Abstract
The C4′-oxidized abasic site (C4-AP), which is produced by a variety of damaging agents has significant consequences on DNA. The lesion is highly mutagenic and reactive, resulting in interstrand cross-links. The base excision repair of DNA containing independently generated C4-AP was examined. C4-AP is incised by Ape1 ~12-fold less efficiently than an apurinic/apyrimidinic lesion. DNA polymerase β induces the β-elimination of incised C4-AP in ternary complexes, duplexes, and single stranded substrate. However, excision from a ternary complex is most rapid. In addition, the lesion inactivates the enzyme after ~7 turnovers on average by reacting with one or more lysine residues in the lyase active site. Unlike 5′-(2-phosphoryl-1,4-dioxobutane) which very efficiently irreversibly inhibits Pol β, the lesion is readily removed by strand displacement synthesis carried out by the polymerase in conjunction with flap endonuclease 1. DNA repair inhibition by C4-AP may be a partial cause of the cytotoxicity of drugs that produce this lesion.
Keywords: DNA damage, base excision repair, inhibition, oxidized abasic site
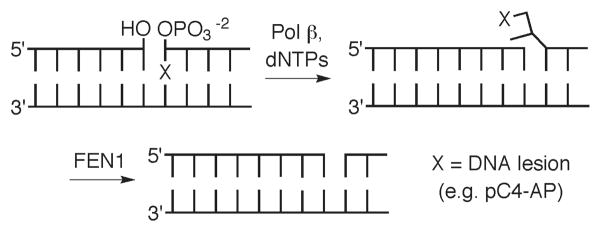
Hydrogen atom abstraction from the 2′-deoxyribose rings of nucleotides in DNA gives rise to several oxidized abasic lesions (1, 2). One of these, the C4′-oxidized abasic site (C4-AP, Chart 1) is produced by a variety of DNA damaging agents, including γ-radiolysis and antitumor antibiotics (3–5). Its frequent occurrence is attributed to the high accessibility of the C4′-hydrogen atom to diffusible species, and the relatively low bond dissociation energy of the respective carbon-hydrogen bond (6, 7). C4-AP is efficiently incised by the endonucleases in E. coli that are responsible for AP incision (8). In addition, previous studies using C4-AP produced by bleomycin indicated that the lesion is a substrate for mammalian BER enzymes, including Ape1 and Pol β (9). Ape1 incision and subsequent Pol β excision are the first two steps in BER of AP lesions (Scheme 1). However, this pathway does not efficiently excise all abasic lesions. DOB is an oxidized abasic lesion that is produced by a variety of DNA damaging agents. Recent experiments revealed that DOB very efficiently inhibits DNA polymerase β irreversibly (10). C4-AP contains the 1,4-dicarbonyl functional group that is crucial for irreversible Pol β inhibition by DOB. The structural similarity between these two lesions, the central role played by Pol β in BER, and its increased expression level in some cancer cells motivated us to investigate C4-AP repair using synthetic oligonucleotides in which the lesion was independently generated at a defined position (11–15).
Chart 1.
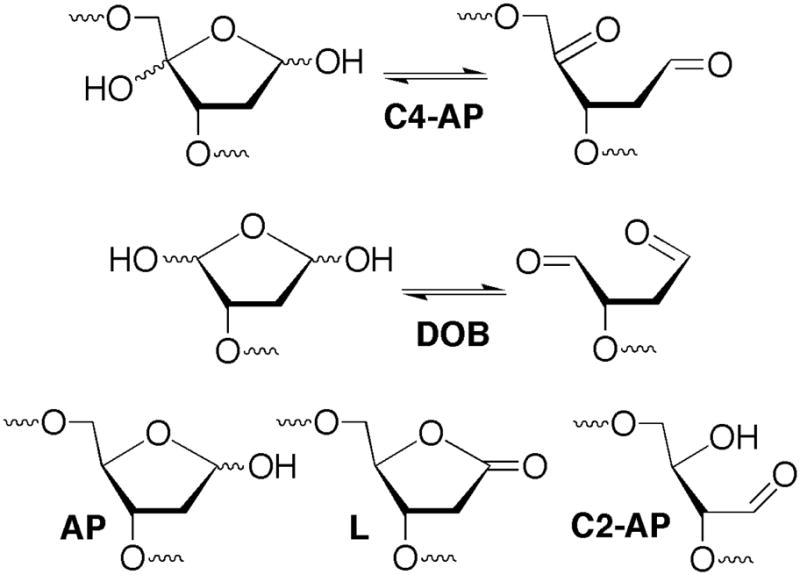
DNA lesions.
Scheme 1.
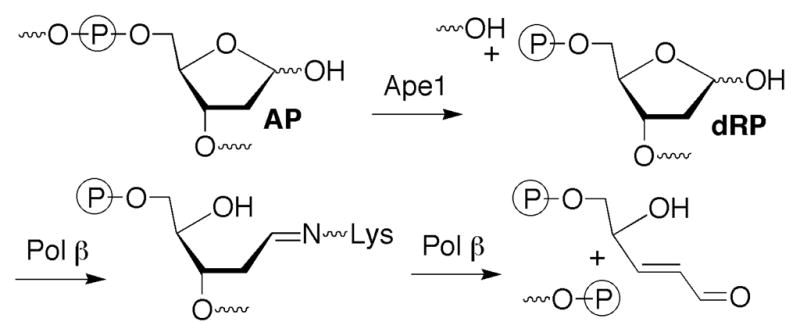
Role of Pol β in AP site repair.
Efficient C4-AP repair is likely important for protecting cells against oxidative stress. The lesion is highly mutagenic in E. coli where it exhibits the distinctive property of giving rise to 3-nucleotide deletions (16). The 1,4-dicarbonyl moiety in C4-AP is also highly reactive with nucleophiles, including those in DNA. Interstrand cross-links ascribable to C4-AP have been detected in cellular DNA and their formation was verified using synthetic oligonucleotides (17–19). The significance of these interstrand cross-links was demonstrated by their misrepair by the bacterial nucleotide excision repair proteins, UvrABC, which resulted in their transformation into double strand breaks (20). In addition, model studies and inferential experiments suggest that C4-AP reacts with the amines in proteins (21–23). This reactivity with amines could present challenges to BER proteins such as Pol β that utilize Schiff base formation to carry out their designated function. In view of this possibility we characterized C4-AP’s repair by Ape1, Pol β, and FEN1 because if left unrepaired the lesion could impart such deleterious consequences on cellular DNA.
Materials and Methods
Materials and General Methods
Oligonucleotides were prepared on an Applied Biosystems Inc. 394 DNA synthesizer. Commercially available DNA synthesis reagents, including 5′-phosphorylation agent were obtained from Glen Research Inc. Oligonucleotides containing the photolabile C4-AP precursor were synthesized as previously described (18, 24). All others were synthesized and deprotected using standard protocols. Oligonucleotides containing AP or dRP were obtained via UDG treatment of the corresponding DNA complex containing dU. T4 polynucleotide kinase, UDG, terminal deoxytransferase, typsin, and GluC were obtained from New England Biolabs. DNA Pol β and FEN1 were obtained from Trevigen. Radionuclides were obtained from Perkin Elmer. ZipTips were from Millipore. Single turnover experiments were carried out using a KinTek rapid quench instrument. Quantitative analysis of radiolabeled oligonucleotides was carried out using a Storm 840 Phosphorimager and ImageQuant 5.1 software. UPLC was carried out using an Agilent Infinity 1290 system. MALDI-TOF MS data were obtained on a Bruker AutoFlex spectrometer. MALDI-TOF mass spectrometry was performed in reflective positive mode. Laser power was varied, starting at lower values and increasing until signal was ~103–104 units for 103 shots (at 100 Hz). The detection range was varied, but was commonly set at 440–2000 m/z, and the instrument was programmed to perform a “partial sample” random walk to get appropriate signal coverage. Please note that when radiolabeling was used in the experiments described below either the 3′-terminus of the strand containing the modified nucleotide or the 5′-terminus of the flanking oligonucleotide (when appropriate) was labeled.
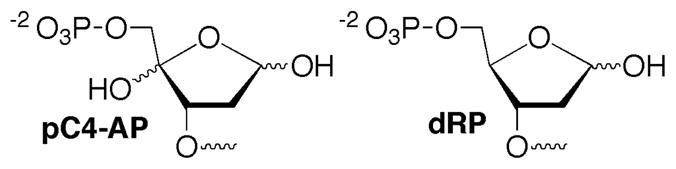
Structures pC4-AP and dRP
Preparation of Nucleic Acids Containing C4-AP, pC4-AP, and dRP Lesions
The lesions were unmasked after hybridizing (if appropriate) with the corresponding oligonucleotide(s). Hybridization was carried out at 90 °C (5 min), followed by slow cooling to room temperature in the appropriate buffer. The radiolabeled oligonucleotide was hybridized with 2 equivalents of the template strand and flanking oligonucleotide (when appropriate). The corresponding oligonucleotides or oligonucleotide complexes containing C4-AP precursor were photolyzed in clear eppendorf tubes using a Rayonet photoreactor (RPR-100) equipped with 16 lamps having an output maximum at 350 nm. The tubes were placed ~10 cm from the lamps and rotated, maintaining the samples at room temperature with ventilation. The volume of the solutions varied, but was never more than 100 μL (in a 600 μL tube). For most reactions the concentration of labeled oligonucleotide was 1 μM during photolysis. For intact C4-AP precursors, the samples were photolyzed for 20 min. For pC4-AP precursors, a 5 min photolysis was sufficient. Complexes containing dU were treated with UDG (1 U/pmol dU) in UDG reaction buffer (100 μL, 20 mM Tris-HCl, pH 8.0, 1 mM DTT, 1 mM EDTA) for 10 min at 37 °C, cooled in an ice bath and used immediately (25).
Ape1 Incision Kinetics on C4-AP
The radiolabeled duplex 5′-32P-17 (3–60 nM), was treated with Ape1 (150 pM) in a total of 10 μL at room temperature for 3 min by mixing a 2 × solution of substrate (5μL) containing 100 mM KCl and 20 mM MgCl2 with a 2 × enzyme solution (5 μL) in 50 mM Hepes- KOH (pH 7.5), 200 μg/mL BSA, 10% glycerol, 0.05% Triton X-100. The reactions were quenched with an equal volume of 90% formamide loading buffer containing EDTA (1 mM) and analyzed by 20% denaturing PAGE. All experiments were carried out in triplicate. Control experiments without enzyme were carried out under identical conditions to establish the amount of background cleavage. The percent cleavage in the reaction was determined by subtracting the amount of cleavage in the control from that in the reactions.
Steady State Kinetics of the Lyase Activity of Polymerase β
Ternary complexes (3′-32P-14 or 3′- 32P-16) were used in reactions (30 μL) containing pol β (10 nM) and varying concentrations of substrate (100–1000 nM) in HM buffer (50 mM Hepes, pH 7.4, 5 mM MgCl2). Reactions were initiated by adding a 2 × enzyme solution (15 μL) to a 2 × DNA solution (15 μL) and incubated at room temperature. At time-points of 3, 6, 10, 30 and 60 min, an aliquot (3 μL) was removed and quenched with 1 μL NaBH4 (400 mM). The quenching reaction was nearly instantaneous, but letting the NaBH4 sit 1 hour prevented further reaction with the polyacrylamide gel. Aliquots were removed from a control reaction containing all of the above ingredients except enzyme were quenched at the same time-points. Products were separated by denaturing 20% PAGE (pre-run at least 3 h, xylene cyanol run ~30 cm at 3000 V for best separation). The percent cleavage in the reaction was determined by subtracting the amount of cleavage in the control from the reaction at each time point.
Preincubation Assay
Unlabeled pC4-AP ternary complex (4 pmol, 16) was incubated with pol β (0.4 pmol) in a 24 μL reaction containing HM buffer for 15 min at room temperature. Ternary dRP complex (3′-32P-14, 20 pmol) in HM buffer was added to dilute the reaction to 40 μL, and yielding final concentrations of 100 nM 16, 5 nM pol β, and 500 nM 3′-32P-14. The reaction was incubated at room temperature and aliquots (5 μL) were removed at 5, 30, and 60 min, and then quenched with 1 μL NaBH4 (500 mM). Products were separated by 20% denaturing PAGE. Control reactions with initial incubations of unlabeled dRP ternary complex (14) or single-stranded C4-AP oligo (18) were carried out in the same manner using the same concentrations of reactants.
Single-Turnover Kinetics of Polymerase β
The respective substrates (3′-32P-2, -4, -6, -8, -10, - 12, -14, -16) were prepared as described above. Working solutions (2 ×) of 200 nM Pol β (460 μL) and 40 nM DNA substrate (400 μL) in HM buffer were prepared and stored on ice. A portion of the solution (~120 uL) was loaded in a sterile disposable 1 mL syringe, and attached to the rapid quench instrument. The buffer syringes of the instrument were loaded with HM buffer and the quench syringe was loaded with methanol. During the experimental cycle, the reaction loop was first rinsed with methanol and evacuated for 30 s by vacuum pump. Aliquots of substrate and enzyme (~15 μL each) were loaded from their respective inlets, and reacted for a defined period before methanol quench. Products released from the rapid quench (~300 μL) were immediately stabilized with 1 M NaBH4 (20 μL pre-added in a 1.6 mL eppendorf tube) and cooled to 0 °C. To each sample was added a solution (20 μL) containing NaOAc (3 M) and calf thymus DNA (200 μg/mL), and the sample was incubated at least 10 min at room temperature (to quench the NaBH4). The precipitation was completed with ethanol (1 mL), resuspended in formamide loading buffer (10 μL) and the products were separated by 20% denaturing PAGE (with the xylene cyanol running ~30 cm, products migrated ~33 cm at 3000 V to achieve optimum resolution).
The cycle was repeated for 7 time-points (usually 0.02, 0.05, 0.1, 0.2, 0.5, 1, and 2 s for ternary substrates 14 and 16, or 0.1, 0.2, 0.5, 1, 2, 5, and 10 s for other substrates). After a control reaction (no enzyme) was quenched, the loading syringes were replaced with new syringes containing fresh solutions. The 7 time-points were performed in triplicate, and the fraction cleaved was plotted against time. The resulting graph was fit to the exponential equation Faction cleaved = (Max. fraction cleaved)(1 − e− kobs*time).
Identification of the pC4-AP DNA Polymerase β Adduct
For the GluC digestion, a solution (100 μL) of pol β (5 μM) was incubated with or without ternary complex 16 (25 μM) in HM buffer at room temperature for 20 min. The solution was diluted with 47.5 μL GluC buffer (50 mM Tris-HCl, pH 8.0, 0.5 mM Glu-Glu), and 2.5 μL of resuspended GluC (10 mM), and then incubated at 37 °C for 4 h. For the trypsin digestion, a solution (100 μL) of pol β (5 μM) was incubated with or without ternary complex 16 (25 μM) in HM buffer at room temperature for 20 min. The solution was diluted with 47.5 μL trypsin buffer (50 mM Tris-HCl, pH 8.0, 20 mM CaCl2), and 2.5 μL resuspended TPCK-treated trypsin (10 mM), and then incubated at 37 °C for 4 h. Crude digestions were either separated by UPLC or directly spotted on a MALDI plate.
MALDI-MS Preparation for Crude Pol β Digests
The crude samples were concentrated and desalted by ZipTip (Milipore). The tip was wetted with 50% MeCN (10 μL), and equilibrated with 0.1% TFA (3 × 10 μL). The sample (100 μL) was adjusted to 0.1% TFA (with 2.5% stock solution) and bound to the column by pipeting up and down ~10 times, allowing the full solution to pass through the tip. The tip was then washed with 0.1% TFA (3 × 10 μL) and eluted with 2 μL CHCA matrix (10 mg/mL α-cyano-4-hydroxycinnamic acid in 50% MeCN, 0.1% TFA) directly on to the MALDI plate.
UPLC Analysis of Pol β Digests
Digested samples (100 μL) were diluted with 250 μL water and passed by syringe (1 mL) through a filter (0.22 μm, 4 mm diameter). The samples were further purified by 10k microcon (Milipore), washing with 200 μL water and keeping the filtrate, which was concentrated to a dry residue. The products were resuspended in 20 μL water and injected on a UPLC as 2 μL (analytical) or 6 μL (for collection) samples. The column (Zorbax RRHD SB-C18, 2.1 × 100 mm, 1.8 μm) was heated to 40 °C and equilibrated with 97% A (0.1% TFA in water) and 3% B (0.09% TFA in acetonitrile). The total flow rate was 0.5 mL/min. Both solvents were 0.22 μm filtered and made fresh every 2–3 days. The 3% B was maintained for 2 min after injection, after which it was increased to 6% B at 10 min, followed by a faster gradient that reached 50% B at 25 min. The column was purged with 90% B from 26–30 min, and then re-equilibrated at 3% B. The 214 nm wavelength was monitored for products. A background run consisting of a water injection was subtracted to remove the gradual increase in signal associated with increasing solvent B. Individual peaks were collected from a short splint of tubing off the detector. These peaks were collected in eppendorf tubes and concentrated to dryness in a speedvac before being resuspended in CHCA matrix (2 μL). The products were spotted on a MALDI plate for further characterization.
Strand Displacement Synthesis of pC4-AP with and without FEN1
Primer-template ternary complex 5′-32P-16 was assembled as described above. Reactions (20 μL) consisting of pol β (2 nM) and 5′-32P-16 (200 nM) with HM buffer, dTTP (50 μM), dGTP (50 μM), and 0.1 mg/mL BSA were initiated by mixing a 2 × solution of DNA and dNTPs (10 μL) with a 2 × solution of enzyme (10 μL) and incubated at 37 °C in the presence or absence of FEN1 (10 nM). Aliquots (3 μL) were removed at 2, 5, 10, and 15 min, and quenched with 5 μL formamide loading buffer. Ternary complex 3′-32P-16 (200 nM) was reacted (20 μL) with pol β (2 nM) in HM buffer, along with dTTP (50 μM), dGTP (50 μM), and 0.1 mg/mL BSA at 37 °C only in the presence of FEN1 (10 nM). Aliquots (3 μL) were removed at 2, 5, 10, and 15 min, and quenched with 1 μL NaBH4 (400 μM). After sitting for 1 h, 10 μL formamide loading buffer was added and the products were separated by 20% denaturing PAGE. A no-enzyme control time-course was carried out side by side.
Results
Preparation of DNA substrates containing C4-AP, pC4-AP, or dRP
Oligonucleotides containing 5′-phosphorylated lesions at their termini (e.g. dRP, pC4-AP) were prepared via solid phase synthesis using commercially available 5′-phosphorylation reagent (Chart 2). dRP sites were generated by reaction with UDG in 5′-phosphorylated oligonucleotides containing dU (25). Oligonucleotides containing dU were synthesized via standard oligonucleotide synthesis conditions and reagents. Oligonucleotides containing C4-AP were obtained via photolysis of the respective biopolymer containing NV on an as needed basis (Scheme 2) (18, 24). Duplexes and ternary complexes containing C4-AP were prepared by hybridizing the oligonucleotide containing NV prior to photolysis as previously described (18).
Chart 2.
Modified DNA molecules used in this study.
Scheme 2.
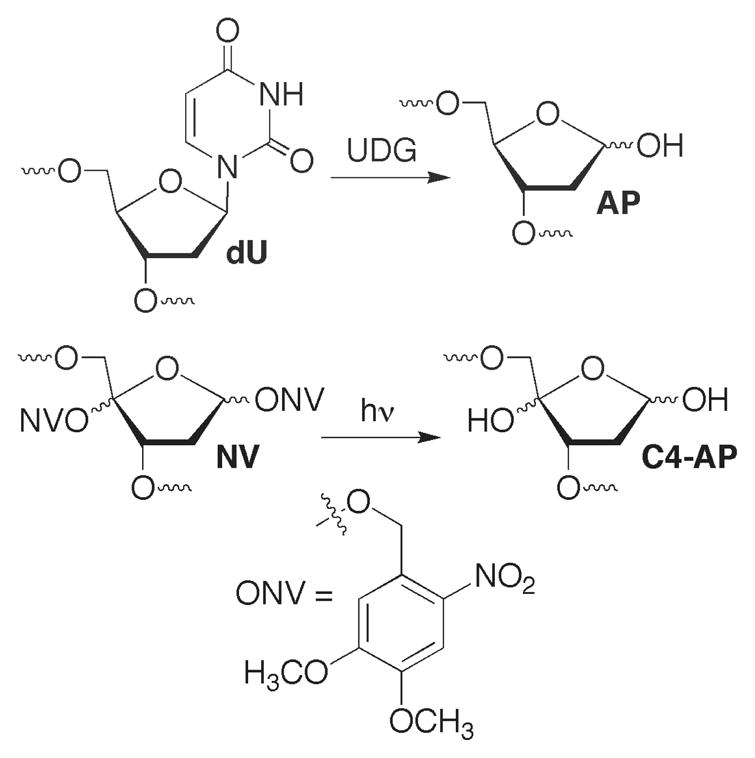
Generation of abasic lesions from synthetic precursors.
Ape1 incision of C4-AP
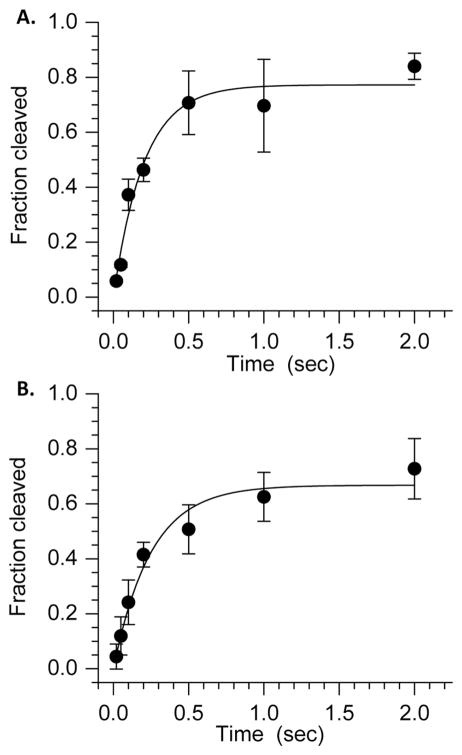
Incision of C4-AP (5′-32P-17) by Ape1 (representative data: Figure 1) was studied under steady-state conditions as previously described for the related oxidized abasic site, C2-AP (26). The kinetic parameters (Km = 28.0 ± 9.0 nM, Vmax = 2.4 ± 0.9 nM·min−1 at 150 pM Ape1, kcat = 16.0 ± 6.0 min−1) are the average of 3 experiments, each carried out using 3 replicates. The kinetic parameters are very similar to those measured for Ape1 incision of C2-AP but are less efficient than Xth incision of C4-AP in a duplex of similar sequence (8, 26). Oligonucleotides and complexes containing 5′-phosphates (e.g. pC4-AP, dRP) were obtained by incorporating the phosphate group using commercially available 5′-phosphorylation reagent.
Figure 1.
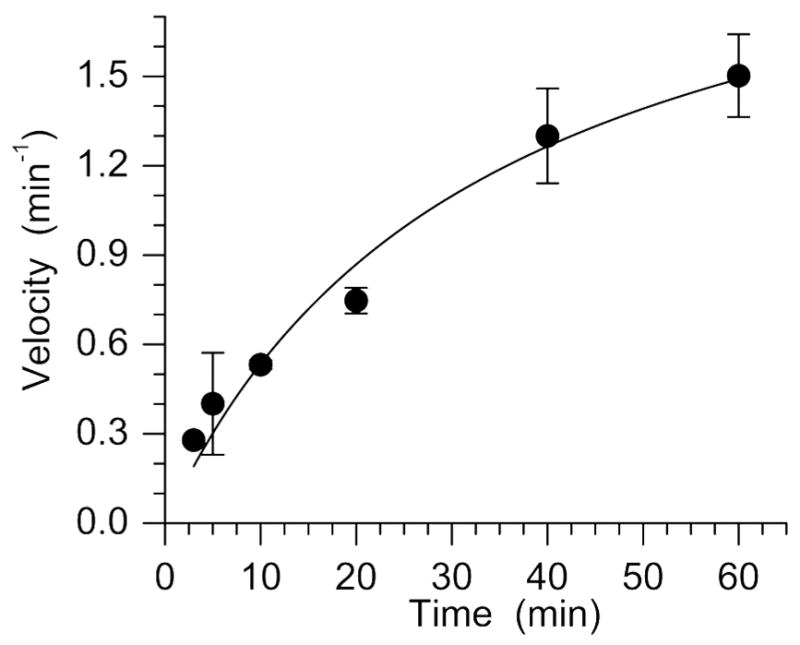
Representative plot of Ape1 incision of C4-AP in 5′-32P-17 (Km = 33.4 ± 10.0 nM, Vmax = 2.3 ± 0.3 min−1).
Single turnover kinetic analysis of the Pol β lyase reaction with dRP and pC4-AP
The lyase reaction of dRP is known to be the rate-determining step in BER of AP lesions (25, 27). Although qualitative experiments show that pC4-AP is a substrate for Pol β lyase activity, we took a more comprehensive, quantitative approach due to the lesion’s structural similarity to DOB (9, 10). pC4-AP and DOB are both 1,4-dicarbonyl containing molecules that readily react with amines. Given that DOB efficiently irreversibly inhibits DNA Pol β, we investigated whether pC4-AP also inactivates Pol β. Irreversible inhibition of Pol β by C4-AP would bias steady-state measurements (vide infra). Hence, pC4-AP excision in various structural situations was determined under single-turnover conditions (Table 1). For comparison, the rate constants for dRP excision were measured in otherwise identical substrates. dRP was excised from a ternary complex (14) less than twice as fast as pC4-AP (16, Figure 2). In addition, both lesions were excised by Pol β more rapidly from a ternary complex than when incorporated in single stranded (2, 4) or either duplex substrates. The double stranded substrates included those in which the 5′-phosphorylated lesions were present in a recessed region (6, 8) or an overhang (10, 12).
Table 1.
Rate constants for pC4-AP and dRP excision by Pol β under single turnover conditions.
| KObs (S−1) X: | pC4-APa | dRPa |
|---|---|---|
|
14, 16 |
3.6 ± 0.9 (6) | 6.2 ± 2.2 (5) |
|
6, 8 |
1.8 ± 0.9 (3) | 0.9 ± 0.6 (4) |
|
10, 12 |
0.4 ± 0.1 (3) | 0.4 ± 0.2 (2) |
|
2, 4 |
0.8 ± 0.2 (3) | 2.8 ± 1.2 (3) |
Number of replicates in parentheses. Each replicate consisted of 3 independent samples.
Figure 2.
Representative Pol β excision of ternary complexes containing (A) dRP (3′-32P-14) and (B) pC4-AP (3′-32P-16) under single turnover conditions.
Steady-state analysis of the Pol β lyase reaction with pC4-AP and dRP
Kinetic constants were extracted by measuring initial velocities in reactions containing various concentrations of ternary complex containing dRP (3′-32P-14) with Pol β (10 nM) (See Supporting Information). The kinetic parameters measured (Km = 82 nM, kcat = 2.1 min−1) were similar to those reported previously for excising dRP in a different sequence (25). The observed Km is ~5 times smaller and the kcat is about one-half as fast previously reported in a different sequence. Attempts to determine Michaelis-Menten parameters for excision of pC4-AP (3′-32P-16) were unsuccessful (data not shown). Unlike time course studies of dRP excision which showed reaction progress over the course of an hour, reactions between pC4-AP and Pol β ceased before 30 min regardless of the ratio of substrate to enzyme. The number of turnovers as a function of time at concentrations of 3′-32P-16 from 0.1 – 1.0 μM converged at between 6 and 7 even when the substrate was in 100-fold excess of enzyme (Figure 3). In contrast, more than 40 turnovers were observed when dRP was present in sufficiently large excess (See Supporting Information).
Figure 3.
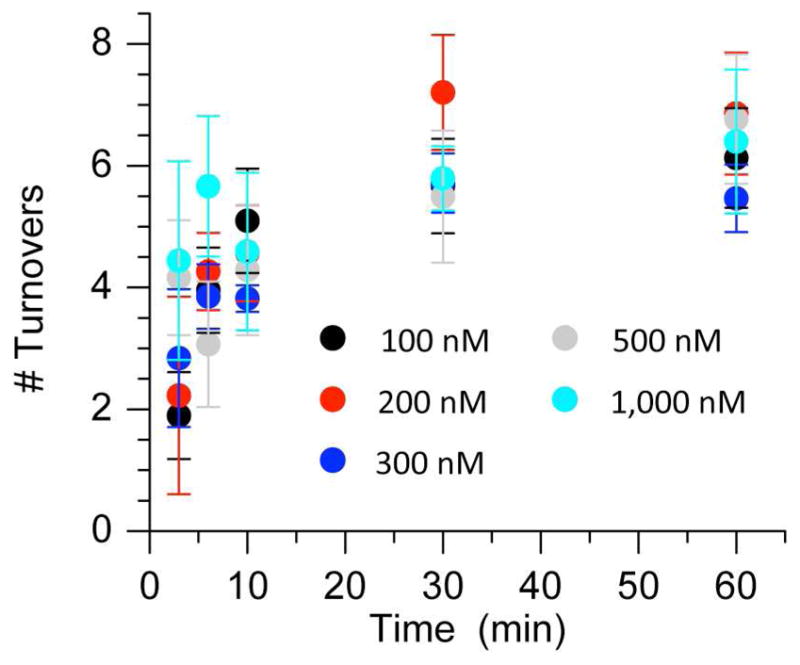
Number of Pol β (10 nM) turnovers during excision of various concentrations (100 – 1,000 nM) of pC4-AP (3′-32P-16) as a function of time.
Pol β inhibition by pC4-AP
The premature ceasing of Pol β lyase activity and lack of dependence of turnover number on pC4-AP concentration (Figure 3) suggested that the oxidized abasic lesion irreversibly inhibited the enzyme. Further support for pC4-AP inactivation of Pol β was garnered by preincubating the enzyme (10 nM) with 16 (100 nM) for 15 min prior to measuring the excision of dRP from 3′-32P-14 (500 nM) (Figure 4). dRP excision was significantly reduced compared to samples in which the enzyme was preincubated with either unlabeled 14 (100 nM) or a single stranded oligonucleotide containing C4-AP (18, 100 nM). However, unlike experiments with DOB we were unable to kinetically characterize irreversible inhibition of Pol β by pC4-AP. This was attributed to the large concentration of pC4-AP required to observe this effect. When the concentration of pC4-AP was less than 10-fold higher than Pol β, the moderate pC4-AP turnover masked its inhibition of the dRPase activity.
Figure 4.
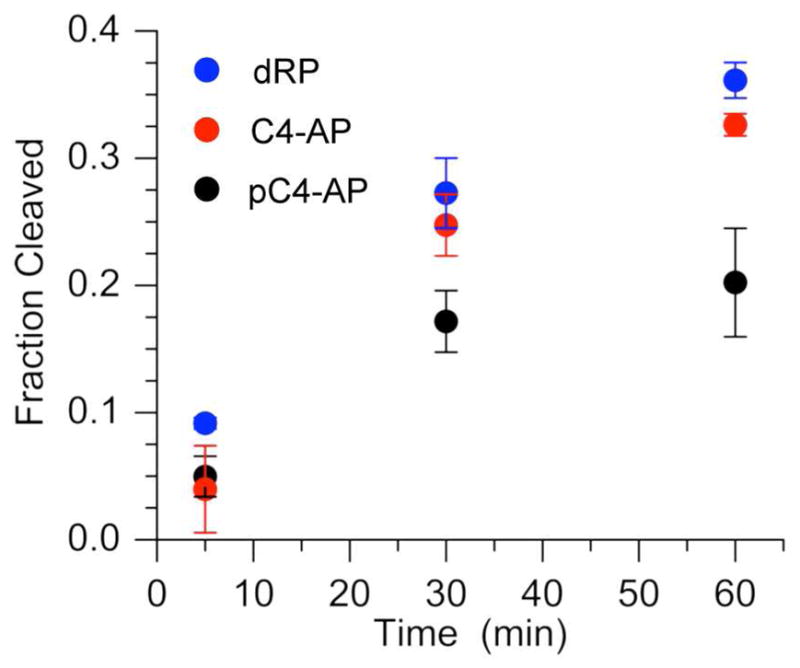
Pol β (10 nM) excision of dRP in 3′-32P-14 (500 nM) after preincubation of the enzyme with 100 nM of dRP (14), pC4-AP (16), or single stranded DNA containing C4-AP (18).
Despite the absence of kinetic data for the irreversible Pol β inhibition by pC4-AP, qualitative support for this process provided the impetus to examine the enzyme for modification. We anticipated that one or more of the nucleophilic lysine residues in the Pol β lyase active site would be modified. Direct evidence for Pol β modification was obtained by comparing GluC protein digests with or without prior incubation with pC4-AP (16). GluC hydrolyzes peptides at the C-terminus of glutamic acid residues. Several peptide fragments, including an ion that corresponds to amino acids 76–85 (m/z = 1275.9) were identified via MALDI-TOF MS analysis of the crude Pol β digest. This ion was not detected in the GluC digest carried out following incubation of Pol β with 5 equivalents of 16. Instead, a fragment corresponding to the 76–85 amino acid peptide fragment + 78 a.m.u. (m/z = 1353.8) was observed (See Supporting Information). This fragment contains two lysine residues but only Lys84 is accessible to the lyase binding site (28). Adduct (19) is consistent with Pol β structure and the type of product previously observed in reactions between C4-AP and primary amines (22, 23). More inferential evidence for protein modification was obtained by comparing trypsin digests of Pol β to trypsin digests of the enzyme after pC4-AP incubation (16) (See Supporting Information). Trypsin hydrolyzes the C- termini of lysine residues. The digestion of Pol β in the absence of pC4AP yielded a peak in the UPLC chromatogram whose molecular ion (m/z = 993.4) corresponded to residues 73–81. This peptide results from cleavage at Lys72, the residue shown to form a Schiff base with dRP, and Lys81 (29). The intensity of this peptide was greatly reduced in the digest of enzyme that was incubated with pC4-AP (16). This is consistent with modification of Lys72, which prevents hydrolysis by trypsin at this site. However, a corresponding longer peptide containing a sugar fragment was not observed.
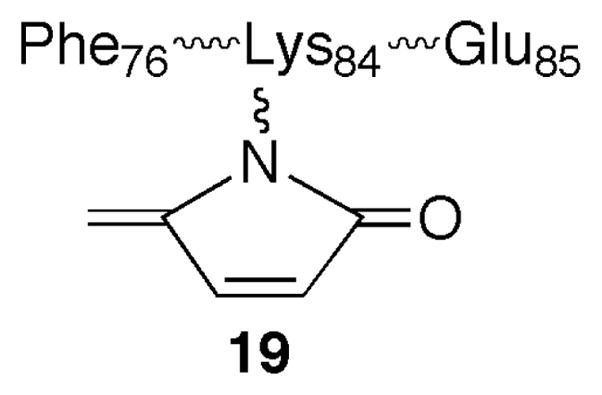
Structure 19
pC4-AP removal by long patch BER
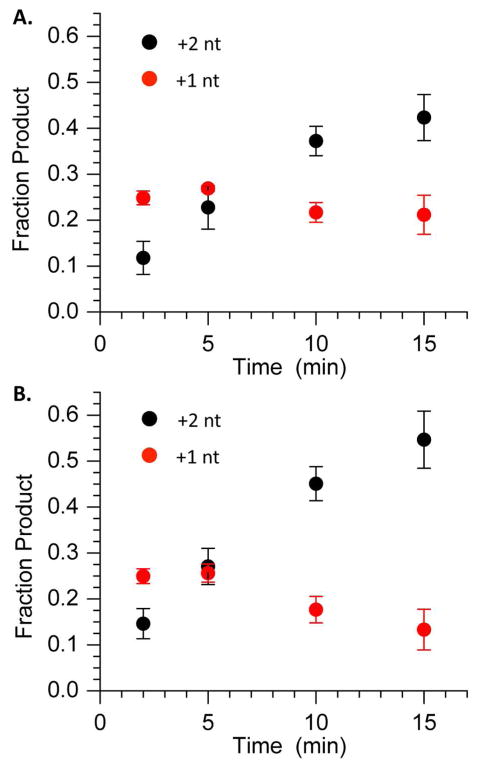
Pol β contributes to long patch BER by carrying out strand displacement synthesis. During this process, Pol β extends the 3′-terminus at the point of incision in the DNA and in the process displaces the incised lesion and one or more of the 3′-adjacent nucleotides (Scheme 3). The displaced segment of DNA (“flap”) is cleaved by FEN1 producing a nicked duplex that is poised for completion of repair by DNA ligase. Extension of 5′-32P-16 by Pol β (2 nM) was monitored in the presence of dTTP (50 μM) and dGTP (50 μM) in the absence and presence of FEN1 (10 nM) (Figure 5). One and two nucleotide extension products were observed. Although dGTP was present in order to enable addition of a third nucleotide, none of this product was observed. In addition, the amount of +1 nucleotide product reaches a maximum in ~5 minutes and then begins to decrease, whereas the amount +2 nucleotide product continues to increase throughout the course of the reaction. The overall amount of extension was insignificantly affected by FEN1, but the fraction of 2-nucleotide extension product increased modestly at the expense of single nucleotide addition in the presence of the endonuclease. The amount of 2-nucleotide cleavage product observed from 3′-32P-16, which results from FEN1 cleavage of the flap produced as a result of strand displacement, subjected to the same reaction conditions was comparable to the yield of 2-nucleotide extension product (Figure 6).
Scheme 3.
Long patch BER by Pol β and FEN1.
Figure 5.
Extension of 5′-32P-16 (200 nM) by Pol β (2 nM) in the presence of dTTP (50 μM) and dGTP (50 μM) in (A) absence or (B) presence of FEN1 (10 nM).
Figure 6.
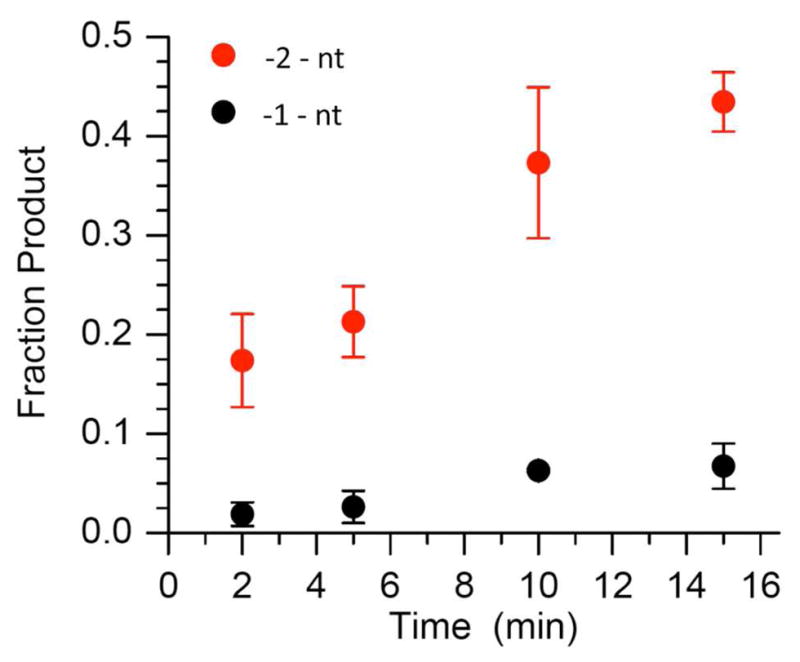
Cleavage of 3′-32P-16 (200 nM) by FEN1 (10 nM) in the presence of Pol β (2 nM), dTTP (50 μM), and dGTP (50 μM).
Discussion
AP sites are produced spontaneously via depurination, as a result of oxidative stress, and as intermediates in BER of damaged nucleotides. They are efficiently repaired by the BER pathway in which the first two steps involve Ape1 incision, followed by removal of the 5′-(2′-deoxyribose)phosphate (dRP) fragment by the lyase activity of Pol β. Oxidized abasic sites such as L and C2-AP are substrates for Ape1 but pose obstacles to Pol β that require alternative repair processes (26, 30, 31). Previous qualitative studies showed that the oxidized abasic lesion, C4-AP was a substrate for Ape1 and Pol β, two principle enzymes responsible for the BER of AP lesions (9). However, kinetic parameters were not reported. Given the structural similarity (Chart 1) between C4-AP and the potent irreversible inhibitor of Pol β, DOB, we took advantage of our ability to chemically synthesize oligonucleotides containing C4-AP or pC4-AP to reinvestigate its repair (10, 18).
C4-AP is incised ~12-fold less efficiently than AP by Ape1 and is a slightly poorer substrate than 2-deoxyribonolactone (L) (31). However, due to an ~2-fold lower Km and 2-fold higher kcat, Ape1 incises C4-AP ~4-fold more efficiently than C2-AP (26). Incision by Ape1 is a necessary first step for BER of an abasic site. These data indicate that C4-AP is a viable candidate for this pathway. Excision of dRP or pC4-AP by Pol β is the next step in short patch BER. Single turnover kinetic experiments (Table 1) indicate that the polymerase’s lyase activity removes pC4-AP almost as rapidly as it excises dRP. Excision of either lesion when it is part of a single strand or at the 5′-terminus of a recessed or overhanging duplex was considerably less efficient (Table 1). From these data alone it is unclear whether Pol β plays a significant role as a housekeeping enzyme to clean up DNA termini of single and double strand breaks in preparation for religation (32, 33). However, steady-state experiments reveal that pC4-AP excision by Pol β is different than that of dRP in a ternary complex of the type formed upon Ape1 hydrolysis. pC4-AP excision under multiple turnover conditions showed that this lesion irreversibly inhibits Pol β. Previous studies, which were carried out with relatively large concentrations of Pol β did not uncover this mode of action (9). Interestingly, our data show that on average each Pol β molecule turns over 6–7 times before being inactivated. The earlier study was conducted using less than 6-fold excess substrate. Hence, previous investigators would not have detected this inhibition under their reaction conditions. Pol β turns over a greater number of times when excising pC4-AP than when removing DOB (2–4 turnovers) from the 5′-terminus of DNA. A previous study on DOB indicated that the 1,4-dicarbonyl component of the lesion was required for efficient irreversible inhibition (34). We attribute the less efficient Pol β inhibition by pC4-AP than DOB to the reduced electrophilicity of the former. pC4-AP contains the requisite 1,4-dicarbonyl functionality, but the ketone group at the C4 position is less reactive than the aldehyde in DOB at the comparable position.
Although quantitative analysis of irreversible Pol β inhibition by pC4-AP was not carried out, GluC digestion and subsequent MALDI-TOF MS analysis provided direct evidence for Lys modification in the peptide fragment containing residues 76–86. Two Lys residues are present in this fragment, but the X-ray crystal structure of Pol β complexed with a stable dRP analogue indicates that only Lys84 is present in the lyase active site (28). The modified fragment could result from liberated sugar fragment and/or trapping by pC4-AP and subsequent decomposition. Although Lys84 may play an active role in the lyase reaction, Lys72 is believed to be responsible for the majority of Schiff base formation en route to dRP excision (35–37). We did not detect a modified peptide fragment by MS containing this residue but inferential support for the Lys72 modification during inactivation of Pol β by pC4-AP was evident from analysis of the trypsin digest. Disappearance of the amino acid 73–81 fragment is consistent with modification of Lys72 (29).
The less efficient inactivation of Pol β by pC4-AP compared to DOB is also evident in long patch repair of the incised lesion (Scheme 3). Whereas Pol β achieved insignificant amounts of strand displacement synthesis when acting on DNA containing DOB, Pol β readily extended the strand flanking pC4-AP by two nucleotides (34). Pol β mediated displacement of pC4-AP and the 3′-adjacent nucleotide was accompanied by comparable amounts of 2-nucleotoide excision by FEN1 in the strand containing the lesion. The level of long patch BER of pC4-AP was similar to that observed for C2-AP (26). We attribute the more effective long patch BER of pC4-AP than that of DOB to the less efficient Pol β inactivation by the former, which enables this process to proceed.
Conclusions
The C4′-oxidized abasic site is a commonly produced lesion, which exhibits effects on base excision repair that are intermediate between those of structurally related AP and DOB lesions. The lesion is not repaired as efficiently as an AP lesion. Moreover, like the DOB lesion it irreversibly inhibits Pol β, a critical enzyme involved in BER, albeit less efficiently. The less efficient inactivation of Pol β also enables this enzyme in conjunction with FEN1 to excise C4-AP through long patch BER. These data suggest that C4-AP is not as toxic as DOB but that its ability to inhibit BER may contribute to the cytotoxicity of drugs that produce it.
Supplementary Material
Abbreviations
- AP
apurinic/apyrimidinic site
- L
2-deoxyribonolactone
- C2-AP
C2′-oxidized abasic site
- C4-AP
C4′-oxidized abasic site
- pC4-AP
5′-phosphorylated C4′-oxidized abasic site
- BER
base excision repair
- DOB
5′-(2-phosphoryl-1,4-dioxobutane)
- dsb
double strand break
- dRP
5′-deoxyribose phosphate
- Pol β
DNA polymerase β
- ssb
single strand break
- FEN1
flap endonuclease 1
- Ape1
apurinic endonuclease 1
Footnotes
We are grateful for support of this research by the National Institute of General Medical Science (GM- 063028).
Supporting Information Available. Plots of Pol β excision of dRP, GluC and trypsin digests of Pol β following reaction with pC4-AP. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pitie M, Pratviel G. Activation of DNA Carbon‚ Hydrogen Bonds by Metal Complexes. Chem Rev. 2010;110:1018–1059. doi: 10.1021/cr900247m. [DOI] [PubMed] [Google Scholar]
- 2.Dedon PC. The Chemical Toxicology of 2-Deoxyribose Oxidation in DNA. Chem Res Toxicol. 2008;21:206–219. doi: 10.1021/tx700283c. [DOI] [PubMed] [Google Scholar]
- 3.von Sonntag C. The Chemical Basis of Radiation Biology. Taylor & Francis; London: 1987. [Google Scholar]
- 4.Rabow LE, Stubbe J, Kozarich JW. Identification and Quantitation of the Lesion Accompanying Base Release in Bleomycin-Mediated DNA Degradation. J Am Chem Soc. 1990;112:3196–3203. [Google Scholar]
- 5.Dhar S, Kodama T, Greenberg MM. Selective Detection and Quantification of Oxidized Abasic Lesions in DNA. J Am Chem Soc. 2007;129:8702–8703. doi: 10.1021/ja073014e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balasubramanian B, Pogozelski WK, Tullius TD. DNA Strand Breaking by the Hydroxyl Radical Is Governed by the Accessible Surface Areas of the Hydrogen Atoms of the DNA Backbone. Proc Nat Acad Sci USA. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li MJ, Liu L, Wei K, Fu Y, Guo QX. Significant Effects of Phosphorylation on Relative Stabilities of DNA and RNA Sugar Radicals: Remarkably High Susceptibility of H-2′ Abstraction in RNA. J Phys Chem B. 2006;110:13582–13589. doi: 10.1021/jp060331j. [DOI] [PubMed] [Google Scholar]
- 8.Greenberg MM, Weledji YN, Kim J, Bales BC. Repair of Oxidized Abasic Sites by Exonuclease III, Endonuclease IV, and Endonuclease III. Biochemistry. 2004;43:8178–8183. doi: 10.1021/bi0496236. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Kim EY, Demple B. Excision of C-4′-Oxidized Deoxyribose Lesions from Double-Stranded DNA by Human Apurinic/Apyrimidinic Endonuclease (Ape1 Protein) and DNA Polymerase B. J Biol Chem. 1998;273:28837–28844. doi: 10.1074/jbc.273.44.28837. [DOI] [PubMed] [Google Scholar]
- 10.Guan L, Greenberg MM. Irreversible Inhibition of DNA Polymerase Beta by an Oxidized Abasic Lesion. J Am Chem Soc. 2010;132:5004–5005. doi: 10.1021/ja101372c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto Y, Kim K. Excision of Deoxyribose Phosphate Residues by DNA Polymerase · During DNA Repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 12.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2. ASM Press; Washington, D.C: 2006. [Google Scholar]
- 13.Husain I, Arteaga CL, Srivastava DK, Wilson SH. DNA Polymerase Beta Expression Differences in Selected Human Tumors and Cell Lines. Carcinogenesis. 1999;20:1049–1054. doi: 10.1093/carcin/20.6.1049. [DOI] [PubMed] [Google Scholar]
- 14.Albertella MR, Lau A, O’Connor MJ. The Overexpression of Specialized DNA Polymerases in Cancer. DNA Repair. 2005;4:583–593. doi: 10.1016/j.dnarep.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Starcevic D, Dalal S, Sweasy JB. Is There a Link between DNA Polymerase β and Cancer? Cell Cycle. 2004;3:998–1001. [PubMed] [Google Scholar]
- 16.Kroeger KM, Kim J, Goodman MF, Greenberg MM. Effects of the C4′-Oxidized Abasic Site on Replication in Escherichia Coli. An Unusually Large Deletion Is Induced by a Small Lesion. Biochemistry. 2004;43:13621–13627. doi: 10.1021/bi048337r. [DOI] [PubMed] [Google Scholar]
- 17.Regulus P, Duroux B, Bayle PA, Favier A, Cadet J, Ravanat JL. Oxidation of the Sugar Moiety of DNA by Ionizing Radiation or Bleomycin Could Induce the Formation of a Cluster DNA Lesion. Proc Nat Acad Sci USA. 2007;104:14032–14037. doi: 10.1073/pnas.0706044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sczepanski JT, Jacobs AC, Greenberg MM. Self-Promoted DNA Interstrand Cross-Link Formation by an Abasic Site. J Am Chem Soc. 2008;130:9646–9647. doi: 10.1021/ja8030642. [DOI] [PubMed] [Google Scholar]
- 19.Sczepanski JT, Jacobs AC, Majumdar A, Greenberg MM. Scope and Mechanism of Interstrand Cross-Link Formation by the C4′-Oxidized Abasic Site. J Am Chem Soc. 2009;131:11132–11139. doi: 10.1021/ja903404v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sczepanski JT, Jacobs AC, Van Houten B, Greenberg MM. Double Strand Break Formation During Nucleotide Excision Repair of a DNA Interstrand Cross-Link. Biochemistry. 2009;48:7565–7567. doi: 10.1021/bi901006b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett RAO, Swerdlow PS, Povirk LF. Spontaneous Cleavage of Bleomycin-Induced Abasic Sites in Chromatin and Their Mutagenicity in Mammalian Shuttle Vectors. Biochemistry. 1993;32:3188–3195. doi: 10.1021/bi00063a034. [DOI] [PubMed] [Google Scholar]
- 22.Aso M, Usui K, Fukuda M, Kakihara Y, Goromaru T, Suemune H. Photochemical Generation of C4′-Oxidized Abasic Site Containing Oligodeoxynucleotide and Its Efficient Amine Modification. Org Lett. 2006;8:3183–3186. doi: 10.1021/ol060987v. [DOI] [PubMed] [Google Scholar]
- 23.Usui K, Aso M, Fukuda M, Suemune H. Photochemical Generation of Oligodeoxynucleotide Containing a C42-Oxidized Abasic Site and Its Efficient Amine Modification: Dependence on Structure and Microenvironment. J Org Chem. 2008;73:241–248. doi: 10.1021/jo702080r. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Gil JM, Greenberg MM. Synthesis and Characterization of Oligonucleotides Containing the C4′-Oxidized Abasic Site Produced by Bleomycin and Other DNA Damaging Agents. Angew Chem Int Ed. 2003;42:5882–5885. doi: 10.1002/anie.200352102. [DOI] [PubMed] [Google Scholar]
- 25.Prasad R, Beard WA, Strauss PS, Wilson SH. Human DNA Polymerase β Deoxyribose Phosphate Lyase. J Biol Chem. 1998;273:15263–15270. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 26.Wong RS, Sczepanski JT, Greenberg MM. Excision of a Lyase-Resistant Oxidized Abasic Lesion from DNA. Chem Res Toxicol. 2010;23:766–770. doi: 10.1021/tx9003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava DK, Vande Berg BJ, Prasad R, Molina JT, Beard WA, Tomkinson AE, Wilson SH. Mammalian Abasic Site Base Excision Repair. Identification of the Reaction Sequence and Rate-Determining Steps. J Biol Chem. 1998;273:21203–21209. doi: 10.1074/jbc.273.33.21203. [DOI] [PubMed] [Google Scholar]
- 28.Prasad R, Batra VK, Yang XP, Krahn JM, Pedersen LC, Beard WA, Wilson SH. Structural Insight into the DNA Polymerase β Deoxyribose Phosphatase Lyase Mechanism. DNA Repair. 2005;4:1347–1357. doi: 10.1016/j.dnarep.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 29.Deterding LJ, Prasad R, Mullen GP, Wilson SH, Tomer KB. Mapping of the 5′-2-Deoxyribose-5-Phosphate Lyase Active Site in DNA Polymerase Beta by Mass Spectrometry. J Biol Chem. 2000;275:10463–10471. doi: 10.1074/jbc.275.14.10463. [DOI] [PubMed] [Google Scholar]
- 30.DeMott MS, Beyret E, Wong D, Bales BC, Hwang JT, Greenberg MM, Demple B. Covalent Trapping of Human DNA Polymerase β by the Oxidative DNA Lesion 2-Deoxyribonolactone. J Biol Chem. 2002;277:7637–7640. doi: 10.1074/jbc.C100577200. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y-J, DeMottt MS, Hwang JT, Greenberg MM, Demple B. Action of Human Apurinic Endonuclease (Ape1) on C1′-Oxidized Deoxyribose Damage in DNA. DNA Repair. 2003;2:175–185. doi: 10.1016/s1568-7864(02)00194-5. [DOI] [PubMed] [Google Scholar]
- 32.Pascucci B, Maga G, Hubscher U, Bjoras M, Seeberg E, Hickson ID, Villani G, Giordano C, Cellai L, Dogliotti E. Reconstitution of the Base Excision Repair Pathway for 7,8-Dihydro-8-Oxoguanine with Purified Human Proteins. Nucleic Acids Res. 2002;30:2124–2130. doi: 10.1093/nar/30.10.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP Endonuclease-Independent DNA Base Excision Repair in Human Cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Guan L, Bebenek K, Kunkel TA, Greenberg MM. Inhibition of Short Patch and Long Patch Base Excision Repair by an Oxidized Abasic Site. Biochemistry. 2010 doi: 10.1021/bi101533a. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prasad R, Beard WA, Chyan JY, Maciejewski MW, Mullen GP, Wilson SH. Functional Analysis of the Amino-Terminal 8-kDa Domain of DNA Polymerase β as Revealed by Site-Directed Mutagenesis. J Biol Chem. 1998;273:11121–11126. doi: 10.1074/jbc.273.18.11121. [DOI] [PubMed] [Google Scholar]
- 36.Feng J-A, Crasto CJ, Matsumoto Y. Deoxyribose Phosphate Excision by the N- Terminal Domain of the Polymerase β: The Mechanism Revisited. Biochemistry. 1998;37:9605–9611. doi: 10.1021/bi9808619. [DOI] [PubMed] [Google Scholar]
- 37.Matsumoto Y, Kim K, Katz DS, Feng J-a. Catalytic Center of DNA Polymerase β for Excision of Deoxyribose Phosphate Groups. Biochemistry. 1998;37:6456–6464. doi: 10.1021/bi9727545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.