4.5 half-lives for IL-8), and in fact this was observed in these experiments, with the greatest difference in transcript retention between APP- and TNFα-treated cells being observed 90m after transcription was inhibited via actinomycin D. Since the greatest change in transcript stability was observed 90m after transcript blockade, we chose this time point when validating our methods.
We performed three additional experiments to address potential artifact in these transcript stability experiments. First, to address the potential that the observed transcript stabilization was an artifact of the MOLT14 cell line, we tested primary cultures for increased stabilization in APP-treated cultures. Bovine PBMC cultures were selected because they contain a higher percentage of responding cells than human or mouse PBMCs. As shown in Figure 3D, primary PBMC cultures treated with APP had more stable GM-CSF transcripts than TNFα – reated cultures. This demonstrated that the transcript stabilization in MOLT14 cells was not an artifact of the cell line. Secondly, to control for artifact due to actinomycin D in the t=30, 90, and 240, but not t=0 RNA preparations, we added actinomycin D to all samples, including t=0, prior to collection. Similar to previous experiments (Figures 3A and 3C), we observed rapid degradation in TNFα-, but not APP-treated cultures (Supplemental Figure 2A). Finally, since procyanidins bind with high avidity to proteins and other compounds10, we questioned whether the observed transcripts could be a result of APP-induced actinomycin D precipitation and thereby incomplete blockade of transcription. To address this potential artifact, we tested whether removal of APP at the point of actinomycin D treatment had an effect on transcript maintenance. To this end, we treated bovine PBMCs with APP for 4h, washed twice with medium to remove free procyanidin, and added fresh medium containing actinomycin D with either APP or PBS. Ninety minutes after the fresh media were added, RNA was collected from the cells and GM-CSF transcripts were assayed by RT-PCR. As expected, due to the outbred nature of calves as well as varying ages of individuals, there were differences in GM-CSF transcript levels from animal to animal. However, there was no difference in transcript levels when actinomycin D was incubated in the presence or absence of APP (Supplemental Figure 2B), thereby indicating APP did not affect actinomycin D activity.
The ERK1/2 pathway is activated in MOLT-14 cells treated with APP
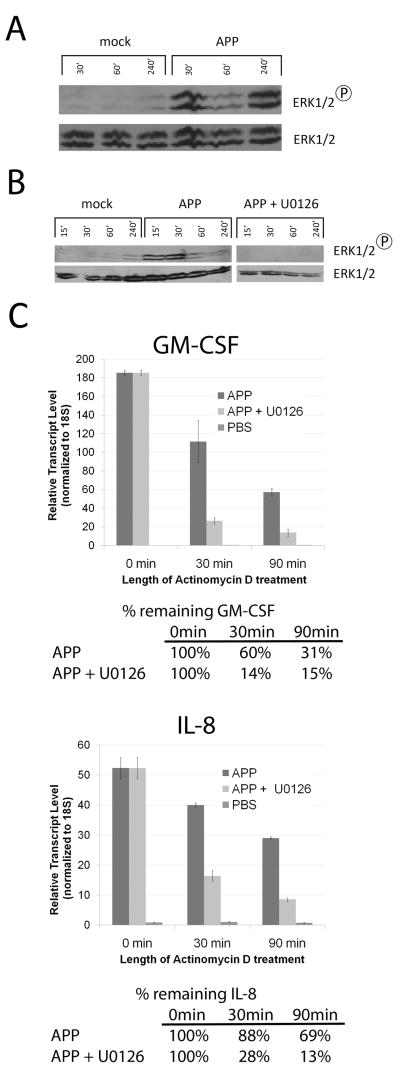
We next performed experiments to identify the pathway(s) that could be responsible for the transcript stabilization observed in cells treated with procyanidins. We began by investigating the possible activation of a MAPK pathway, as signaling through these modules is important in transcript stability32–35. MOLT-14 cells were treated with PBS or APP for 30m, 60m, or 4h, whereupon cell lysates were prepared and subjected to SDS-PAGE and Western immunoblot using antibodies specific for phosphorylated ERK1/2 (p44/p42), p38, as well as JNK MAP Kinases. Blots were subsequently stripped and re-probed using antibodies specific for the unphosphorylated form of the corresponding protein to verify equal loading between lanes. As shown in Figure 4A, the ERK1/2 molecules were phosphorylated to high levels in cells treated with APP, but not in those treated with PBS alone. p38 MAPK was not phosphorylated in response to APP, whereas APP did result in the phosphorylation of JNK1 and JNK2 (Supplemental Figure 3), suggesting activation of the JNK/SAPK MAPK pathway.
Figure 4. APP activates the ERK1/2 pathway, which is required for GM-CSF and IL-8 stabilization.
A: MOLT-14 cells were treated with PBS or APP in cRPMI for the time indicated in the figure. Cell lysates were prepared and subjected to Western immunoblot for phospho-ERK1/2 or total ERK1/2. Data are representative of three independent experiments. B: Cells were treated with APP, APP and 20μM U0126, or mock and harvested at 15, 30, 60, and 240 min. Cells were then lysed and analyzed for phospho-ERK1/2 or ERK1/2 via western blot. C: MOLT-14 cells were treated with PBS or APP in cRPMI for two hours, followed by treatment with U0126 for two hours, and then with actinomycin D for the time indicated in the figure. RNA was extracted from each sample and subjected to quantitative RT-PCR for 18S, GM-CSF, and IL-8. Data represent mean of triplicate values +/− SD and are presented as the normalized amount of each transcript to 18S. The percentage of each transcript that remained following treatment was calculated after subtracting background (PBS t=90m). Data are representative of three independent experiments.
The ERK1/2 MAPK pathway is involved in GM-CSF and IL-8 transcript stabilization in MOLT-14 cells treated with APP
Since the ERK and p38 pathways have been reported to stabilize AURE-containing transcripts, and the p38 pathway was shown to not be activated during procyanidin culture, we next tested whether the ERK1/2 MAPK pathway plays a role in the increased stability of the GM-CSF and IL-8 transcripts in APP-treated MOLT-14 cells. To this end, we employed an ERK1/2 inhibitor in stability experiments. U0126 inhibits MEK1/2, the upstream kinase of ERK1/2, and thus inhibits any signaling events downstream of ERK1/2 phosphorylation36. In these experiments, MOLT-14 cells were treated with APP for 2h to induce gene transcription, and then with 20 μM U0126 for an additional 2h. This treatment condition was determined to inhibit ERK1/2 phosphorylation in MOLT-14 cells by Western immunoblot (Figure 4B). Cultures stimulated for 4h as above with PBS, APP, or APP/U0126 were then treated with actinomycin D, and RNA samples were harvested at 0m, 30m and 90m. As shown in Figure 4C, treatment with APP induced stability of the GM-CSF transcript as previously seen, but treatment with U0126 prior to actinomycin D treatment abolished this effect (presence of the transcript was reduced to 14% of the initial total by 30m and to 7% by 90m in the presence of U0126, compared to 60% and 31%, respectively, when cells were treated with APP alone). A similar observation was made for the IL-8 transcript in cells pre-treated with U0126, where its presence was reduced to 54% and 39% of the initial concentration after 30m and 90m of actinomycin D exposure, respectively. In contrast, APP-treated cells not treated with U0126 retained 89% and 73% of IL-8 transcript after 30m and 90m, respectively (Figure 4C). These data suggest that the ERK1/2 pathway is at least partially responsible for the induced stability of these transcripts.
We next tested the ability of U0126 to regulate the decay rate of c-jun, a quickly degraded (half-life of ~20m37) transcript containing a class III AURE (as opposed to the class II AURE from IL-8 and GM-CSF). MOLT-14 cells were treated with APP for 2h, then with the U0126 inhibitor for 2h, followed by treatment with actinomycin D. RNA was harvested at 0m, 30m, and 90m post-actinomycin D treatment, and RT-PCR was performed as previously described. As shown in Figure 5, c-jun transcript expression was induced in cells treated with APP at time 0, as expected. The presence of the c-jun transcript decreased over time in cells treated with APP. When cells were pre-treated with the U0126 inhibitor prior to actinomycin D treatment, the decay rate of the c-jun transcript was not accelerated. In fact, treatment with the inhibitor potentially stimulated transcript production despite the presence of actinomycin D. Importantly, this observation is distinct from what was observed for both the GM-CSF and IL-8 transcripts, and suggests that APP-induced ERK1/2 activation is specific for these class II AURE-containing transcripts. Together, these data suggest that transcript stabilization may be a mechanism for γ δT cell priming in response to procyanidins.
Figure 5. The ERK1/2 pathway inhibitor U0126 does not prevent c-jun transcript stabilization in MOLT-14 cells treated with APP.

MOLT-14 cells were treated with PBS or APP in cRPMI for two hours, followed by treatment with U0126 for two hours, and then with actinomycin D for the time indicated in the figure. RNA was extracted from each sample and subjected to quantitative RT-PCR for 18S and c-jun. Data represent mean of triplicate values +/−SD and are presented as the normalized amount of each transcript to 18S. The percentage of each transcript that remained following treatment was calculated after subtracting background (PBS t=90min). Data are representative of three independent experiments.
Discussion
This report begins to define the mechanisms that are involved in our previous reports of procyanidin-induced γ δ T cell priming. Many transcripts associated with inflammation, including IL-8 and GM-CSF, were found to be increased in expression upon procyanidin stimulation. However, the effect on γ δ T cells is unlike TCR stimulation in that IFNγ transcript levels are unchanged7. We show in vivo relevance for the procyanidin-induced cytokine response with the observation that injection of APP results in a rapid neutrophil influx into both the peritoneum and blood. Furthermore, these studies show that GM-CSF and IL-8 transcripts are stabilized in cells treated with procyanidins and this likely occurs via ERK1/2 MAPK signaling. Together, these data describe a mechanism where cytokines involved in the generation and recruitment of neutrophils are induced and stabilized by APP procyanidins. The microarray analyses performed on γ δT cells indicate that these responses are similar to PAMP- induced γ δT cell priming8. These PAMP responses are characterized as mild inflammatory responses, leading to increased responses upon secondary stimulation. This may prove a useful alternative to PAMP agonists currently being tested in clinical applications as these candidates have undesired side-effects.
These microarray results also confirmed our previous reports7 that GM-CSF is dramatically up-regulated during APP treatment (85.2-fold increase). When tested in mice, procyanidins, including purified PC1, elicited similar responses to those defined in humans and cattle. Since oral delivery increasesγ δT cell populations in the gut11, our confirmation of a mouse response is not surprising, however characterization of a phenotype in these different tissues demonstrates that the effect of procyanidins is not limited to the intestinal mucosa. Furthermore, using this peritonitis model, we were also able to confirm the importance of the secreted cytokine CXCL1/KC (functional equivalent to IL-8).
Using CXCL1/KC (IL-8) and GM-CSF as key cytokines in the procyanidin response, we looked for a mechanism that could explain the γ δT cell priming response. Both of these transcripts contain an AURE in their 3’ UTR14,15, which suggested that the presence of these elements play a role in the reduced degradation rate of these mRNAs in APP-treated MOLT-14 cells. These studies demonstrated that both the GM-CSF and IL-8 transcripts were stabilized in cells treated with APP. AURE-containing transcript degradation is often regulated and associated with p38 and/or ERK1/2 MAPK activation. These results indicate that ERK1/2 but not p38 activity is important for the increased half-life of the GM-CSF and IL-8 transcripts in APP-treated MOLT-14 cells. This finding is uncommon, but has previously been described in the context of transcript stability17. Conversely, the p38 MAPK pathway is commonly implicated in stabilizing transcripts in other systems31,33,34,38, but APP-mediated transcript stability appears independent of the p38 kinase, since its phosphorylation was not observed (data not shown). Perhaps the fact that the p38 MAPK pathway is not activated in APP-treated cells and is instead regulated by the ERK1/2 MAPK accounts for the unique inflammatory phenotype we detect. This conclusion is supported by prior studies that observe p38 MAPK activation in response to cellular stress, while the ERK1/2 MAPK module primarily responds to mitogenic stimuli39–42. Previously, we found that APP enhanced γ δ T cell proliferation in response to either IL-2 or IL-158. Selective activation of the ERK1/2 MAPK pathway is consistent with these observations. Likewise, activation of the ERK1/2 MAPK pathway results in an anti-inflammatory cellular environment43 and could explain why, even in the face of robust induction of some inflammatory cytokines, expression of the prototypic inflammatory cytokines of γ δ T cells, such as IFNγ , is not seen. Investigation into the role of the JNK MAPK pathway in APP-treated MOLT-14 cells will also shed light on APP-mediated mechanisms, since the JNK module, like p38, signals in response to events such as inflammation and apoptosis44–46.
Although it is not clear how MAP kinases regulate AURE degradation, studies have revealed that they can both phosphorylate and regulate the activity of at least some AURE-binding proteins. There are many immunologically-relevant AURE-BPs that can affect transcript stability and can have opposing roles in transcript degradation based on the presence, absence, or phosphorylation state these proteins. Commonly-described AURE-BPs include the following: TTP, BRF1, BRF2, Roquin, and CUGBP1, which promote transcript degradation, HuR, NF90, YB-1, and AUF2, which prevent degradation, and AUF1 and KSRP, which can either induce or prevent transcript degradation. Many of these AURE-BPs have been shown to regulate GM-CSF and/or IL-8 (CXCL1/KC) transcript stability including AUBF47, YB-148, TTP15,49, HuR50, and KSRP15. Since the GM-CSF, IL-8, and CXCL1/KC transcripts contain Class II AUREs14,15,51,52, it is possible that regulation of their decay is related to the activity of a common protein. Further experiments are underway to fully understand MAP kinase activity and AURE-BP interactions during procyanidin treatment.
These studies shed some light on the apparent dichotomy existing with respect to the function of procyanidin molecules. Procyanidins are proposed for the treatment of inflammatory diseases53 as well as in the clearance of infection54,55, indicating that they have both anti-inflammatory and pro-inflammatory properties. For example, some procyanidins can inhibit NFκB activity56–61 and play a role in inhibiting the development of food allergies11 and allergic rhinitis62. Likewise, procyanidins from grape seeds have anti-tumor activity63. In contrast, procyanidins from cocoa have a pro-inflammatory effect on PBMCs by inducing cytokine release64. Our data point to an inflammatory phenotype for γ δ T cells treated with APP, as evidenced by the nature of the induced cytokines such as GM-CSF and IL-8. However, it is clearly a restricted phenotype in that overt activation markers, such as IFNγ , are not induced, and activation of the ERK1/2 and JNK1/2 MAPK pathways but not p38 are detected. Perhaps this limited inflammatory response contributes to downstream regulatory responses that ultimately dampen or control expression of other inflammatory molecules. Alternatively, it seems other AURE-containing transcripts could be stabilized during procyanidin treatment. If this were to occur, it would stand to reason that the production of these cytokines, should the cell be producing them, would similarly increase. This could lead to an improvement in the response, should it be inflammatory or suppressive. Clearly, these questions remain to be answered, however the demonstration that mice respond to procyanidins will provide an accessible model system to test these hypotheses under both inflammatory and infectious conditions.
Collectively, the data reported herein divulge a mechanism by which APP rapidly and robustly induces a unique response in vitro and in vivo. Importantly, we have begun to define a cross-species mechanism by which γ δ T cells, thought to be a bridge between the innate and adaptive immune systems, are primed in response to APP. Our model predicts that this priming response could enhance either pro-inflammatory or immunoregulatory activities of γ δT cells, which would be dictated by the nature of a secondary stimulus. Although we are focused on γ δT cells, these mechanisms could also be involved in procyanidin effects on other lymphocytes, such as NK cells and subsets of α β T cells that also respond, as shown in our earlier study8. Further defining these mechanisms and their impact in vivo will provide insight into optimizing the use of these types of compounds to treat inflammatory diseases53, as well as enhancing the clearance of various infections54,55.
Supplementary Material
A: MOLT-14 cells were stained with an anti-pan γ δ TCR antibody (BD Bioscences), followed by a goat anti-mouse IgG-PE conjugated secondary antibody as described in Materials and Methods. B: MOLT-14 cells were treated with APP at 54 μg/mL or PBS for 24 hours, and then supernatant fluids were harvested and subjected to GM-CSF ELISA as described in Materials and Methods.
A. To control for potential artifact due the absence of actinomycin D in t=0 RNA preparations, XVIVO-activated MOLT-14 cells were cultured at 1 × 107 cells/mL for 4h with APP (54μg/mL) or TNFα (50ng/mL) to induce GM-CSF transcription. At 4h, all cultures, including t=0, received 5mg/mL actinomycin D. Cells were harvested immediately (t=0) or at 30, 90, and 240 minutes later and GM-CSF transcripts, relative to actin, were analyzed. Data represent the percentage of GM-CSF transcripts at time=0 post-actinomycin treatment. B. To control for APP-induced inhibition of actinomycin D activity, Bovine PBMCs from 3 calves were cultured in XVIVO for 4h with APP, and then medium was washed and replaced with fresh media containing actinomycin D plus PBS or APP. Cells were harvested 90m after the addition of these fresh media to determine the amount of remaining GM-CSF transcript. There was no significant difference in the amount of GM-CSF in relation to secondary media treatment (P>0.2). Variability between calves was significant (P<0.001). Analyses were performed using a 2-way ANOVA with 3 replicates for each treatment/block. Data from APP-treated PBMCs replaced with medium containing PBS and actinomycin D were also used as part of the data set in Figure 3D.
MOLT-14 cells were cultured for 30, 60, or 240 min, with APP, anisomycin (aniso; 20μg/mL), or mock. Cells were harvested, lysed, and western blots performed for phospho-p38 or phospho-JNK1/2; figures A and B, respectively. Nitrocellulose was stripped, and re-probed for total p38 or JNK1/2 as appropriate.
Acknowledgments
Dr. Ed Schmidt and Kate McInnerney (Montana State University) are acknowledged for critical review of the manuscript and technical assistance with microarray experiments, respectively.
Footnotes
This study was supported by funding from the National Institutes of Health (NIH) (NCCAM AT0004986-01), NIAID Contract (HHSN26620040009C/N01-AI-40009), NIH COBRE, and support from the Murdock Charitable Trust.
AURE; also referred to as an ARE; however, to avoid confusion with antioxidant response elements, whichare also targeted by procyanidins1, the AURE nomenclature will be used herein
Conflict of Interest
M.A.J. holds shares in LigoCyte Pharmaceuticals, Inc., which together with Montana State University, held a National Institutes of Health contract that partially funded this work. All other authors have no financial conflict of interest.
Reference List
- 1.Holtmeier W, Kabelitz D. γδ T cells link innate and adaptive immune responses. Chem Immunol Allergy. 2005;86:151–83. doi: 10.1159/000086659. [DOI] [PubMed] [Google Scholar]
- 2.Sicard H, et al. In vivo immunomanipulation of V gamma 9V delta 2 T cells with a synthetic phosphoantigen in a preclinical nonhuman primate model. J Immunol. 2005;175:5471–5480. doi: 10.4049/jimmunol.175.8.5471. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka Y, et al. Nonpeptide ligands for human gamma delta T cells. Proc Natl Acad Sci U S A. 1994;91:8175–8179. doi: 10.1073/pnas.91.17.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukowski JF, Morita CT, Brenner MB. Human gamma delta T cells recognize alkylamines derived from microbes, edible plants, and tea: implications for innate immunity. Immunity. 1999;11:57–65. doi: 10.1016/s1074-7613(00)80081-3. [DOI] [PubMed] [Google Scholar]
- 5.Brandes M, Willimann K, Moser B. Professional antigen-presentation function by human gammadelta T Cells. Science. 2005;309:264–268. doi: 10.1126/science.1110267. [DOI] [PubMed] [Google Scholar]
- 6.Collins RA, et al. Gammadelta T cells present antigen to CD4+ alphabeta T cells. J Leukoc Biol. 1998;63:707–714. doi: 10.1002/jlb.63.6.707. [DOI] [PubMed] [Google Scholar]
- 7.Graff JC, Jutila MA. Differential regulation of CD11b on gammadelta T cells and monocytes in response to unripe apple polyphenols. J Leukoc Biol. 2007;82:603–607. doi: 10.1189/jlb.0207125. [DOI] [PubMed] [Google Scholar]
- 8.Holderness J, et al. Select plant tannins induce IL-2Ralpha up-regulation and augment cell division in gammadelta T cells. J Immunol. 2007;179:6468–6478. doi: 10.4049/jimmunol.179.10.6468. [DOI] [PubMed] [Google Scholar]
- 9.Jutila MA, Holderness J, Graff JC, Hedges JF. Antigen-independent priming: a transitional response of bovine gammadelta T-cells to infection. Anim Health Res Rev. 2008;9:47–57. doi: 10.1017/S1466252307001363. [DOI] [PubMed] [Google Scholar]
- 10.Holderness J, et al. Response of gammadelta T Cells to plant-derived tannins. Crit Rev Immunol. 2008;28:377–402. doi: 10.1615/critrevimmunol.v28.i5.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akiyama H, et al. Dietary unripe apple polyphenol inhibits the development of food allergies in murine models. FEBS Lett. 2005;579:4485–4491. doi: 10.1016/j.febslet.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 12.Hedges JF, Lubick KJ, Jutila MA. Gamma delta T cells respond directly to pathogen-associated molecular patterns. J Immunol. 2005;174:6045–6053. doi: 10.4049/jimmunol.174.10.6045. [DOI] [PubMed] [Google Scholar]
- 13.Baggiolini M, Clark-Lewis I. Interleukin-8, a chemotactic and inflammatory cytokine. FEBS Lett. 1992;307:97–101. doi: 10.1016/0014-5793(92)80909-z. [DOI] [PubMed] [Google Scholar]
- 14.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winzen R, et al. Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets. Mol Cell Biol. 2007;27:8388–8400. doi: 10.1128/MCB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 17.Clark A, Dean J, Tudor C, Saklatvala J. Post-transcriptional gene regulation by MAP kinases via AU-rich elements. Front Biosci. 2009;14:847–871. doi: 10.2741/3282. [DOI] [PubMed] [Google Scholar]
- 18.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann E, Dittrich-Breiholz O, Holtmann H, Kracht M. Multiple control of interleukin-8 gene expression. J Leukoc Biol. 2002;72:847–855. [PubMed] [Google Scholar]
- 20.Thomas RS, et al. ETS1, NFkappaB and AP1 synergistically transactivate the human GM-CSF promoter. Oncogene. 1997;14:2845–2855. doi: 10.1038/sj.onc.1201125. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins F, Cockerill PN, Bohmann D, Shannon MF. Multiple signals are required for function of the human granulocyte-macrophage colony-stimulating factor gene promoter in T cells. J Immunol. 1995;155:1240–1251. [PubMed] [Google Scholar]
- 22.Hedges JF, Cockrell D, Jackiw L, Meissner N, Jutila MA. Differential mRNA expression in circulating gammadelta T lymphocyte subsets defines unique tissue-specific functions. J Leukoc Biol. 2003;73:306–314. doi: 10.1189/jlb.0902453. [DOI] [PubMed] [Google Scholar]
- 23.Hedges JF, et al. Mucosal lymphatic-derived gammadelta T cells respond early to experimental Salmonella enterocolitis by increasing expression of IL-2R alpha. Cell Immunol. 2007;246:8–16. doi: 10.1016/j.cellimm.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lahmers KK, et al. Comparative gene expression by WC1+ gammadelta and CD4+ alphabeta T lymphocytes, which respond to Anaplasma marginale, demonstrates higher expression of chemokines and other myeloid cell-associated genes by WC1+ gammadelta T cells. J Leukoc Biol. 2006;80:939–952. doi: 10.1189/jlb.0506353. [DOI] [PubMed] [Google Scholar]
- 25.Graff JC, Behnke M, Radke J, White M, Jutila MA. A comprehensive SAGE database for the analysis of gammadelta T cells. Int Immunol. 2006;18:613–626. doi: 10.1093/intimm/dxl001. [DOI] [PubMed] [Google Scholar]
- 26.Ramiro-Puig E, et al. Intestinal immune system of young rats influenced by cocoa-enriched diet. J Nutr Biochem. 2008;19:555–565. doi: 10.1016/j.jnutbio.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 27.McColl SR, Clark-Lewis I. Inhibition of murine neutrophil recruitment in vivo by CXC chemokine receptor antagonists. J Immunol. 1999;163:2829–2835. [PubMed] [Google Scholar]
- 28.Wengner AM, Pitchford SC, Furze RC, Rankin SM. The coordinated action of G-CSF and ELR + CXC chemokines in neutrophil mobilization during acute inflammation. Blood. 2008;111:42–49. doi: 10.1182/blood-2007-07-099648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tebo J, et al. Heterogeneity in control of mRNA stability by AU-rich elements. J Biol Chem. 2003;278:12085–12093. doi: 10.1074/jbc.M212992200. [DOI] [PubMed] [Google Scholar]
- 30.Akashi M, Hachiya M, Koeffler HP, Suzuki G. Irradiation increases levels of GM-CSF through RNA stabilization which requires an AU-rich region in cancer cells. Biochem Biophys Res Commun. 1992;189:986–993. doi: 10.1016/0006-291x(92)92301-d. [DOI] [PubMed] [Google Scholar]
- 31.Winzen R, et al. Distinct domains of AU-rich elements exert different functions in mRNA destabilization and stabilization by p38 mitogen-activated protein kinase or HuR. Mol Cell Biol. 2004;24:4835–4847. doi: 10.1128/MCB.24.11.4835-4847.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang SW, et al. Cytokine mRNA decay is accelerated by an inhibitor of p38-mitogen-activated protein kinase. Inflamm Res. 1999;48:533–538. doi: 10.1007/s000110050499. [DOI] [PubMed] [Google Scholar]
- 33.Winzen R, et al. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 1999;18:4969–4980. doi: 10.1093/emboj/18.18.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frevel MA, et al. p38 Mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol Cell Biol. 2003;23:425–436. doi: 10.1128/MCB.23.2.425-436.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esnault S, Malter JS. Extracellular signal-regulated kinase mediates granulocyte-macrophage colony-stimulating factor messenger RNA stabilization in tumor necrosis factor-alpha plus fibronectin-activated peripheral blood eosinophils. Blood. 2002;99:4048–4052. doi: 10.1182/blood.v99.11.4048. [DOI] [PubMed] [Google Scholar]
- 36.Favata MF, et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 37.Chauhan D, et al. Regulation of c-jun gene expression in human T lymphocytes. Blood. 1993;81:1540–1548. [PubMed] [Google Scholar]
- 38.Henness S, et al. IL-17A acts via p38 MAPK to increase stability of TNF-alpha-induced IL-8 mRNA in human ASM. Am J Physiol Lung Cell Mol Physiol. 2006;290:L1283–L1290. doi: 10.1152/ajplung.00367.2005. [DOI] [PubMed] [Google Scholar]
- 39.Cobb MH, Goldsmith EJ. How MAP kinases are regulated. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 40.Freshney NW, et al. Interleukin-1 activates a novel protein kinase cascade that results in the phosphorylation of Hsp27. Cell. 1994;78:1039–1049. doi: 10.1016/0092-8674(94)90278-x. [DOI] [PubMed] [Google Scholar]
- 41.Han J, Lee JD, Bibbs L, Ulevitch RJ. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 42.Derijard B, et al. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 43.Madonna S, Scarponi C, De Pita O, Albanesi C. Suppressor of cytokine signaling 1 inhibits IFN-gamma inflammatory signaling in human keratinocytes by sustaining ERK1/2 activation. FASEB J. 2008;22:3287–3297. doi: 10.1096/fj.08-106831. [DOI] [PubMed] [Google Scholar]
- 44.Ip YT, Davis RJ. Signal transduction by the c-Jun N-terminal kinase (JNK)--from inflammation to development. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 45.Lewis TS, Shapiro PS, Ahn NG. Signal transduction through MAP kinase cascades. Adv Cancer Res. 1998;74:49–139. doi: 10.1016/s0065-230x(08)60765-4. [DOI] [PubMed] [Google Scholar]
- 46.Robinson MJ, Cobb MH. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 47.Rajagopalan LE, Malter JS. Modulation of granulocyte-macrophage colony-stimulating factor mRNA stability in vitro by the adenosine-uridine binding factor. J Biol Chem. 1994;269:23882–23888. [PubMed] [Google Scholar]
- 48.Capowski EE, Esnault S, Bhattacharya S, Malter JS. Y box-binding factor promotes eosinophil survival by stabilizing granulocyte-macrophage colony-stimulating factor mRNA. J Immunol. 2001;167:5970–5976. doi: 10.4049/jimmunol.167.10.5970. [DOI] [PubMed] [Google Scholar]
- 49.Datta S, et al. Tristetraprolin regulates CXCL1 (KC) mRNA stability. J Immunol. 2008;180:2545–2552. doi: 10.4049/jimmunol.180.4.2545. [DOI] [PubMed] [Google Scholar]
- 50.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tebo JM, et al. Interleukin-1-mediated stabilization of mouse KC mRNA depends on sequences in both 5'-and 3'-untranslated regions. J Biol Chem. 2000;275:12987–12993. doi: 10.1074/jbc.275.17.12987. [DOI] [PubMed] [Google Scholar]
- 52.Xu N, Chen CY, Shyu AB. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol Cell Biol. 2001;21:6960–6971. doi: 10.1128/MCB.21.20.6960-6971.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blazso G, Gabor M, Rohdewald P. Antiinflammatory activities of procyanidin-containing extracts from Pinus pinaster Ait. after oral and cutaneous application. Pharmazie. 1997;52:380–382. [PubMed] [Google Scholar]
- 54.Cheshier JE, et al. Immunomodulation by pycnogenol in retrovirus-infected or ethanol-fed mice. Life Sci. 1996;58:L–96. doi: 10.1016/0024-3205(95)02303-8. [DOI] [PubMed] [Google Scholar]
- 55.Liu FJ, Zhang YX, Lau BH. Pycnogenol enhances immune and haemopoietic functions in senescence-accelerated mice. Cell Mol Life Sci. 1998;54:1168–1172. doi: 10.1007/s000180050245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terra X, et al. Grape-seed procyanidins act as antiinflammatory agents in endotoxin-stimulated RAW 264.7 macrophages by inhibiting NFkB signaling pathway. J Agric Food Chem. 2007;55:4357–4365. doi: 10.1021/jf0633185. [DOI] [PubMed] [Google Scholar]
- 57.Mackenzie GG, et al. Epicatechin, catechin, and dimeric procyanidins inhibit PMA-induced NF-kappaB activation at multiple steps in Jurkat T cells. FASEB J. 2004;18:167–169. doi: 10.1096/fj.03-0402fje. [DOI] [PubMed] [Google Scholar]
- 58.Oh GS, et al. Penta-O-galloyl-beta-D-glucose inhibits phorbol myristate acetate-induced interleukin-8 [correction of intereukin-8] gene expression in human monocytic U937 cells through its inactivation of nuclear factor-kappaB. Int Immunopharmacol. 2004;4:377–386. doi: 10.1016/j.intimp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 59.Yang F, et al. The green tea polyphenol (-)-epigallocatechin-3-gallate blocks nuclear factor-kappa B activation by inhibiting I kappa B kinase activity in the intestinal epithelial cell line IEC-6. Mol Pharmacol. 2001;60:528–533. [PubMed] [Google Scholar]
- 60.Sharma SD, Meeran SM, Katiyar SK. Dietary grape seed proanthocyanidins inhibit UVB-induced oxidative stress and activation of mitogen-activated protein kinases and nuclear factor-kappaB signaling in in vivo SKH-1 hairless mice. Mol Cancer Ther. 2007;6:995–1005. doi: 10.1158/1535-7163.MCT-06-0661. [DOI] [PubMed] [Google Scholar]
- 61.Williams RJ, Spencer JP, Rice-Evans C. Flavonoids: antioxidants or signalling molecules? Free Radic Biol Med. 2004;36:838–849. doi: 10.1016/j.freeradbiomed.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 62.Enomoto T, Nagasako-Akazome Y, Kanda T, Ikeda M, Dake Y. Clinical effects of apple polyphenols on persistent allergic rhinitis: A randomized double-blind placebo-controlled parallel arm study. J Investig Allergol Clin Immunol. 2006;16:283–289. [PubMed] [Google Scholar]
- 63.Zhang XY, et al. Proanthocyanidin from grape seeds potentiates anti-tumor activity of doxorubicin via immunomodulatory mechanism. Int Immunopharmacol. 2005;5:1247–1257. doi: 10.1016/j.intimp.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 64.Kenny TP, Keen CL, Schmitz HH, Gershwin ME. Immune effects of cocoa procyanidin oligomers on peripheral blood mononuclear cells. Exp Biol Med (Maywood) 2007;232:293–300. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: MOLT-14 cells were stained with an anti-pan γ δ TCR antibody (BD Bioscences), followed by a goat anti-mouse IgG-PE conjugated secondary antibody as described in Materials and Methods. B: MOLT-14 cells were treated with APP at 54 μg/mL or PBS for 24 hours, and then supernatant fluids were harvested and subjected to GM-CSF ELISA as described in Materials and Methods.
A. To control for potential artifact due the absence of actinomycin D in t=0 RNA preparations, XVIVO-activated MOLT-14 cells were cultured at 1 × 107 cells/mL for 4h with APP (54μg/mL) or TNFα (50ng/mL) to induce GM-CSF transcription. At 4h, all cultures, including t=0, received 5mg/mL actinomycin D. Cells were harvested immediately (t=0) or at 30, 90, and 240 minutes later and GM-CSF transcripts, relative to actin, were analyzed. Data represent the percentage of GM-CSF transcripts at time=0 post-actinomycin treatment. B. To control for APP-induced inhibition of actinomycin D activity, Bovine PBMCs from 3 calves were cultured in XVIVO for 4h with APP, and then medium was washed and replaced with fresh media containing actinomycin D plus PBS or APP. Cells were harvested 90m after the addition of these fresh media to determine the amount of remaining GM-CSF transcript. There was no significant difference in the amount of GM-CSF in relation to secondary media treatment (P>0.2). Variability between calves was significant (P<0.001). Analyses were performed using a 2-way ANOVA with 3 replicates for each treatment/block. Data from APP-treated PBMCs replaced with medium containing PBS and actinomycin D were also used as part of the data set in Figure 3D.
MOLT-14 cells were cultured for 30, 60, or 240 min, with APP, anisomycin (aniso; 20μg/mL), or mock. Cells were harvested, lysed, and western blots performed for phospho-p38 or phospho-JNK1/2; figures A and B, respectively. Nitrocellulose was stripped, and re-probed for total p38 or JNK1/2 as appropriate.