Abstract
Purpose
To explore if functional single nucleotide polymorphisms (SNPs) of base-excision repair genes are predictors of radiation treatment-related pneumonitis (RP), we investigated associations between functional SNPs of ADPRT, APEX1, and XRCC1 and RP development.
Methods and Materials
We genotyped SNPs of ADPRT (rs1136410 [V762A]), XRCC1 (rs1799782 [R194W], rs25489 [R280H], and rs25487 [Q399R]), and APEX1 (rs1130409 [D148E]) in 165 patients with non-small cell lung cancer (NSCLC) who received definitive chemo-radiation therapy. Results were assessed by both Logistic and Cox regression models for RP risk. Kaplan-Meier curves were generated for the cumulative RP probability by the genotypes.
Results
We found that SNPs of XRCC1 Q399R and APEX1 D148E each had a significant effect on the development of grade ≥2 RP (XRCC1: AA vs. GG, adjusted hazard ratio [HR] = 0.48, 95% confidence interval [CI] 0.24–0.97; APEX1: GG vs. TT, adjusted HR = 3.61, 95% CI 1.64–7.93) in an allele-dose response manner (Trend tests: P = 0.040 and 0.001, respectively). The number of the combined protective XRCC1 A or APEX1 T alleles (from 0 to 4) also showed a significant trend of predicting RP risk (P = 0.001).
Conclusions
SNPs of the base-excision repair genes may be biomarkers for susceptibility to RP. Larger prospective studies are needed to validate our findings.
Keywords: Radiation pneumonitis, Polymorphism, Non-small cell lung cancer
INTRODUCTION
Radiation therapy is one of the major treatment modalities for unresectable non-small cell lung cancer (NSCLC) and has been shown to improve local control and overall survival. However, such treatment is associated with a risk of developing radiation treatment-related pneumonitis (RP), resulting from the irradiation and inflammation of the normal lung tissues. RP has been identified as the most common dose-limiting complication of thoracic radiation, with 10% to 20% of patients experiencing moderate or severe RP (1). Several therapeutic and patient-related factors have been reported to influence susceptibility to RP, including patient performance status, dosimetric parameters, smoking, and plasma inflammatory cytokine levels (2). Recently, we demonstrated that a single nucleotide polymorphism (SNP) of the TGFβ1 gene (rs1982073 [T869C]) independently predicted RP risk in patients with NSCLC (3), suggesting that genetic predisposition may be involved in RP development. Considering the relatively sparse reports of RP investigation from a genetic perspective, we were interested in identifying additional genetic factors that are likely to predict RP susceptibility.
Theoretically, RP is a dysregulated wound-healing inflammatory reaction to normal lung tissue injury, characterized by lympholytic alveolitis (2). Genetic factors that could extend the wound-healing process or cause suboptimal repair of irradiated normal lung tissues presumably could lead to prolonged and intensified inflammatory reactions. Therefore, the genes involved in repair of damaged DNA in the irradiated normal lung tissues are potential candidates to predict susceptibility to RP. This hypothesis was reinforced by the evidence that patients with ataxia telangiectasis, Fanconi’s anemia, or Nijmegen breakage syndrome are at increased risk of severe RP as a result of germline mutations in DNA repair genes (4). However, these syndromes, all of which involve increased sensitivity to ionizing radiation, are very rare and thus the mutations associated with them may not influence the development of RP among patients with NSCLC that are otherwise unselected from the general population.
Base excision repair (BER) is the major system responsible for the removal of damaged single DNA bases and for efficient repair of DNA single-strand breaks generated extensively by radiation therapy (5, 6). Several key repair genes play important roles in BER, of which the APEX1 (apurinic/apyrimidinic endonuclease 1), ADPRT (adenosine diphosphate ribosyl transferase), and XRCC1 (x-ray repair cross-complementing 1) genes are the most extensively investigated, commonly for their effects on genetic susceptibility to cancer risk (7, 8) but rarely for tissue sensitivity to radiation therapy (9). It has been shown that a low BER capacity may lead to increased tissue radiosensitivity and perhaps more severe radiation toxicity (6, 10). Since SNPs can contribute directly to disease predisposition by modifying a gene’s function, or they can serve as genetic markers for nearby disease-causing variants through association or linkage disequilibrium (LD), we hypothesized that functional SNPs of the BER genes are biomarkers for predicting susceptibility to RP among patients with lung cancer treated with radiation. To test this hypothesis, we performed a case-only study, seeking associations between RP risk and common functional variants of ADPRT, XRCC1, and APEX1 in patients who received definitive radiation therapy, with or without chemotherapy, for NSCLC.
METHODS AND MATERIALS
Patient population
We identified 261 patients with DNA samples available from a data set of 576 patients with NSCLC treated with definitive radiation at The University of Texas M. D. Anderson Cancer Center between 1999 and 2005. Among these 261 patients, 172 patients had documented information on RP with complete follow-up information and radiation dosimetric data. Since stage IV patients normally have limited life expectancy and only receive a palliative dose of radiation, which often do not cause significant toxicity, we excluded these 7 patients, and the final sample pool consisted of 165 patients, including 120 whites, 32 blacks, 9 Hispanics, 1 Asian and 3 patients whose ethnicities were not self-reported. The median total radiation dose was 63 Gy (range, 50.4 to 84.0 Gy) given at 1.2 to 2.0 Gy/fraction. Of all patients, 19 patients received radiation only, 145 received radiation in combination with chemotherapy, and one received radiation but other therapies were unknown. Details of the radiation treatment planning, follow-up schedule and tests, guidelines for RP scoring, and dosimetric data analysis have been described elsewhere (11, 12). Briefly, all patients were examined by their treating radiation oncologists weekly during concurrent chemoradiotherapy and 4–6 weeks after completion of treatment. The patients were then followed every 3 months for the first 3 years and every 6 months thereafter unless they had symptoms that required immediate examination or intervention. Radiographic examination by chest X-ray or CT was performed at each follow-up visit after completion of chemoradiotherapy. For this analysis, we reviewed all relevant dictated clinical notes by the treating physicians and all radiographic images for every patient. Radiation treatment-related pneumonitis was diagnosed by clinical presentation and any of the following radiographic abnormalities: ground-glass opacity, attenuation, or consolidation changes within the radiation field. Pneumonitis was assessed and scored using the Common Terminology Criteria for Adverse Events version 3.0 (13). The time to end point development was calculated from the beginning of the radiation therapy; patients not experiencing the end point were censored at the last follow-up. This study was approved by the appropriate M. D. Anderson Cancer Center institutional review board.
Genotyping methods
We selected five common, well-studied functional variants of ADPRT, XRCC1, and APEX1 genes involved in BER: ADPRT (rs1136410 [V762A]), XRCC1 (rs1799782 [R194W], rs25489 [R280H], and rs25487 [Q399R]), and APEX1 (rs1130409 [D148E]). These SNPs were selected because they cause nonsynonymous amino acid changes that have been reportedly associated with cancer risk (7). Blood collection and sample preparation were described previously (3). Genotypes were generated by the polymerase chain reaction restriction fragment-length polymorphism method. The primer sequences, restriction enzymes, and polymerase chain reaction conditions used for the experiments are available upon request.
Statistical analysis
Patients were grouped according to their genotypes. Statistical analysis was performed using the SAS 9.2 statistical software (SAS Inc, Chicago, IL). Cox proportional hazards analysis was performed to calculate the hazard ratios (HRs) and 95% confidence intervals (CIs) to evaluate the influence of genotypes on RP risk. Kaplan-Meier analysis was performed to estimate the cumulative RP probability. In addition, multivariate Cox regression was performed to adjust for other covariates. Likelihood ratio tests were performed for each multivariate Cox regression to assess the goodness of fit. Chi-square test was used to calculate the Hardy-Weinberg equilibrium. P values of 0.05 or less were considered statistically significant.
Finally, we created a classification and regression tree (CART) to identify higher-order interactions between clinical factors and genetic variants using the rpart package in S-PLUS Version 8.0.4 (TIBCO, Palo Alto, CA). CART creates a decision tree to identify how well each genotype or environmental factor variable predicts the class (e.g. RP). Specifically, the recursive procedure starts at the root node and uses a log-rank statistic to determine the first optimal split and each subsequent split of the data set. This process continues until the terminal nodes have no subsequent statistically significant splits or the terminal nodes reach a pre-specified minimum size (n > 10).
RESULTS
Patient characteristics and association with radiation pneumonitis
Table 1 lists the characteristics of the 165 patients (93 men and 72 women with a median age of 63 years [range, 35 to 88 years]), of whom 72.7% were white, 76.4% had stage III disease, 88.5% were treated with a combination of chemotherapy and radiation therapy, and 96.8% received radiation doses between 60 and 70 Gy. To determine if any confounding factors were influencing the risk of RP, we analyzed the associations between RP and clinicopathologic characteristics, including age, sex, race, Karnofsky Performance Score, disease stage, tumor histology, smoking history, use of chemotherapy, radiation dose, mean lung dose (MLD) and V20. The median follow-up time for RP of any grade after radiation therapy, with or without chemotherapy, was 21 months (range, 1 to 97 months). We found that disease stage and smoking history were significantly associated with RP (grade ≥2) in the univariate analysis (HR for stage I/II vs. stage III 0.35, 95% CI 0.15–0.82, P = 0.016; and HR for former vs. current smokers 2.07, 95% CI 1.07–4.00, P = 0.031). The traditional RP risk factors, MLD and V20, were also confirmed in the current study (HR for MLD ≥ 19.8 vs. MLD < 19.8 1.91, 95% CI 1.16–3.12, P = 0.011; and HR for V20 ≥ 0.33 vs. V20 < 0.33 2.10, 95% CI 1.27–3.48, P = 0.004). None of the other clinicopathologic characteristics were associated with RP risk in this study population.
Table 1.
Clinical characteristics of 165 NSCLC patients included in the study of radiation pneumonitis
| Parameter | Patient No. (%) | HR | 95% CI | P† |
|---|---|---|---|---|
| Sex | ||||
| Female | 72 (43.6) | 1.00 | ||
| Male | 93 (56.4) | 1.25 | 0.76–2.07 | 0.378 |
| Age (years) | ||||
| <63 | 79 (47.9) | 1.00 | ||
| ≥63 | 86 (52.1) | 0.90 | 0.55–1.47 | 0.672 |
| Race | ||||
| White | 120 (72.7) | 1.00 | ||
| Black | 32 (19.4) | 1.08 | 0.56–2.08 | 0.819 |
| Other | 13 (7.9) | 1.49 | 0.64–3.49 | 0.357 |
| KPS | ||||
| <80 | 24 (14.5) | 1.00 | ||
| ≥80 | 141 (85.1) | 1.31 | 0.60–2.87 | 0.505 |
| Disease stage | ||||
| III | 126 (76.4) | 1.00 | ||
| I, II | 39 (23.6) | 0.35 | 0.15–0.82 | 0.016 |
| Tumor histology | ||||
| Adenocarcinoma | 54 (32.7) | 1.00 | ||
| NSCLC, NOS | 56 (33.9) | 0.73 | 0.40–1.35 | 0.317 |
| Squamous cell | 54 (32.7) | 0.82 | 0.46–1.46 | 0.503 |
| Missing | 1 (0.7) | |||
| Smoking | ||||
| Current | 39 (23.6) | 1.00 | ||
| Former | 111 (67.3) | 2.07 | 1.07–4.00 | 0.031 |
| Never | 14 (8.5) | 2.28 | 0.79–6.58 | 0.128 |
| Missing | 1 (0.6) | |||
| Chemotherapy | ||||
| No | 19 (11.5) | 1.00 | ||
| Yes | 145 (88.5) | 1.71 | 0.74–3.98 | 0.210 |
| Radiation dose (Gy) | ||||
| <63 | 32 (19.4) | 1.00 | ||
| ≥63 | 133 (80.6) | 0.60 | 0.34–1.07 | 0.081 |
| MLD (Gy) | ||||
| <19.8 | 86 (52.1) | 1.00 | ||
| ≥19.8 | 79 (47.9) | 1.91 | 1.16–3.12 | 0.011 |
| V20 | ||||
| <0.33 | 83 (50.3) | 1.00 | ||
| ≥0.33 | 82 (49.7) | 2.10 | 1.27–3.48 | 0.004 |
NSCLC, NOS: non-small cell lung cancer, not otherwise specified
P values were obtained from the univariate Cox hazard model
Table 2 shows the frequency distribution of the five SNPs among the patients. Notably, the minor allele frequency of the XRCC1 R194W and R280H polymorphisms was 0 in this patient group, and their rarity was consistent with some reported data from the National Center for Biotechnology Information’s SNP database of nucleotide sequence variation (dbSNP; http://www.ncbi.nlm.nih.gov/projects/SNP/). The genotype distribution of ADPRT V762A, XRCC1 Q399R, and APEX1 D148E met the Hardy-Weinberg equilibrium.
Table 2.
Allele and genotype frequency of the polymorphisms of candidate BER genes used in the study of radiation pneumonitis in NSCLC patients
| Gene and Variant | No. of Subjects | Wild-type | Hetero-variant | Homo-variant | q* | P† |
|---|---|---|---|---|---|---|
| XRCC1 Q399R | 165 | 49 | 72 | 44 | 0.48 | 0.104 |
| XRCC1 R280H | 165 | 165 | 0 | 0 | 0 | |
| XRCC1 R194W | 165 | 165 | 0 | 0 | 0 | |
| ADPRT V762A | 159‡ | 117 | 35 | 7 | 0.15 | 0.051 |
| APEX1 D148E | 165 | 44 | 87 | 34 | 0.47 | 0.454 |
q: frequency of minor allele.
P: P values for Hardy-Weinberg equilibrium.
Genotyping failed in 6 patients for the ADPRT V762A SNP.
Radiation pneumonitis and genotype association
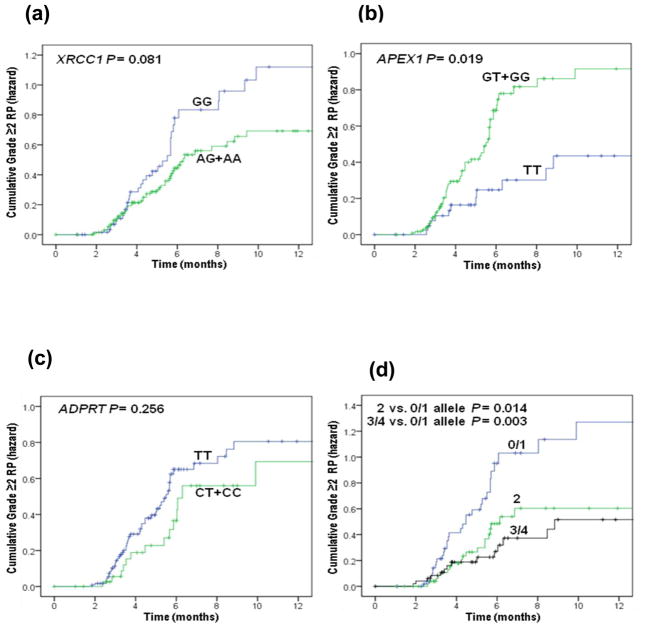
Table 3 shows the results of univariate and multivariate analyses of the associations between genetic polymorphisms and grade ≥2 RP by the Cox proportional hazard model. In the univariate analysis, there was a trend effect for having the increasing number of XRCC1 A allele with a decreasing HR (P = 0.049) and for having the increasing number of APEX1 G allele with an increasing HR (P = 0.008); however, the association was statistically significant only for having the APEX1 G allele (crude HR for GT+GG vs. TT 2.18, 95% CI 1.14–4.18, P = 0.019). In multivariate analyses performed with adjustment of confounding factors shown in Table 1, we found that the XRCC1 Q399R AA genotype was associated with a reduced HR of RP (adjusted HR for AA vs.GG 0.48, 95% CI 0.24–0.97, P = 0.041), whereas having the APEX1 D148E GG genotype was associated with an increased hazard of RP (adjusted HR for GG vs. TT 3.61, 95% CI 1.64–7.93, P = 0.001) (Table 3), and both the trend effects for the XRCC1 A and APEX1 G alleles remained (P = 0.040 and 0.001, respectively). Furthermore, patient with increased number of the combined protective A or T alleles showed a dose-response effect of RP reduction in both univariate and multivariate analyses (adjusted HR = 0.48, 95% CI 0.27–0.86 for 2 vs. 0/1 alleles, P = 0.013; adjusted HR = 0.35, 95% CI 0.18–0.68 for 3/4 vs. 0/1 alleles, P = 0.002 and P = 0.001 for the trend test) (Table 3). Fig. 1 plots the incidence of RP grade ≥2 as a function of time since radiation therapy according to these three genetic polymorphisms. We further assessed these associations in the white patients only (n=120) and found similar tendency of associations, although significance level was not reached for the XRCC1 Q399R polymorphism (Supplementary Table 1). None of these significant findings were observed for the ADPRT V762A SNP (Table 3).
Table 3.
Analysis of BER genotypes and grade ≥2 radiation pneumonitis in patients after treatment for NSCLC
| Genotypes | No. of Patient | Event | HR | Crude HR |
P* | HR | Adjusted HR |
P† |
|---|---|---|---|---|---|---|---|---|
| 95% CI | 95% CI | |||||||
| XRCC1 (rs25487 A>G Q399R) | ||||||||
| GG | 49 | 25 | 1.00 | 1.00 | ||||
| AG | 72 | 28 | 0.73 | 0.42–1.25 | 0.248 | 0.76 | 0.44–1.31 | 0.322 |
| AA | 44 | 13 | 0.52 | 0.26–1.01 | 0.054 | 0.48 | 0.24–0.97 | 0.041 |
| Trend test | P = 0.049 | P = 0.040 | ||||||
| AG+AA | 116 | 41 | 0.64 | 0.39–1.06 | 0.081 | 0.65 | 0.39–1.09 | 0.100 |
| APEX1 (rs1130409 G>T D148E) | ||||||||
| TT | 44 | 11 | 1.00 | 1.00 | ||||
| GT | 87 | 38 | 1.99 | 1.01–3.91 | 0.046 | 1.97 | 1.00–3.88 | 0.050 |
| GG | 34 | 17 | 2.76 | 1.29–5.89 | 0.009 | 3.61 | 1.64–7.93 | 0.001 |
| Trend test | P = 0.008 | P = 0.001 | ||||||
| GT+GG | 121 | 55 | 2.18 | 1.14–4.18 | 0.019 | 2.26 | 1.17–4.35 | 0.017 |
| ADPRT (rs1136410 T>C V762A) | ||||||||
| TT | 117 | 50 | 1.00 | 1.00 | ||||
| CT | 35 | 12 | 0.76 | 0.41–1.43 | 0.396 | 0.67 | 0.35–1.29 | 0.664 |
| CC | 7 | 2 | 0.55 | 0.13–2.26 | 0.408 | 0.58 | 0.14–2.42 | 0.459 |
| Trend test | P = 0.232 | P = 0.172 | ||||||
| CT+CC | 42 | 14 | 0.71 | 0.39–1.28 | 0.256 | 0.64 | 0.35–1.19 | 0.162 |
| XRCC1 399A + APEX1 148T | ||||||||
| 0/1 | 57 | 32 | 1.00 | 1.00 | ||||
| 2 | 57 | 20 | 0.49 | 0.28–0.87 | 0.014 | 0.48 | 0.27–0.86 | 0.013 |
| 3/4 | 51 | 14 | 0.39 | 0.21–0.73 | 0.003 | 0.35 | 0.18–0.68 | 0.002 |
| Trend test | P = 0.001 | P = 0.001 | ||||||
HR, hazard ratio; CI, confidence interval
0 (1, 2, 3, 4): indicates the number of combined A or T alleles in individual patients.
P values were obtained from the univariate Cox hazard model.
P values were obtained from the Cox hazard model with adjustment for disease stage, smoking history, mean lung dose, and V20.
Fig. 1.
Cumulative probability of grade ≥ 2 radiation pneumonitis in 165 patients with NSCLC as a function of time from the start of radiation therapy by genotypes. (a) XRCC1 Q399R; (b) APEX1 D148E; (c) ADPRT V762A; (d) XRCC1 399A and APEX1 148T combined alleles.
Association of multiple factor interaction with RP risk (CART analysis)
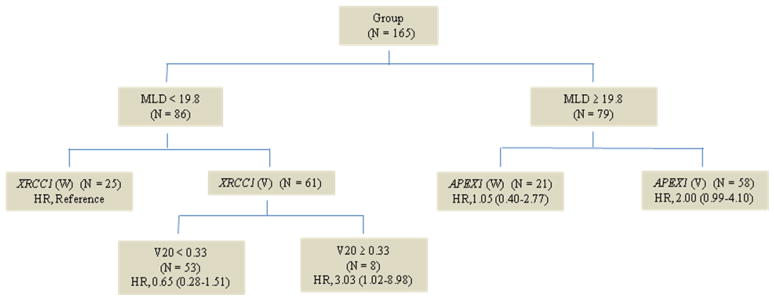
We built a decision tree using clinicopathological variables (disease stage, smoking, MLD and V20) and two SNPs (XRCC1 Q399R and APEX1 D148E) that were found to be significantly associated with RP risk in the 165 patients. In the CART analysis, the first split in the decision tree was MLD, indicating that MLD was the strongest risk factor for RP among the factors considered. Further inspection of the tree structure showed distinct patterns between low MLD (< 19.8Gy) and high MLD (≥ 19.8Gy) groups (Fig. 2). In the low MLD group, compared with those with the wild-type XRCC1 GG genotype, patients carrying XRCC1 AG/AA genotypes had the highest risk of RP when V20 ≥ 0.33 (HR, 3.03, 95% CI 1.02–8.98). In the high MLD group, patients carrying APEX1 GT/GG genotypes had the highest risk of RP (HR, 2.00, 95% CI 0.99–4.10).
Fig. 2.
Classification and regression tree analysis for predictors of grade ≥ 2 radiation pneumonitis in 165 patients with NSCLC. SNPs were classified as wild type (W) and variant genotype (V).
DISCUSSION
To the best of our knowledge, this is the first report of an association between putatively functional SNPs of the BER genes and risk of developing severe RP in lung cancer patients treated with radiotherapy. Although due to our small sample size we could not detect two rare functional polymorphisms of XRCC1 R194W and R280H, nor we had enough study power to evaluate the effect of the ADPRT V762A, our findings indicated that polymorphisms of XRCC1 Q399R and APEX1 D148E both independently and collectively influenced RP development in patients treated with radiotherapy, and thus these polymorphisms, if validated, could be used as biomarkers for susceptibility to RP in patients under consideration of being given definitive radiation therapy for NSCLC.
Thoracic irradiation is a common approach of controlling lung malignancies to improve survival and reduce symptoms; however, thoracic irradiation can also cause severe adverse effects by agitating electrons and producing tissue-damaging free radicals, which limit the thoracic radiation doses and volumes that can be safely delivered and thus restrict its effective doses (14). Clinical RP can be life-threatening, despite aggressive treatment with steroids, especially in patients with preexisting pulmonary diseases (15). Irradiation of normal lung tissues induces immediate damage through intracellular protein denaturation, membrane disruption, and alterations of DNA. RP is the primary consequence of cellular DNA injury that appears in the second generation of cells, usually occurring 4–12 weeks after the completion of radiation (16). Detailed information on radiation-induced DNA damage and responses to it can be found elsewhere (17).
Ionizing radiation can produce a wide variety of lesions in cellular DNA, including rupture of strands, alteration of bases, destruction of sugars, crosslinks and formation of dimers. It is estimated that a dose of ~1 Gy of x rays produces about 1,000 single-strand breaks and 50–100 double-strand breaks in the DNA of a typical mammalian cell, which leads to the death of 50% those cells (18, 19). BER is one of the major pathways for repairing DNA base lesions, AP sites, and single-strand breaks caused by ionizing radiation; this pathway requires the normal function of four major proteins—a DNA glycosylase, an AP endonuclease, a DNA polymerase, and a DNA ligase, of which APEX1, ADPRT, and XRCC1 have been shown to play key roles (20). APEX1 processes the abasic sites left from the incision of the damaged base by cleaving the DNA backbone at the 5′ side to the abasic site, leaving a 3′-hydroxyl group and a 5′-deoxyribose phosphate group flanking the nucleotide gap, and initiates the BER process (21); ADPRT specifically binds to DNA strand breaks and participates in the long-patch repair process (22); and XRCC1 interacts with a complex of DNA repair proteins, including poly (ADP-ribose) polymerase, DNA ligase III, and DNA polymerase β, and coordinates the gap-sealing process in the short-patch BER (23).
In our study, XRCC1 Q399R and APEX1 D148E each showed a significant association with RP development in patients undergoing thoracic irradiation for NSCLC, independent of patient-, tumor-, and therapy-related factors. The XRCC1 Q399R polymorphism is situated within the BRCT-1 region harboring the ADPRT binding domain that may affect the interaction between XRCC1 and ADPRT (24); however, its functional significance remains controversial; some studies showed an association of the A allele with measurably reduced DNA repair capacity (25, 26), which is likely to lead to an increased radiosensitivity, whereas others showed negative results (27–29). Since reduced DNA repair capacity may lead to an increased radiosensitivity in patients with NSCLC (30, 31), our current results indicating a protective effect against RP by the A allele requires further mechanistic investigation. As for the APEX1 D148E polymorphism, although it does not affect the endonuclease activity (32), cells having the variant G allele may have a higher sensitivity to ionizing radiation (33), which may cause an increased RP risk as observed in our study.
Our CART analysis is an explorative, non-parametric approach that requires no assumption of a genetic model. Through this decision tree-based data mining, we identified the mean lung dose to be the most important risk factor. XRCC1 399AG/AA genotypes with V20 ≥ 0.33 and MLD < 19.8 Gy and APEX1 GT/GG genotypes with MLD ≥ 19.8 Gy were most likely to be associated with RP, which suggest that those patients with such genotypes may require close surveillance after thoracic radiotherapy. Besides these positive results, our study did not support an association between the ADPRT V762A polymorphism and RP risk, although the functional relevance of this polymorphism with the BER capacity has been reported previously (22). In addition, the increased RP risk among former smokers (HR for former vs. current smokers 2.07, 95% CI 1.07–4.00) did not seem to result from the different tobacco consumption, because further examination did not reveal higher smoking pack-years in the former smokers compared with the current smokers (50.9 vs. 56.4). It is possible that the increased RP risk in former smokers may result from an unknown selection bias or misclassification of these two subgroups due to recall bias. Finally, we did not adjust the p-values for multiple tests because this was an exploratory study. We plan to confirm current findings in more stringent conditions in future larger studies.
In summary, our study indicated that polymorphisms of the BER genes XRCC1 and APEX1 each had a significant effect on the risk of RP in patients who received definitive radiation therapy for NSCLC, and therefore these polymorphisms could be useful biomarkers for susceptibility to RP. However, larger independent studies are needed to validate these findings. An ongoing cohort study among patients with NSCLC who had received radiation therapy more recently at our institution than those included in the present study will be used to validate the current findings in the near future.
Supplementary Material
Acknowledgments
This study was in part supported by National Institutes of Health grants R01 ES011740 and CA131274 (to Q. W.) and Cancer Center Core Grant CA016672 (to M. D. Anderson Cancer Center). We thank Kejing Xu, Jianzhong He and Min Zhao for laboratory assistance, and Christine F. Wogan for manuscript editing.
Footnotes
Conflict of interest: none.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roach M, 3rd, Gandara DR, Yuo HS, et al. Radiation pneumonitis following combined modality therapy for lung cancer: analysis of prognostic factors. J Clin Oncol. 1995;13:2606–2612. doi: 10.1200/JCO.1995.13.10.2606. [DOI] [PubMed] [Google Scholar]
- 2.Provatopoulou X, Athanasiou E, Gounaris A. Predictive markers of radiation pneumonitis. Anticancer Res. 2008;28:2421–2432. [PubMed] [Google Scholar]
- 3.Yuan X, Liao Z, Liu Z, et al. Single nucleotide polymorphism at rs1982073:T869C of the TGFbeta 1 gene is associated with the risk of radiation pneumonitis in patients with non-small-cell lung cancer treated with definitive radiotherapy. J Clin Oncol. 2009;27:3370–3378. doi: 10.1200/JCO.2008.20.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentzen SM. Preventing or reducing late side effects of radiation therapy: radiobiology meets molecular pathology. Nat Rev Cancer. 2006;6:702–713. doi: 10.1038/nrc1950. [DOI] [PubMed] [Google Scholar]
- 5.Mourgues S, Lomax ME, O’Neill P. Base excision repair processing of abasic site/single-strand break lesions within clustered damage sites associated with XRCC1 deficiency. Nucleic Acids Res. 2007;35:7676–7687. doi: 10.1093/nar/gkm947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhry MA. Base excision repair of ionizing radiation-induced DNA damage in G1 and G2 cell cycle phases. Cancer Cell Int. 2007;7:15. doi: 10.1186/1475-2867-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li C, Hu Z, Lu J, et al. Genetic polymorphisms in DNA base-excision repair genes ADPRT, XRCC1, and APE1 and the risk of squamous cell carcinoma of the head and neck. Cancer. 2007;110:867–875. doi: 10.1002/cncr.22861. [DOI] [PubMed] [Google Scholar]
- 8.Yin M, Tan D, Wei Q. Genetic variants of the XRCC1 gene and susceptibility to esophageal cancer: a meta-analysis. Int J Clin Exp Med. 2009;2:26–35. [PMC free article] [PubMed] [Google Scholar]
- 9.Zschenker O, Raabe A, Boeckelmann IK, et al. Association of single nucleotide polymorphisms in ATM, GSTP1, SOD2, TGFB1, XPD and XRCC1 with clinical and cellular radiosensitivity. Radiother Oncol. 2010 doi: 10.1016/j.radonc.2010.01.016. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 10.Burri RJ, Stock RG, Cesaretti JA, et al. Association of single nucleotide polymorphisms in SOD2, XRCC1 and XRCC3 with susceptibility for the development of adverse effects resulting from radiotherapy for prostate cancer. Radiat Res. 2008;170:49–59. doi: 10.1667/RR1219.1. [DOI] [PubMed] [Google Scholar]
- 11.Wang S, Liao Z, Wei X, et al. Analysis of clinical and dosimetric factors associated with treatment-related pneumonitis (TRP) in patients with non-small-cell lung cancer (NSCLC) treated with concurrent chemotherapy and three-dimensional conformal radiotherapy (3D-CRT) Int J Radiat Oncol Biol Phys. 2006;66:1399–1407. doi: 10.1016/j.ijrobp.2006.07.1337. [DOI] [PubMed] [Google Scholar]
- 12.Jin H, Tucker SL, Liu HH, et al. Dose-volume thresholds and smoking status for the risk of treatment-related pneumonitis in inoperable non-small cell lung cancer treated with definitive radiotherapy. Radiother Oncol. 2009;91:427–432. doi: 10.1016/j.radonc.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Program CTE. Common terminology criteria for adverse events, version 3. Bethesda, MD: National Cancer Institute; 2003. [Google Scholar]
- 14.Mehta V. Radiation pneumonitis and pulmonary fibrosis in non-small-cell lung cancer: pulmonary function, prediction, and prevention. Int J Radiat Oncol Biol Phys. 2005;63:5–24. doi: 10.1016/j.ijrobp.2005.03.047. [DOI] [PubMed] [Google Scholar]
- 15.Makimoto T, Tsuchiya S, Hayakawa K, et al. Risk factors for severe radiation pneumonitis in lung cancer. Jpn J Clin Oncol. 1999;29:192–197. doi: 10.1093/jjco/29.4.192. [DOI] [PubMed] [Google Scholar]
- 16.Choi YW, Munden RF, Erasmus JJ, et al. Effects of radiation therapy on the lung: radiologic appearances and differential diagnosis. Radiographics. 2004;24:985–997. doi: 10.1148/rg.244035160. discussion 998. [DOI] [PubMed] [Google Scholar]
- 17.Jeggo P, Lobrich M. Radiation-induced DNA damage responses. Radiat Prot Dosimetry. 2006;122:124–127. doi: 10.1093/rpd/ncl495. [DOI] [PubMed] [Google Scholar]
- 18.Nikjoo H, O’Neill P, Wilson WE, et al. Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat Res. 2001;156:577–583. doi: 10.1667/0033-7587(2001)156[0577:cafdts]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 19.Ward JF. The yield of DNA double-strand breaks produced intracellularly by ionizing radiation: a review. Int J Radiat Biol. 1990;57:1141–1150. doi: 10.1080/09553009014551251. [DOI] [PubMed] [Google Scholar]
- 20.Hung RJ, Hall J, Brennan P, et al. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am J Epidemiol. 2005;162:925–942. doi: 10.1093/aje/kwi318. [DOI] [PubMed] [Google Scholar]
- 21.Tell G, Damante G, Caldwell D, et al. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 22.Lockett KL, Hall MC, Xu J, et al. The ADPRT V762A genetic variant contributes to prostate cancer susceptibility and deficient enzyme function. Cancer Res. 2004;64:6344–6348. doi: 10.1158/0008-5472.CAN-04-0338. [DOI] [PubMed] [Google Scholar]
- 23.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 24.Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604–608. [PubMed] [Google Scholar]
- 25.Lunn RM, Langlois RG, Hsieh LL, et al. XRCC1 polymorphisms: effects on aflatoxin B1-DNA adducts and glycophorin A variant frequency. Cancer Res. 1999;59:2557–2561. [PubMed] [Google Scholar]
- 26.Matullo G, Palli D, Peluso M, et al. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22:1437–1445. doi: 10.1093/carcin/22.9.1437. [DOI] [PubMed] [Google Scholar]
- 27.Taylor RM, Thistlethwaite A, Caldecott KW. Central role for the XRCC1 BRCT I domain in mammalian DNA single-strand break repair. Mol Cell Biol. 2002;22:2556–2563. doi: 10.1128/MCB.22.8.2556-2563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pastorelli R, Cerri A, Mezzetti M, et al. Effect of DNA repair gene polymorphisms on BPDE-DNA adducts in human lymphocytes. Int J Cancer. 2002;100:9–13. doi: 10.1002/ijc.10463. [DOI] [PubMed] [Google Scholar]
- 29.Hou SM, Ryk C, Kannio A, et al. Influence of common XPD and XRCC1 variant alleles on p53 mutations in lung tumors. Environ Mol Mutagen. 2003;41:37–42. doi: 10.1002/em.10128. [DOI] [PubMed] [Google Scholar]
- 30.Duchesne GM. Fundamental bases of combined therapy in lung cancer: cell resistance to chemotherapy and radiotherapy. Lung Cancer. 1994;10 (Suppl 1):S67–72. doi: 10.1016/0169-5002(94)91668-3. [DOI] [PubMed] [Google Scholar]
- 31.Guo WF, Lin RX, Huang J, et al. Identification of differentially expressed genes contributing to radioresistance in lung cancer cells using microarray analysis. Radiat Res. 2005;164:27–35. doi: 10.1667/rr3401. [DOI] [PubMed] [Google Scholar]
- 32.Hadi MZ, Coleman MA, Fidelis K, et al. Functional characterization of Ape1 variants identified in the human population. Nucleic Acids Res. 2000;28:3871–3879. doi: 10.1093/nar/28.20.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu JJ, Smith TR, Miller MS, et al. Amino acid substitution variants of APE1 and XRCC1 genes associated with ionizing radiation sensitivity. Carcinogenesis. 2001;22:917–922. doi: 10.1093/carcin/22.6.917. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.