Abstract
Background
Thalassaemia is a genetic disease in which there is a relative or complete lack of alpha or beta globin chains. Patients with moderate to severe forms of thalassaemia need transfusions from the early years of life. Antibody production against blood group antigens may cause many problems in preparing compatible blood units for transfusion. The identification of definite blood group phenotypes by the haemagglutination method can be difficult because of the mixed population of red blood cells from the donor and recipient.
Materials and methods
Forty multiply transfused thalassaemic patients and ten healthy controls with no history of blood transfusion were enrolled in this study. Allele-specific oligonucleotide polymerase chain reaction (ASO-PCR) and haemagglutination methods were used to determine the presence of Rhesus (Rh) C, c, E and e antigens.
Results
In this study four primer sets were used for ASO-PCR amplification of RhC/c and RhE/e. Although PCR assays for RhC/c and RHE/e genotyping have been described previously, in this study we used a new condition for PCR by decreasing the annealing temperature from 63 °C to 58 °C in order to amplify all four genes in the same condition. In order to evaluate this single run molecular method, we used the haemagglutination test as the standard method and compared the results from the two methods. We found discrepancies between phenotype and genotype results among patients with beta thalassaemia, but complete agreement between phenotype and genotype in the control group.
Conclusions
The advantage of this new ASO-PCR method compared to a restriction fragment length polymorphism (RFLP) PCR method is that with the former all four genes can be amplified at the same time by PCR, and electrophoresis can be performed immediately to determine individual antigen profiles. The simplicity of the ASO-PCR method makes it suitable for routine use in medical centres and it is also cheaper than RFLP-PCR. Furthermore, as shown by previous studies, the results of haemagglutination and PCR tests often differ because the existence of donor red blood cells in the patient’s circulation can interfere with the interpretation of the haemagglutination test.
Keywords: Rh typing, polymerase chain reaction, thalassaemia, allele-specific oligonucleotide polymerase chain reaction, ASO-PCR
Introduction
Thalassaemia is a genetic disease in which there is a relative or complete lack of alpha or beta globin chains. Patients with moderate to severe forms of thalassaemia need transfusions from the early years of life. Regular blood transfusion is necessary for the satisfactory growth and development of thalassaemic patients, to improve their living conditions during childhood and to sustain a good quality of life during adulthood1. Repeated blood transfusions are, however, associated with a risk of alloimmunisation and the incidence of this complication in multiply transfused patients is about 30%2,3. Alloimmunisation may cause many problems with regards to long-term management and transfusion. It can lead to haemolytic transfusion reactions as well as create difficulty and delays in finding compatible blood units4,5.
The Rhesus (Rh) blood group system has the highest prevalence of polymorphisms among human blood group systems and is clinically significant in transfusion medicine. The genes coding for the Rh antigens are located on the short arm of chromosome 1 (1p34-p36) and consist of two adjacent gene structures, RHCcEe and RHD. The Rh blood group system comprises more than 50 different antigens with very high polymorphism. Proteins C, c, E and e are detected as immunological isoforms2,6. Nowadays only compatibility for RhD is tested serologically on blood units before transfusion. Antigen compatibility for the other systems, such as C, c, E and e, is not tested routinely until the patient shows alloantibodies against them. The production of alloantibodies against these antigens will, however, cause problems in preparing compatible blood units for transfusion. In multiply transfused patients, donors’ red blood cells circulate in the recipients’ vessels for a few weeks and a precise diagnosis of blood group phenotypes is rendered difficult by the presence of the mixed population of donors’ and recipients’ red blood cells, especially when patients’ pre-transfusion blood samples are not available8. Since anti-RhC/c and anti-RhE/e may also cause transfusion reactions, their identification in multiply transfused patients, such as those with thalassaemia, is important9.
Materials and methods
Forty multitransfused thalassaemic patients and ten healthy controls with no history of transfusion were enrolled in this study. Peripheral blood samples (4 mL) were drawn into test-tubes containing EDTA. The Rh phenotypes were determined by the haemagglutination method (CE-Immunodiagnostika, Germany).
Genomic DNA was isolated from whole blood using a commercial kit (KIAsorb purification kit). The primers that were used for RhC/c and Rh E/e genotyping are shown in Table I as described by Tanaka M. et al.10. Polymerase chain reaction was conducted using 100 ng of genomic DNA, 1 μM of each primer, 200 μM of each dNTP, 2.0 U Taq DNA in buffer, provided polymerase, and 1.5 mM MgCl2 by the polymerase manufacturer, in a final volume of 50 μL. The amplification reaction were carried out in a thermal cycler (Gradient, Eppendorf). After initial denaturation at 94 ºC for 10 min, 30 three-step cycles at 94 ºC for 30 sec, 58 ºC for 45 sec and 72 ºC for 45 sec were performed, followed by 5 minutes of elongation at 72 ºC (Table II). PCR products were analysed by electrophoresis in 3% agarose.
Table I.
PCR primers for RhC/c and RhE/e ASO-PCR.
| Primer name | Sequence 5′ to 3′ | Product size | Allele specificity |
|---|---|---|---|
| TRH 1 | CGCTGCCTGCCCCTCTGC | 118 | C |
| TRH 2 | TTGATAGGATGCCACGAGCC | ||
| TRH 3 | CTTGGGCTTCCTCACCTCAAA | 107 | c |
| TRH 4 | AAGCCGTCCAGCAGGATTGC | ||
| TRH 5 | TGGCCACGTGTCAACTCTC | 143 | E |
| TRH 7 | CATGCTGATCTTCCTTTGGG | ||
| TRH 6 | TGGCCACGTGTCAACTCTG | 143 | e |
| TRH 7 | CATGCTGATCTTCCTTTGGG |
Table II.
PCR conditions for amplification.
| Stage | Time | Temperature (°C) | |
|---|---|---|---|
| 10 min | 94 | ||
| 30 cycles | Denaturation | 30 sec | 94 |
| Annealing | 45 sec | 58 | |
| Extension | 45 sec | 72 | |
| Final elongation | 5 min | 72 |
Results
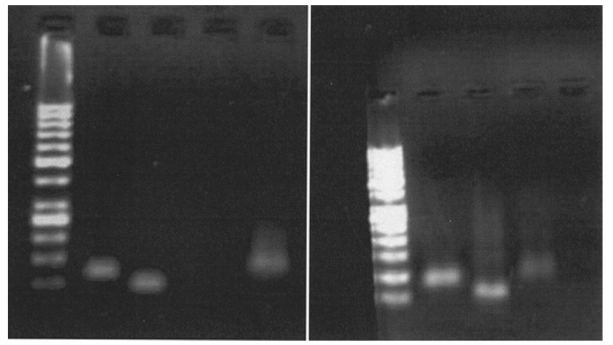
In the present study four primer sets were used for allele-specific oligonucleotide (ASO)-PCR amplification of RhC/c and RhE/e in two genomic regions: exon 1 and exon 5. Primers TRH1/2 and TRH3/4 were used for RhC/c ASO-PCR and primers TRH5/7 and TRh6/7 were used for RhC/c ASO-PCR. Although PCR assays for RhC/c and RHE/e genotyping have been described previously, in this study we used a new condition for PCR by decreasing the annealing temperature from 63 °C to 58 °C in order to amplify all four genes in the same condition (Figure 1). We were, therefore, able to amplify all four genes in each sample in a single run. A 118 bp fragment was amplified with primers TRH1/2 for the RhC allele in RhC-positive samples (CC or Cc). A 107 bp product was amplified with primers TRH3/4 for the Rhc allele in Rhc samples. A 143 bp product was amplified with primer sets TRH5/7 for the RhE allele in RhE-positive samples.
Figure 1.
RhC/c and RhE/e genotyping by PCR for two patients. Left: a patient typed RhCcee; right: a patient with RhCcEE.
Finally another 143 bp product was amplified with primer set TRH6/7 from DNA samples containing the Rhe allele. In order to evaluate this single run molecular method, we used the haemagglutination test as the standard method and the results were compared (Table III). There were some discrepancies between phenotype and genotype in patients with beta thalassaemia, but complete agreement between phenotype and genotype was observed in the control group.
Table III.
Random sampling study of RHCE genotyping by ASO-PCR and phenotyping by the haemagglutination test.
| Genotyping | Phenotyping | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C | C | E | e | C | c | E | E | |||
| C 1* | P | P | N | P | Ccee | P | P | N | P | Ccee |
| C 2 | P | N | P | P | CCEe | P | N | P | P | CCEe |
| C 3 | P | P | P | P | CcEe | P | P | P | P | CcEe |
| C 4 | P | N | P | P | CCEe | P | N | P | P | CCEe |
| C 5 | P | P | P | P | CcEe | P | P | P | P | CcEe |
| C 6 | N | P | P | N | ccE | N | P | P | N | ccEE |
| C 7 | P | N | P | P | CCEe | P | N | P | P | CCEe |
| C 8 | P | P | P | P | CcEe | N | P | P | P | ccEe |
| C 9 | P | P | N | CCEE | P | N | P | N | CCEE | |
| C 10 | P | P | P | N | CceEE | P | P | P | N | CcEE |
| T1** | P | P | P | P | CcEe | P | N | P | P | CCEe |
| T2 | P | P | P | N | CcEE | P | P | P | P | CcEe |
| T3 | P | P | P | P | CcEe | P | P | P | P | CcEe |
| T4 | P | P | N | P | Ccee | P | P | P | N | CcEE |
| T5 | P | N | P | P | CcEe | P | N | P | N | CCEE |
| T6 | P | P | P | N | CcEE | N | P | N | P | Ccee |
| T7 | P | P | P | P | CcEe | P | P | P | N | CcEE |
| T8 | P | P | N | P | Ccee | P | P | P | P | CcEe |
| T9 | P | P | P | P | CcEe | P | P | P | P | CcEe |
| T10 | P | P | P | N | CcEE | P | P | P | P | CcEe |
| T11 | P | N | P | N | CCEE | P | P | N | P | Ccee |
| T12 | N | P | N | P | Ccee | P | N | P | P | CCEe |
| T13 | P | P | P | N | CcEE | P | P | P | P | CcEe |
| T14 | P | P | P | P | CcEe | N | P | P | P | ccEe |
| T15 | P | P | P | P | CcEe | P | N | P | N | CCEE |
| T16 | P | P | N | P | Ccee | P | P | N | P | Ccee |
| T17 | P | P | P | P | CcEe | P | N | N | P | Ccee |
| T18 | P | P | P | P | CcEe | P | N | P | P | CCEe |
| T19 | P | P | N | P | Ccee | P | P | P | N | CcEE |
| T20 | P | P | P | P | CcEe | P | N | P | P | CCEe |
| T21 | P | P | N | P | Ccee | P | P | P | P | CcEe |
| T22 | P | P | P | P | CcEe | P | N | P | N | CCEE |
| T23 | P | N | P | N | CCEE | P | N | P | N | CCEE |
| T24 | P | P | P | P | CcEe | P | P | P | P | CcEe |
| T25 | P | P | N | P | Ccee | P | P | N | P | Ccee |
| T26 | P | N | P | N | CCEE | P | P | P | P | CcEe |
| T27 | P | P | N | P | Ccee | P | N | P | N | CcEe |
| T28 | P | N | N | P | CCee | P | N | P | P | CCEe |
| T29 | P | N | P | P | CCEe | N | P | P | P | ccEe |
| T30 | P | N | P | N | CCEE | P | P | P | N | CcEE |
| T31 | P | N | P | P | CCEe | P | P | P | P | CcEe |
| T32 | P | P | P | P | CcEe | P | N | N | P | Ccee |
| T33 | N | P | P | P | ccEe | P | P | P | N | CcEE |
| T34 | P | P | P | N | CcEE | P | N | P | P | CCEe |
| T35 | P | N | P | P | CCEe | P | P | P | P | CcEe |
| T36 | P | P | P | P | CcEe | P | N | P | N | CCEE |
| T37 | P | P | N | P | Ccee | P | N | P | P | CCEe |
| T38 | P | N | P | P | CCEe | P | P | P | P | CcEe |
| T39 | P | P | P | P | CcEe | P | P | P | P | CcEe |
| T40 | N | P | P | P | ccEe | P | P | N | P | Ccee |
C1 to C10: control group;
T1 to T40: thalassaemic patients.
Discussion
The incidence of alloimmunisation in transfusion-dependent patients is higher than that in other groups of patients. The reported incidence of alloimmunisation in multiply transfused patients varies from 8% to 76%, generally tending to increase with transfusion load and age11–15. As shown in previous studies, serological methods are not suitable for the correct determination of blood groups in multiply transfused patients. However, over the last decades the molecular bases of almost all of the clinically significant polymorphisms have been elucidated and it is now possible to determine blood group genotypes.
The development of high-throughput molecular methods means that it may soon be feasible to test large numbers of blood donors for all clinically important blood group polymorphisms16–20.
In this study we attempted to define a feasible, reliable and low-cost molecular method for Rh genotyping that could be used in clinical laboratories in order to decrease alloimmunisation reactions in multitransfused patients. Many polymorphisms of blood groups are due to point mutations10,11. Between the RhC and Rhc alleles, a single nucleotide substitution at position 48 in exon 1 and five base changes at positions 150, 178, 201, 203 and 307 in exon 2 of the RhCE gene have been detected. These six nucleotide substitutions lead to four amino acid changes. Among these amino acids, one at residue 103 is the outer leaflet of the plasma membrane and is, therefore, of importance. The polymorphism at nucleotide 307 leads to the Rhc phenotype and Rhe alleles differ by a single nucleotide substitution (C to G) at position 676 along exon 5 of the RHCE gene, which results in a proline to alanine substitution. Several PCR assays for the Rh alleles have been developed. Faas et al., relying on the polymorphism at nucleotide 676, introduced an allele-specific primer amplification (ASPA) method21. Tanaka et al. designed an ASO-PCR method to distinguish RhC and Rhc genes in separate runs10. A real-time PCR method has also been used for the detection of RhCE alleles by Legler et al.16. Castilho et al. found phenotype and genotype discrepancies in thalassaemia patients using a restriction fragment length polymorphism (RFLP)-PCR method19. Shayegan et al., using RFLP-PCR on 44 thalassemic patients and 20 healthy controls, reported that most discrepancies between phenotype and genotype were seen in the Rh blood group21.
In conclusion, in this study we introduced an ASO-PCR method for simultaneous genotyping of RhC/c and RhE/e alleles which could be performed in a single procedure within a short time. The advantage of this method compared to the RFLP-PCR method is that all four genes are amplified under the same conditions; it is also a fast and low-cost procedure, making it suitable for use in medical centres and by researchers.
References
- 1.McPherson RA, Pincus MR, editors. Henry’s Clinical Diagnosis and Management by Laboratory Methods. 21st ed. Elsevier; 2007. [Google Scholar]
- 2.Singer ST, Wu V, Mignacca R, et al. Alloimmunization and erythrocyte autoimmunization in transfusion dependent thalassemia patients of predominantly Asian descent. Blood. 2000;96:3369–73. [PubMed] [Google Scholar]
- 3.Castro O, Sandler SG, Houston-Yu P, Rana S. Predicting the effect of transfusing only phenotype-matched RBCs to patients with sickle cell disease: theoretical and practical implications. Transfusion. 2002;42:684–90. doi: 10.1046/j.1537-2995.2002.00126.x. [DOI] [PubMed] [Google Scholar]
- 4.Lee GR, Foerster J, Lukens J, et al., editors. Wintrobe’s Clinical Hematology. 10th ed. Lippincott Williams & Wilkins; 1999. pp. 830–2. [Google Scholar]
- 5.Ho HK, Ha SY. Alloimmunization in Hong Kong southern Chinese transfusion dependent thalassemia patients. Blood. 2001;97:3999–4000. doi: 10.1182/blood.v97.12.3999. [DOI] [PubMed] [Google Scholar]
- 6.Mouro I, Colin Y, Cherif-Zahar B, et al. Molecular genetic basis of the human Rhesus blood group system. Nature Genet. 1993;5:62–5. doi: 10.1038/ng0993-62. [DOI] [PubMed] [Google Scholar]
- 7.Spanos T, Karageorga M, Ladis V, et al. Red cell alloantibodies in patients with thalassemia. Vox Sang. 1990;58:50. doi: 10.1111/j.1423-0410.1990.tb02055.x. [DOI] [PubMed] [Google Scholar]
- 8.Reid ME, Yazdanbakhsh K. Molecular insights into blood groups and implications for blood transfusions. Curr Opin Hematol. 1998;5:93–102. doi: 10.1097/00062752-199803000-00003. [DOI] [PubMed] [Google Scholar]
- 9.Avent ND. Human erythrocyte antigen expression: its molecular bases. Br J Biomed Sci. 1997;54:16–37. [PubMed] [Google Scholar]
- 10.Tanaka M, Yamashita N, Takahashi J, et al. RHC/c genotyping based on polymorphism in the promoter region of the RHCE gene. Leg Med (Tokyo) 2001;3 :205–12. doi: 10.1016/s1344-6223(01)00035-9. [DOI] [PubMed] [Google Scholar]
- 11.Pellegrino J, Jr, Castilho L, Rios M, De Souza CA. Blood group genotyping in a population of highly diverse ancestry. J Clin Lab Anal. 2001;15:8–13. doi: 10.1002/1098-2825(2001)15:1<8::AID-JCLA2>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schonewille HL, Haak HL, Van Zijl AM. Alloimmunization after blood transfusion in patients with hematologic and oncologic disease. Transfusion. 1999;39:763–71. doi: 10.1046/j.1537-2995.1999.39070763.x. [DOI] [PubMed] [Google Scholar]
- 13.Ansari S, Voosogh P, Moshtaghian S. Assessment of frequency of alloimmunization and erythrocyte autoimmunization in transfusion dependent thalassemia patients. Acta Med Iran. 2008;46:137–40. [Google Scholar]
- 14.Wang LY, Liang DC, Liu HC, et al. Alloimmunization among patients with transfusion-dependent thalassemia in Taiwan. Transfus Med. 2006;16:200–3. doi: 10.1111/j.1365-3148.2006.00656.x. [DOI] [PubMed] [Google Scholar]
- 15.Poole J, Daniels G. Blood group antibodies and their significance in transfusion medicine. Transfus Med Rev. 2007;21:58–71. doi: 10.1016/j.tmrv.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Legler TJ, Lynen R, Mass JH, et al. Prediction of fetal Rh D and Rh CcEe phenotype from maternal plasma with real-time polymerase chain reaction. Transfus Apher Sci. 2002;27:217–23. doi: 10.1016/s1473-0502(02)00068-x. [DOI] [PubMed] [Google Scholar]
- 17.Castilho L, Rios M, Bianco C, et al. DNA-based typing of blood groups for management of multiply-transfused sickle cell disease patients. Transfusion. 2002;42 :232–8. doi: 10.1046/j.1537-2995.2002.00029.x. [DOI] [PubMed] [Google Scholar]
- 18.Castilho L, Rios M, Pellegrino J, Jr, et al. Blood group genotyping facilitates transfusion of beta-thalassemia patients. J Clin Lab Anal. 2002;16:216–20. doi: 10.1002/jcla.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Van Kim C, Mouro I, Colin C, Brossaed Y, Cartron JP. PCR based determination of Rhc and RhE status of fetuses at risk of Rhc and RhE haemolytic disease. Br J Haematol. 1994;88:193–5. doi: 10.1111/j.1365-2141.1994.tb04996.x. [DOI] [PubMed] [Google Scholar]
- 20.Faas BHW, Simsek S, Bleeker PM, et al. Rh E/e genotyping by allele-specific primer amplification. Blood. 1995;85:829–32. [PubMed] [Google Scholar]
- 21.Shayegan M, Samei S, Azarkeyvan A, et al. Molecular genotyping of blood groups in patients with thalassemia in thalassemia clinic for adults in Tehran. Blood Quarterly. 2008;6(2):107–15. [Google Scholar]