Abstract
Animal pigment patterns are important for a range of functions, including camouflage and communication. Repeating pigment patterns, such as stripes, bars and spots have been of particular interest to developmental and theoretical biologists, but the genetic basis of natural variation in such patterns is largely unexplored. In this study, we identify a difference in a periodic pigment pattern among juvenile threespine sticklebacks (Gasterosteus aculeatus) from different environments. Freshwater sticklebacks exhibit prominent vertical bars that visually break up the body shape, but sticklebacks from marine populations do not. We hypothesize that these distinct pigment patterns are tuned to provide crypsis in different habitats. This phenotypic difference is widespread and appears in most of the freshwater populations that we sampled. We used quantitative trait locus (QTL) mapping in freshwater–marine F2 hybrids to elucidate the genetic architecture underlying divergence in this pigmentation pattern. We identified two QTL that were significantly associated with variation in barring. Interestingly, these QTL were associated with two distinct aspects of the pigment pattern: melanophore number and overall pigment level. We compared the QTL locations with positions of known pigment candidate genes in the stickleback genome. We also identified two major QTL for juvenile body size, providing new insights into the genetic basis of juvenile growth rates in natural populations. In summary, although there is a growing literature describing simple genetic bases for adaptive coloration differences, this study emphasizes that pigment patterns can also possess a more complex genetic architecture.
Keywords: pigment pattern, camouflage, QTL, fish
Introduction
Appropriate coloration has a critical role in an animal's survival and reproduction. Pigment patterns are essential for both intraspecific communication, such as species recognition, mate choice and dominance, and interspecific communication with predators or prey, including warning coloration, mimicry and camouflage (Protas and Patel, 2008). The genetics of pigmentation in vertebrates has been extensively studied for over a century in a range of laboratory model systems, yielding tremendous insights into the molecular and developmental mechanisms that generate variant colors and patterns (Barsh, 1996; Steingrimsson et al., 2006). In recent decades, advances in molecular genetic technology have expanded the repertoire of animal models of pigmentation, facilitating the discovery of genetic contributions to variation in pigmentation resulting from both artificial (Kelsh, 2004; Steingrimsson et al., 2006; Candille et al., 2007; Cook et al., 2008; Fang et al., 2009; Eizirik et al., 2010) and natural selection (Gratten et al., 2007; Steiner et al., 2007; Protas and Patel, 2008; Anderson et al., 2009; Roberts et al., 2009; Wittkopp and Beldade, 2009).
The evolution of vertebrate coloration occurs in various forms, including overall color change, alterations in the location, size or color of distinct patches or localized regions on the body, as well as changes in the nature of repeating patterns of spots, bars, stripes or ‘saddles' across the body. Much of what is known about the genetic basis for pigment evolution in vertebrates relates to overall or localized color changes. There are numerous examples of single genes that have large effects on coloration in vertebrates (reviewed in the studies by Hoekstra (2006) and Protas and Patel (2008)). In particular, one gene, the melanocortin-1 receptor (Mc1r) is known to underlie color change in multiple taxa (Hoekstra, 2006; Gross et al., 2009; Uy et al., 2009). Despite the large number of examples of single locus effects on pigment evolution, multilocus contributions to natural color variation have also been identified (Steiner et al., 2007; Tripathi et al., 2008, 2009).
The evolutionary genetics of repeating pigment patterns has been substantially less explored than other types of pigment changes. Periodic patterns have long been of interest to a range of biologists, not only because of their important roles in animal ecology and evolution (Caro, 2005; Stevens and Merilaita, 2009a) but also because of the intricate processes contributing to their development and maintenance (Widelitz et al., 2006; Kondo et al., 2009). Several developmental mechanisms are hypothesized to underlie pattern formation, including the formation of early prepatterns and autonomous generation of patterns by reaction-diffusion processes (Widelitz et al., 2006; Kondo et al., 2009; Werner et al., 2010). Altering patterns created by these types of mechanisms may require modifications to different genetic and developmental pathways than those required to simply change color. Furthermore, the evolution of more complex patterns might also require a more complex genetic architecture than has been observed for overall color change. An appreciation of the genetic basis of pigment patterns in various systems is required to gain insights into these issues.
Recent studies have started to explore the genetic basis of variation in repeating patterns in both mammals (Candille et al., 2007; Kerns et al., 2007; Eizirik et al., 2010) and fish (Kelsh, 2004; Parichy, 2006; Boulding et al., 2008; Kondo et al., 2009; Roberts et al., 2009). Zebrafish (Danio rerio) mutants have proved to be particularly useful for elucidating genes and developmental mechanisms involved in periodic pattern formation. Pigmentation takes on added complexity in fish and other poikilotherms, because these animals have three or more classes of pigment cells compared with a single pigment cell type in birds and mammals (Kelsh, 2004; Parichy, 2006). Despite this developmental complexity, laboratory studies have shown that single gene mutations can dramatically alter the zebrafish pigment pattern: eliminating stripes, generating broken or wavy stripes and even changing stripes into spots (Kelsh, 2004; Parichy, 2006; Kondo et al., 2009).
However, the question still remains as to whether natural variations in repeating patterns and mutants identified in the laboratory share a similar genetic basis. In some cases, the genetic basis may not be conserved. For example, a zebrafish mutant (panther) has a phenotype similar to a closely related species (Danio albolineatus); both have lost the characteristic horizontal stripes of D. rerio. A clever use of complementation crosses revealed that these similar phenotypes are caused by changes in the same pathway, but further experiments suggested that the mutations are not in the same gene (Quigley et al., 2005). In addition, the full array of genetic changes that underlie the evolutionary difference in pigment patterns between D. rerio and D. albolineatus has not yet been explored (Quigley et al., 2005). Whether pigment pattern evolution is widely mediated by single or multilocus changes is not yet understood, but two recent studies have provided evidence for both simple (Roberts et al., 2009) and complex (Boulding et al., 2008) contributions to alterations in repeating patterns.
The threespine stickleback (Gasterosteus aculeatus) offers an appealing model system in which to investigate the genetic basis for evolutionary changes in pigment patterns. Sticklebacks experienced a recent adaptive radiation throughout the Northern Hemisphere, wherein migratory (anadromous) marine ancestors invaded new freshwater habitats that were formed after the last ice age, ∼12 000 years ago (Bell and Foster, 1994). These nascent freshwater residents adapted to their new habitats, and now exhibit striking differences in behavior, physiology and morphology, including pigmentation (Bell and Foster, 1994). In addition to this extensive natural phenotypic diversity, an expanding array of genetic and genomic tools is available for sticklebacks (Kingsley and Peichel, 2007). These tools have facilitated recent efforts at identifying the genetic contributions—ranging from genetic architecture to causal genetic mutation—to multiple adaptive traits, including both morphology (Peichel et al., 2001; Cresko et al., 2004; Colosimo et al., 2005; Kimmel et al., 2005; Miller et al., 2007; Albert et al., 2008; Kitano et al., 2009; Chan et al., 2010) and behavior (Kitano et al., 2009).
In this study, we describe a phenotypic difference in the pigment patterns of juvenile threespine sticklebacks from migratory marine vs freshwater-resident populations. We hypothesize that these patterns are important for cryptic coloration in different habitats. We used quantitative trait locus (QTL) mapping to dissect the genetic architecture of these divergent color patterns in freshwater–marine F2 hybrids. We also examined the QTL underlying body size, another trait that is likely to influence the fitness of juveniles.
Materials and methods
Animals
Wild-caught sticklebacks from Hotel Lake and Little Campbell River were collected, and intrapopulation crosses were generated in the laboratory using in vitro fertilization. The young were reared in net breeders for 2 weeks and then released into 110-l tanks. Juveniles from different clutches were collected for analysis at 1–2-week intervals through 8 weeks after hatching. Fish were housed in water containing 0.35% salt (Instant Ocean, Aquarium Systems, Mentor, OH, USA) at 16 h light–8 h dark at 16 °C. Juveniles were fed with brine shrimp nauplii twice a day. All animals were treated in accordance with the Institutional Animal Care and Use Committees at the Fred Hutchinson Cancer Research Center (protocol no. 1575).
To survey the generality of juvenile pigment patterns, adult marine and freshwater sticklebacks were collected from multiple locations throughout the Pacific Northwest and crossed and reared as described above. Populations sampled in British Columbia, Canada included: Garden Bay Lake (GB), Hotel Lake (HL), Little Campbell River Marine (LCM), Little Campbell River Stream Resident (LCS), Paxton Lake Benthic (PB), Paxton Lake Limnetic (PL), North Lake (NL), Salmon River Marine (SR) and Secret Cove Marine (SC). Populations sampled in Washington State, USA, included: Conner Creek (CC), Humptulips Pond (HP), San Juan Island Marine (SJ), Lake Washington (LW), a stream near Clearbrook Road in Sumas (CB) and Willapa Bay Marine (WB). Permits for fish collections were obtained from the Washington State Department of Fish and Wildlife (permit numbers 07-047 and 08-038) and from the British Columbia Ministry of Environment (permit numbers NA/SU06-21454, NA/SU07-31839 and NA/SU08-42033).
Juveniles were photographed between 6 and 8 weeks after hatching. We performed a quantitative measurement of the degree of barring (see below) in juveniles from four freshwater (CC, HL, LW and NL) and four marine (LCM, SJ, SR and WB) populations. For other freshwater and marine populations, we categorized phenotypes as either barred or unbarred, but did not perform quantitative measurements.
Quantification of pigmentation
Fish were anesthetized using 0.025% MS-222 and either photographed immediately (for iridophore quantification) or fixed in ethanol (for melanophore quantification). For iridophore quantification, fish were visualized using incident light and a stereomicroscope (Nikon SMZ1500, Nikon Inc, Melville, NY, USA), and photographed using a scope-mounted CoolPix 4500 camera (Nikon Inc.). For melanophore quantification, fish were immersed in formalin for a minimum of 1 week to reduce iridophore pigmentation, and then visualized using a compound microscope (Nikon Eclipse 80i) and incident light. Photographs were taken using a Nikon DS-Fi1 camera and Nikon NIS Elements software.
Degree of barring (parentals, population survey, F2s)
To quantify barring, we developed a method to measure variation in pigmentation along the flank of each fish. This method (the gray-value coefficient of variation (CV)) quantifies variance in the darkness of pixels (gray-value intensity) from a photograph of the flank (see Supplementary Information, Supplementary Figure S1). As the same portion of each fish is sampled, this method is less biased than other methods that directly target the darkest and lightest portions of the pattern and thereby overestimate the barring of unbarred fish. The pattern of bars is quite variable and can range even in a single fish from true bars to a checkered appearance; hence, we chose to focus on an area close to the midline where the pattern was most consistent. Photographs were cropped to analyze the same portion of each fish: 10 body segments (or myomeres) wide, aligned anteriorly with the start of the dorsal fin and the equivalent of two body segments high, centered on the midline. Images were converted to gray scale. The average gray value (0–255) for each one-pixel column in the image was determined using the ‘Plot Profile' command in ImageJ Software (National Institutes of Health, Bethesda, MD, USA; http://rsbweb.nih.gov/ij/index.html). These values were then ‘smoothed' by averaging the gray value in each pixel column over 50 adjacent pixels. A straight line was fit to each profile (Supplementary Information, Supplementary Figure S1), and the squared residual from each point to this line was calculated. We then summed these squared residuals, divided by the total number of pixels minus one, and took the square root to obtain a ‘standard deviation' of the gray value. To compensate for variable background light levels across photographs, this s.d. was then divided by the average gray value of the image and multiplied by 100 to yield a ‘gray-value CV' across the flank.
Midline melanophore counts within bars (parentals, F2s)
The cropped images described above were used to quantify the relative number of melanophores per segment within the light and dark bars of Hotel Lake fish (‘midline' dark or light bar melanophores; it should be noted that the bars span multiple body segments; see Figure 1). Little Campbell fish are not barred; therefore, melanophores cannot be assigned to dark or light bars and are simply referred to as ‘midline melanophores' (see Table 1). For Little Campbell fish, we counted melanophores in a single segment because more extensive melanophore counts revealed that unbarred fish do not show substantial variation in melanophore number across the flank (average CV of 12 fish=6%). For F2s, we counted melanophores in a single segment within the light bars of barred fish or in an arbitrarily chosen segment of unbarred fish.
Figure 1.
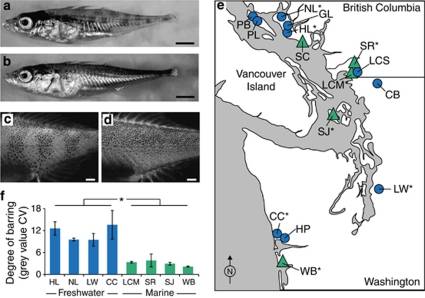
Pigment patterns of juvenile freshwater and marine sticklebacks. Photographs of 6-week-old sticklebacks from (a) a freshwater population from Hotel Lake (HL) and (b) a marine population from Little Campbell River (LCM). Scale bar=2 mm. (c, d) Photomicrographs of melanophore distribution across the flank of fish from (panel c) HL and (panel d) LCM after immersion in formalin to reduce iridophore pigmentation. Scale bar=0.5 mm. (e) A map showing populations surveyed for juvenile pigment pattern phenotype. Marine populations are shown as green triangles, and freshwater populations are shown as blue circles. An asterisk indicates that the population was used for quantitative measurement of the degree of barring. CB, Clearbrook; CC, Conner Creek; GL, Garden Bay Lake; HL, Hotel Lake; HP, Humptulips Pond; LCM, Little Campbell River Marine; LCS, Little Campbell River Stream Resident; LW, Lake Washington; PB, Paxton Lake Benthic; PL, Paxton Lake Limnetic; NL, North Lake; SC, Secret Cove Marine; SJ, San Juan Island Marine; SR, Salmon River Marine; WB, Willapa Bay Marine. (f) Quantification of the degree of barring (mean gray-value CV±s.e.m.) in selected freshwater and marine populations. Asterisk indicates significant difference between fish from marine and freshwater populations (see the ‘Results' section).
Table 1. Phenotypic comparison between parental lines.
| Hotel Lake | Little Campbell Marine | Statistics | |
|---|---|---|---|
| Degree of barring | 12.2±1.1 | 3.3±0.2 | F(1, 22)=62.7; P<0.0001 |
| Dorsal melanophores | 62±5 | 60±3 | F(1, 22)=0.098; P=0.76 |
| Ventral melanophores | 27±3 | 31±2 | F(1, 22)=1.5; P=0.24 |
| Midline dark bar melanophores | 44±3 | NA | NA |
| Midline light bar melanophores | 22±2 | NA | NA |
| Midline melanophores | NA | 42±2 | NA |
| Iridophores | 9.7±1.2 | 26.1±2.7 | F(1, 10)=38.9; P<0.0001 |
Abbreviations: HL, Hotel Lake; LCM, Lake Campbell Marine; NA, not applicable.
The values represent mean±s.e.m. Sample size=12 for both populations, except for iridophore measurements, which were n=7 for HL and n=5 for LCM. Dorsal and ventral melanophores=number of melanophores per segment in the dorsal or ventral regions. Midline dark bar and light bar melanophores=number of melanophores per segment near the midline in the dark and light bars of HL fish. Midline melanophores=number of melanophores per segment near the midline in LCM fish. It should be noted that lake and marine fish have distinct subsets of midline melanophore phenotypes because HL fish have melanophores within the dark or light bars, but melanophores in LCM fish cannot be similarly classified because these fish are not barred.
Dorsal and ventral melanophore counts (parentals, F2s)
We also counted dorsal and ventral melanophores within a larger region of the flank. Similar to measurements of barring, we sampled the same relative portion of each fish, rather than an area of a fixed size. We counted all melanophores per segment in a region centered underneath the dorsal fin. Each image was cropped to 4 segments wide and 4 segment-widths high, centered on the midline. Melanophores in the dorsal and ventral portions of the body were counted independently. To avoid simply detecting the presence of bars, we used the count from the segment with the highest number of melanophores in the QTL analysis.
Degree of melanization (F2s)
The melanin level in melanophores across the flank was rated categorically by eye from 1 to 3. Quantitative measures of individual melanophores were confounded by extensive differences in melanosome aggregation or dispersal among melanophores even from a single fish. We were able to compensate for this variation using this rating system; this measure represents melanin level, regardless of melanosome dispersal. For all fish, we assessed the degree of melanization based on the appearance of the darkest melanophores on the flank. Images were rated twice and the scores averaged.
Iridophores (parentals, F2s)
The percentage of the flank covered by iridophores was quantified from photographs taken of live fish. We cropped images to include the ventral portion of the fish bordered at the top by the midline, excluding the highly reflective skin covering the gut. We analyzed 6 body segments, centered on the anal vent. We used the threshold function in ImageJ to select the reflective guanine platelets. We determined the number of pixels covered by reflective platelets and then divided this value by the total number of pixels to obtain the percentage of the ventral flank covered by iridophores.
QTL analysis
A female from Hotel Lake and an anadromous marine male from the Little Campbell River were used as founders of an F2 intercross. A single F1 female and male sibling were then crossed twice to generate two full-sib clutches (89 and 93 individuals) that were reared in two separate tanks and collected at 6 weeks after hatching. Of these, 176 fish were subjected to whole-genome genotyping using custom-designed Golden Gate single-nucleotide polymorphism (SNP) arrays (Illumina, San Diego, CA, USA). Genotypes were determined using GenomeStudio Software (Illumina). JoinMap 3.0 (Van Ooijen and Voorrips, 2001) was used to construct a linkage map representing all 21 stickleback linkage groups (LGs) using 1 microsatellite marker (Stn194, Peichel et al., 2001) and 279 SNP markers (274 fully polymorphic and 5 polymorphic in only 1 F1 parent). SNP markers are listed in Supplementary Information (Supplementary Table S1) and are accessible on the NCBI website (http://www.ncbi.nlm.nih.gov/projects/SNP/). QTL analysis was performed in R/qtl (Broman and Sen, 2009). Both body size (standard length) and clutch (1 or 2) were included as covariates in the analysis of pigmentation traits. Body size mapped strongly to two LGs (LG17 and LG18; see below). Markers at the peaks of these body-size QTL were also used as covariates for all pigment QTL scans because pigmentation was strongly correlated with overall body size (Table 2), and the residual linkage on LG17 and LG18 due to this correlation was not completely eliminated by inclusion of body size as a covariate. Qualitatively similar results for pigment QTL were obtained when no covariates were included in the analysis (data not shown). Likelihood of odds (LOD) significance thresholds for each trait were determined by permutation testing (1000 permutations). Significant QTL were above the genome-wide threshold (α=0.05). For traits with significant QTL, we also looked for the presence of suggestive QTL that exceeded linkage-group permutation thresholds. The ‘fitqtl' function was used to calculate the percentage variance explained by the peak marker for each QTL.
Table 2. Pairwise correlations between phenotypes used for QTL analysis.
| Standard length | Iridophores | Degree of barring | Degree of melanization | Dorsal melanophores | Ventral melanophores | |
|---|---|---|---|---|---|---|
| Iridophores | 0.660* P<0.00001 | |||||
| Degree of barring | −0.157 P<0.05 | −0.304* P<0.0001 | ||||
| Degree of melanization | −0.229* P<0.01 | −0.212* P<0.01 | 0.488* P<0.00001 | |||
| Dorsal melanophores | 0.400* P<0.00001 | 0.430* P<0.00001 | −0.057 P>0.05 | 0.030 P>0.05 | ||
| Ventral melanophores | 0.292* P<0.0001 | 0.332* P<0.00001 | −0.037 P>0.05 | 0.205* P<0.01 | 0.386* P<0.00001 | |
| Midline light bar melanophores | 0.621* P<0.00001 | 0.482* P<0.00001 | −0.436* P<0.00001 | −0.147 P>0.05 | 0.282 P<0.01 | 0.421* P<0.00001 |
Abbreviation: QTL, quantitative trait locus.
Pearson's correlation coefficient and associated P-value is shown for each pair of traits. Sample size is 176 for all comparisons. Correlations significant after Bonferroni's correction are indicated with an asterisk.
Kit ligand allele genotyping
A previous study identified 2 Kit ligand alleles on LG19 that are associated with variation in melanophore pigmentation across several marine and freshwater stickleback populations (Miller et al., 2007). We genotyped wild-caught Hotel Lake and Little Campbell individuals (n=10 each, including the founders of the F2 intercross) using a microsatellite marker that is diagnostic for these two variant alleles, amplifying a 174-bp product from most populations and a 168-bp product from freshwater populations that have reduced pigmentation (Miller et al., 2007).
Location of pigment candidate genes
A list of pigment candidate genes was compiled from various sources; a complete list of gene names, functions and references is presented as Supplementary Information (Supplementary Table S2). The location of genes in the stickleback genome (Broad S1, February 2006) was determined by using either zebrafish or mouse orthologs in tBLASTn searches or, for annotated genes, by searching the Ensembl genome browser (http://www.ensembl.org/Gasterosteus_aculeatus/Info/Index) for the gene name. Positions on unassembled portions of the genome are listed in reference to the composite ‘ChrUn' contained in the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway?db=gasAcu1). We have not attempted to assign orthology to genes that have multiple paralogs (for example, Kita and Kitb are both called Kit). Ensembl gene predictions are listed in Supplementary Information (Supplementary Table S2) when available.
Results
Variation in juvenile pigment patterns
Observations of laboratory-reared juvenile sticklebacks from a freshwater and an anadromous marine population revealed that these fish display highly divergent pigment patterns during early life. In particular, anadromous marine sticklebacks from the Little Campbell River are silvery in appearance, because of a high density of iridophores containing reflective guanine crystals (Table 1, Figure 1b). In contrast, freshwater-resident sticklebacks from Hotel Lake exhibit prominent vertical bars across their flank and have significantly fewer iridophores (Table 1, Figure 1a). The vertical bars consist of an uneven distribution of black-pigmented melanophores, with twice as many melanophores contained within the dark bars compared with the light bars (Table 1, Figure 1c). The melanophores within the dark bars also appear more darkly pigmented than those within the light bars (Figure 1c). In some Hotel Lake fish, the bars are broken midway between the dorsum and the midline, yielding a ‘checkered' or ‘zebra-striped' appearance (Figure 1a). The pattern of melanophores in Little Campbell marine fish is obscured by the overlying iridophores (Figure 1b), but an even distribution of melanophores is revealed after immersion in formalin to dissolve guanine crystals (Figure 1d). Both populations have more melanophores on the dorsal portion of the flank compared with the ventral regions (Table 1).
The differences in the degree of barring are apparent in fish beginning ∼2 weeks after hatching (>10 mm in length) and continue through several months of life. As growth continues, Hotel Lake fish do develop substantial iridophore pigmentation, which somewhat conceals the persistent underlying barred melanophore pattern after approximately 3–4 months of age (30 mm in length; Supplementary Information, Supplementary Figure S2).
To explore the generality of this pigment pattern difference among independently derived stickleback populations, we characterized the pigmentation phenotypes of juveniles from several locations throughout the Pacific Northwest (Figures 1e and f). At 6 or 8 weeks after hatching, most freshwater lake and stream populations were highly barred. We found one exception to this general trend: the ‘benthic' morph from Paxton Lake did not show strong vertical barring and exhibited a general paucity of melanophores relative to other freshwater populations (data not shown). In contrast, none of the marine populations that we sampled were barred. Quantification of this difference by measuring the gray-value CV (see the ‘Materials and methods' section) across the flank in juveniles obtained from four freshwater and four marine populations revealed a highly statistically significant difference between habitats (Figure 1f; freshwater, n=24; marine, n=27; F(1, 49)=71; P<0.0001).
QTL analysis
To gain insights into the genetic basis for divergent pigmentation, we performed QTL mapping in a Hotel Lake by Little Campbell marine F2 intercross. We genotyped 176 F2 intercross progeny with 279 SNP markers and 1 microsatellite marker. The resulting linkage map consisted of 21 LGs representing the 21 stickleback chromosomes. The total map length was 1217 cM and the average distance between markers was 4.6 c.
QTL analysis of body size
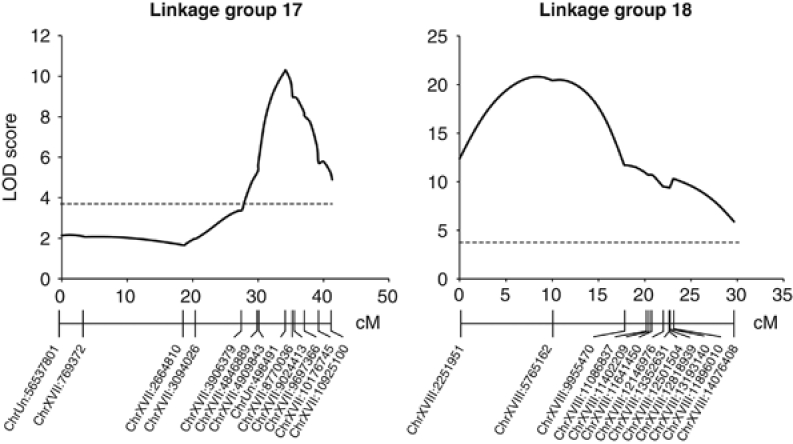
Body size mapped to two LGs: LG17 and LG18. Taken together, these QTL explained >60% of variance in body size (Figure 2, Table 3). In both cases, Hotel Lake alleles were associated with larger body size (Table 3). The peak of the LG17 QTL is at a marker on an unassembled scaffold (scaffold 27). In the genetic map from our cross, this marker fell between markers from two large contiguous scaffolds (namely scaffolds 25 and 18) on chromosome 17 in the current genome assembly. Examination of the 1.5-LOD support intervals flanking the LG17 body-size QTL places it in a region encompassing 9.9 Mb on chromosome 17, between markers chrXVII:4909843 and chrXVII:9697366 (including the unassembled sequence on scaffold 27). The QTL region on chromosome 18 corresponds to 7.2 Mb, between markers chrXVIII:2251951 and chrXVIII:9955470.
Figure 2.
QTL associated with body size. LOD score as a function of distance in cM is shown for LG17 and LG18. Genome-wide significance at α<0.05 is indicated with a dashed line.
Table 3. Location and effect of QTL.
| Trait | LOD | LG | cM | Marker near peak | PVE | Lake | Heterozygous | Marine |
|---|---|---|---|---|---|---|---|---|
| Degree of barring | 4.55 | 1 | 33 | chrI:3310077 | 6.6 | 9.8±0.6 | 8.9±0.4 | 7.1±0.4 |
| Degree of barring | 10.82 | 6 | 55 | chrVI:15780594 | 20.0 | 10.7±0.5 | 8.8±0.4 | 6.4±0.3 |
| Degree of barring | 2.82(s) | 11 | 0 | chrXI:457909 | 6.9 | 10.0±0.5 | 8.6±0.4 | 7.1±0.5 |
| Degree of melanization | 5.88 | 1 | 39 | chrI:4816374 | 11.7 | 2.4±0.1 | 2.0±0.1 | 1.8±0.1 |
| Midline light bar melanophores | 5.14 | 6 | 56 | chrVI:15780594 | 6.3 | 20.9±1.0 | 22.4±0.9 | 26±1.1 |
| Dorsal melanophores | 5.77 | 7 | 22 | chrVII:1728753 | 11.6 | 41.6±0.9 | 36.7±0.8 | 35.6±1.0 |
| Ventral melanophores | 4.25 | 1 | 67 | chrI:21909727 | 8.9 | 17.6±0.6 | 14.6±0.4 | 16±0.6 |
| Standard length | 10.29 | 17 | 34 | chrUn:498491 | 23.6 | 18.9±2.2 | 19.1±2.2 | 14.9±1.2 |
| Standard length | 20.80 | 18 | 8 | chrXVIII:5765162 | 41.1 | 19.8±1.9 | 19.6±1.9 | 16.2±1.7 |
Abbreviations: LG, linkage group; LOD, likelihood of odds; PVE, percentage variance explained; QTL, quantitative trait locus.
For all suggestive and significant QTL detected, the LOD score, LG, marker name, marker position (cM) and PVE is shown. The mean±s.e.m. of the phenotype of F2 progeny for the different genotypic classes is also reported. (s) indicates a suggestive QTL.
QTL analysis of pigment patterns
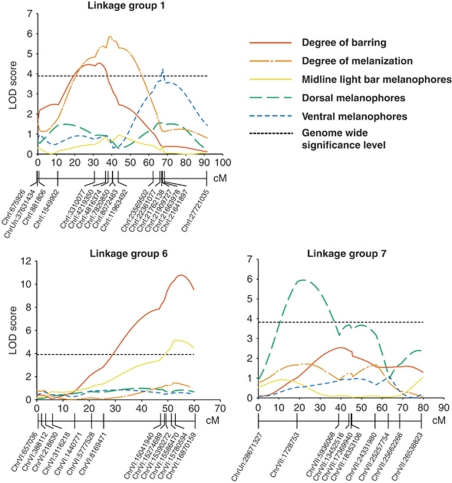
We identified significant QTL for several pigment pattern phenotypes (Figure 3, Table 3). Two significant QTL on LG1 and LG6 explained 26.6% of the variance in the degree of barring (gray-value CV). There was an additional suggestive QTL on LG11 that met the chromosome-wide significance threshold. Hotel Lake alleles were associated with higher amounts of barring at all three QTL, and alleles at all of these QTL seemed to have predominantly additive effects (Figure 4, Table 3). The 1.5-LOD support intervals suggest that the QTL on LG6 is contained within 1.5 Mb of genomic sequence, between markers chrVI:15390272 and chrVI:16870159; the LG1 QTL corresponds to 6 Mb of sequence, between markers chrI:1549902 and chrI:8072483.
Figure 3.
QTL associated with pigmentation traits. LOD score as a function of distance in cM is shown for the three linkage groups that contain significant QTL. Genome-wide significance at α<0.05 is indicated with a thin dashed line. Colored lines represent different traits: red, degree of barring; orange, degree of melanization; yellow, midline light bar melanophore number; green, dorsal melanophore number; blue, ventral melanophore number.
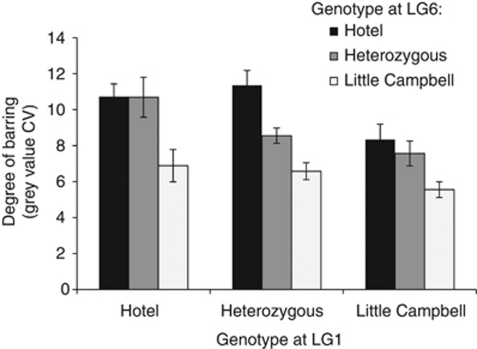
Figure 4.
Effect of LG1 and LG6 QTL on the degree of barring. The mean gray-value CV (±s.e.m) is shown for each two-locus combination of genotypes at markers chrI:3310077 and chrVI:15780594.
Two features of the barred pattern that were linked in parental lines segregated independently in F2s. In Hotel Lake within-population crosses, both the number of melanophores and the degree of melanization within melanophores varied across the light and dark bars (Table 1, Figure 1c). Some F2s appeared similar to Hotel Lake fish; the light bars contained fewer melanophores that were also less melanized than those in the dark bars (Supplementary Information, Supplementary Figure S1a). In such fish, melanophore number and melanization were always coincident; that is, regions of the flank that contained low numbers of melanophores were always less melanized. However, in other barred fish, the melanophore number was relatively constant across the flank, and only the degree of melanization of individual melanophores varied in the light and dark bars (Supplementary Information, Supplementary Figure S1b).
Our measurement of the degree of barring, the gray-value CV, is a composite measure that should capture variation in both the number of melanophores and the overall level of melanin across the flank. We quantified two additional phenotypes to determine whether these distinct pattern features could be genetically dissociated. We counted the number of midline melanophores in the light bars to represent variation in melanophore number, and we rated the overall degree of melanization to assess maximal pigment level. Midline light bar melanophore counts were negatively correlated with the degree of barring: fish that were more extensively barred had lower melanophore counts (Table 2). The degree of barring was also positively correlated with the maximal amount of melanization (Table 2). This correlation suggests that barring is driven in part by an overall increase in the melanization of melanophores in the dark bars of barred fish relative to unbarred fish.
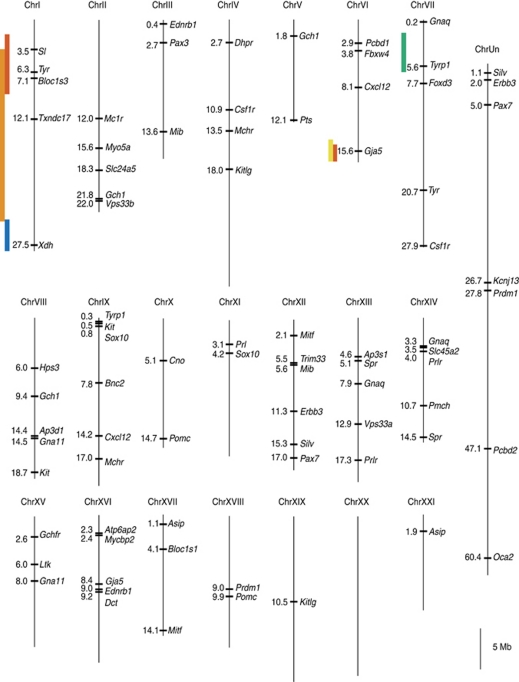
Although both the midline melanophore number and the degree of melanization were correlated with the degree of barring, they were not significantly correlated with one another and they mapped to distinct genomic regions (Figure 3, Tables 2 and 3). The number of melanophores in the light bars mapped strongly to LG6, and shared the same peak marker as the degree of barring phenotype (Figure 3, Table 3). In contrast, the overall degree of melanization mapped significantly to LG1 (Figure 3, Table 3). The degree of barring and the degree of melanization QTL on LG1 did not share the same peak marker (Table 3), but the LOD support intervals overlapped substantially. The LOD support interval for the LG1 degree of melanization QTL extends from chrI:3310077 to chrI:23569502 (Figure 5).
Figure 5.
Map of pigmentation QTL and pigment candidate genes. A plot of the initial stickleback genome assembly showing genomic positions (in Mb) of known candidate genes across 21 chromosomes and 1 large scaffold containing unassembled sequence (ChrUn). A detailed list of the candidate genes can be found in Supplementary Information, Supplementary Table S2. QTL positions including 1.5-LOD support intervals are shown with colored bars on the left side of relevant chromosomes. Different colored bars represent different traits: red, degree of barring; orange, degree of melanization; yellow, midline light bar melanophore number; green, dorsal melanophore number; blue, ventral melanophore number. Scale bar=5 Mb.
The association of a high degree of melanization with overall barring suggests a potential epistatic interaction between loci on LG1 and LG6, such that the barred pattern might simply be masked in fish with low levels of melanin. If this type of epistatic interaction existed, fish with Little Campbell alleles at LG1 would be predicted to be largely unbarred irrespective of the LG6 genotype; however, Hotel Lake LG6 alleles do increase barring in these fish (Figure 4). In addition, there was no statistical evidence for an epistatic interaction between these loci (LG1*LG6, F(4, 167)=1.4; P=0.24), although 176 individuals provide low power to detect epistasis.
In addition to melanophore pigmentation associated with barring, we also examined the number of melanophores in larger dorsal and ventral portions of the flank. The number of melanophores in the dorsal and ventral regions mapped to different genomic locations. The maximum number of dorsal melanophores per segment mapped to LG7, whereas the number of ventral melanophores mapped to LG1. A previous QTL study of pigmentation in sticklebacks found that variation in the Kit ligand gene was associated with differences in overall gill melanization and adult ventral body melanophore number in a marine by freshwater cross (Miller et al., 2007). However, we did not detect an association between juvenile ventral melanophore numbers and markers near Kit ligand in our cross. To further explore these results, we genotyped wild-caught Hotel Lake and Little Campbell River marine fish using a microsatellite marker diagnostic for marine and derived Kit ligand alleles. This marker amplified a product of 174-bp in both populations, suggesting that both of these populations possess the same ancestral ‘marine' Kit ligand allele, rather than the characteristic variant previously associated with reduced pigmentation. As both of the founders of our intercross possessed the same Kit ligand allele, it is not surprising that the F2s did not exhibit phenotypic variation associated with this locus (Miller et al., 2007).
We next asked whether melanophore pigmentation was associated with the other characteristic difference between marine and freshwater pigmentation: the extent of iridophore pigmentation. There was a significant negative correlation between iridophores and the degree of barring in F2s hybrids, but this relationship explained only a small amount of phenotypic variance (R2=0.09; Table 2). There was a stronger correlation between iridophores and the number of midline light bar melanophores (R2=0.23; Table 2), but this correlation is likely somewhat confounded by correlations of both measures with overall body size. We did not identify any significant QTL associated with the extent of iridophore pigmentation.
Discussion
Pigmentation QTL
We identified a striking variation in pigment patterns among different populations of sticklebacks: freshwater sticklebacks are prominently barred, whereas marine sticklebacks are unbarred and exhibit increased silver pigmentation. This divergent pigmentation was associated with several QTL, providing evidence for a more complex genetic architecture than is typically observed for overall color change and suggesting potential differences in the complexity of the underlying biology. In particular, for the degree of barring, there were significant QTL on LG1 and LG6 and there was an additional suggestive QTL on LG11. These two significant QTL were associated with two separable features of the barred pattern: spatial variation in melanophore number (LG6) and degree of melanization within melanophores (LG1). This finding is reminiscent of recent work in the domestic cat, which showed that two loci underlie variation in the classic cat pigment pattern of Tabby striping (Eizirik et al., 2010). The authors hypothesize that one locus, Tabby, regulates development of a striped pattern, whereas a second locus, Ticked, controls how pigment cells regulate pigment synthesis in response to the prepattern established by the Tabby locus (Eizirik et al., 2010). In the future, it will be interesting to determine whether there is a similar hierarchical relationship between the two pattern loci in sticklebacks and what the directionality of such a relationship might be.
There are several aspects of our methodology that should be considered when interpreting our results. First, the modest sample size (176 F2s) leaves open the possibility of overestimating the size of the detected QTL (Beavis, 1998) and also increases the likelihood of underestimating the number of QTL (Doerge, 2002), suggesting that the genetic architecture for barring may be even more complex than the three QTL we identified. Nonetheless, with this sample size, we can rule out the possibility that a single gene of very large effect has a role in the divergent pigment patterns we have studied here. Second, a small sample size, coupled with uneven marker spacing, also contributes to the identification of relatively large regions in the QTL analysis. Additional work will be required to narrow these regions, which will aid in the ultimate identification of causative genes and mutations. Finally, because the pigment phenotypes are highly associated with age and body size, we do not yet know whether these QTL have a role in pigmentation at other developmental stages. Analytical methods have been developed to facilitate detection of QTL across development (Wu and Lin, 2006), and in the future, it would be interesting to compare the genetic architecture of pigment patterns at different life stages.
Despite the large and consistent mean differences in iridophore pigmentation among parental lines, we did not identify any QTL for variation in iridophores in the F2s. This could be attributed to variability in our measurement of iridophore pigmentation. Although a similar measurement was successfully used in another fish QTL study (Nichols et al., 2008), iridophores have traditionally been challenging to study as their appearance changes substantially with light angle and level (Bagnara, 1998). In addition, iridophores must be studied in live fish because their pigment is dissolved after fixation (Lopes et al., 2008). Although we were careful to use a constant level and angle of light during photography, slight variation across photographs could have introduced enough noise into the measurement to confound the QTL analysis. Alternatively, it is possible that a ceiling effect prohibited accurate quantification of increasing numbers of iridophores. Finally, as discussed, our cross might not have sufficient power to detect QTL if many genes of small effect contribute to iridophore pigmentation.
Pigmentation candidate genes
One key advantage of examining pigmentation is the rich literature of genes and genetic pathways that have characterized roles in pigment cell development, pigment synthesis and pigment expression. We collated the location of 56 of these genes in the initial stickleback genome assembly, including 23 with ⩾2 paralogs, to determine whether any of our QTL contained known pigmentation candidate genes (Figure 5). In addition, many QTL studies in fish species that currently lack a complete, annotated genome have relied on synteny between fish chromosomes to identify candidate genes (Protas et al., 2006; Gross et al., 2008; Roberts et al., 2009; Tripathi et al., 2009; Valenzano et al., 2009). Our map of stickleback pigmentation QTL and candidate genes should be a useful resource for facilitating such comparisons.
The barring QTL on LG6 contains the gene Gja5, which encodes a gap junction protein, connexin 41.8. A nonsense mutation in this gene has previously been identified in the zebrafish leopard mutant (Watanabe et al., 2006). This mutation disrupts normal pigment cell interactions and results in an altered melanophore distribution in which spots form in place of the typical horizontal stripes (Watanabe et al., 2006). Strong leopard alleles cause further breakdown of spots, yielding a relatively even distribution of melanophores (Haffter et al., 1996). In an interesting parallel, the barring phenotype that maps to LG6 in sticklebacks also affects variation in melanophore distribution across the flank. Future studies will be required to assess the validity of Gja5 as a candidate for controlling barring in sticklebacks.
The region on LG1 that was associated with both barring and overall melanin level contains two candidate genes that are involved in pigment production. Bloc1s3 is important in the packaging of pigment into organelles (Navarro et al., 2008), and disruption of Bloc1s3 leads to a dilute pigment phenotype in mice (Starcevic and Dell'Angelica, 2004). Mutations of Tyrosinase (Tyr), which encodes a key enzyme in melanin synthesis, eliminate all pigmentation in zebrafish and medaka (Koga et al., 1995; Page-McCaw et al., 2004). In addition, the mosaic expression of Tyr in clonal groups of melanocytes can induce striping in mice (Mintz and Bradl, 1991). A regulatory change affecting either of these genes could plausibly underlie the variation in melanin level across the flank of freshwater vs marine sticklebacks.
QTL for body size
We also performed QTL analysis on juvenile body size, and found that body size maps to two regions: LG17 and LG18. This finding contrasts with previous studies of genetic associations with adult body size in sticklebacks. In three QTL studies on different marine and freshwater stickleback populations, adult body length was mapped to the sex chromosome LG19 (Colosimo et al., 2004; Kitano et al., 2009) and to LG13 (Albert et al., 2008). In our study, body size was assessed during the active growth phase of juveniles, and therefore our measurement might be more reflective of relative growth rate than of final adult body size. It is not surprising that body size at different ages might have distinct genetic architectures (Cheverud et al., 1996). In juveniles, one previous study found significant linkage to the Ectodysplasin locus on LG4 (Barrett et al., 2008), but this linkage was not observed in other populations (Marchinko, 2009). In our cross, we did not see significant linkage to any of the previously identified body-size loci. Taken together, these results suggest that the genetic basis of body size may differ among stickleback populations, as well as between juveniles and adults.
For QTL on both LG17 and LG18, freshwater alleles are associated with a larger body size. We have not investigated differences in growth rate between our parental populations; hence, we do not yet know whether this is the expected direction of effect. Interestingly, a recent study demonstrated that freshwater alleles at the Ectodysplasin locus on LG4 are associated with faster growth in freshwater but not in saltwater (Barrett et al., 2009). Our crosses were reared in freshwater, thus it is possible that the QTL we have identified are also specific to the freshwater environment. In the future, it would be interesting to determine whether different QTL for juvenile body size are found if F2s are reared in different environments.
Possible function of pigmentation differences
We have not yet tested the functional significance of the divergence we observe in juvenile pigmentation; however, we hypothesize that these phenotypes provide crypsis in the distinct environments inhabited by anadromous and freshwater sticklebacks, either during the juvenile period or at later stages after the migration of marines back to oceanic environments. Freshwater sticklebacks typically breed in slow-moving streams or in near-shore lake environments; both habitats have extensive plant cover that provides shelter for juveniles (Hagen, 1967; Foster et al., 1988). Like freshwater sticklebacks, many fish species inhabiting more complex near-shore environments display repeating pigment patterns, such as spots, bars or saddles (Seehausen et al., 1999; Ruxton et al., 2004). Such patterns may be examples of disruptive coloration or background matching, which serve to break up the body form and allow fish to visually recede into intricate backgrounds (Ruxton et al., 2004; Stevens and Merilaita, 2009b). In contrast, the pigmentation of anadromous juveniles may serve a different purpose. Pelagic marine fish of many species exhibit a similar silver coloration, which reflects light and is believed to confuse predators in open-water environments with no background structure (Ruxton et al., 2004; Quinn, 2005). Although our experiments do not directly address the question of function, the fact that these distinctive pigment patterns are highly conserved among different freshwater and marine populations throughout the Pacific Northwest does suggest that there may be selective pressure maintaining these different phenotypes. It will be interesting in the future to use predation experiments to directly test whether these divergent phenotypes are important for avoiding predation (Foster et al., 1988; Marchinko, 2009).
Conclusions
In summary, our QTL analysis revealed that the degree of vertical barring maps to two loci that are associated with separable aspects of this trait, namely melanin level and melanophore number. This result emphasizes the fact that although simple genetic changes have been associated with the evolution of pigmentation, differences in color patterns can also be associated with a more complex genetic architecture. Clearly, additional studies are required to determine whether complex changes in color patterns more often result from changes in few genes of large effect or in many genes of smaller effect. It is a difficult task to find the causative changes that underlie QTL of relatively small effect, but these efforts are required to fully appreciate the complexity of natural variation in biological patterns.
Acknowledgments
We thank Shaun McCann and Amanda Bruner for assistance with fish husbandry and Martin Escandon, Adam Knappe, Tiffany Malek and Kelley Pascoe for technical assistance. Julie Kerns, Craig Miller and Margaret Mills made insightful comments on earlier versions of this manuscript. We thank the Broad Institute Sequencing and Assembly teams for providing stickleback genome sequence information before publication. We also thank Semiahmoo First Nations for permission to collect sticklebacks from the Little Campbell River. This work was supported by a Helen Hay Whitney Foundation Postdoctoral Fellowship to AKG, an Affymetrix Bio-X Stanford Graduate Fellowship to YFC and a National Institutes of Health Center of Excellence in Genomic Science Grant (P50 HG002568) to DMK, RMM and CLP.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Albert AY, Sawaya S, Vines TH, Knecht AK, Miller CT, Summers BR, et al. The genetics of adaptive shape shift in stickleback: pleiotropy and effect size. Evolution. 2008;62:76–85. doi: 10.1111/j.1558-5646.2007.00259.x. [DOI] [PubMed] [Google Scholar]
- Anderson TM, vonHoldt BM, Candille SI, Musiani M, Greco C, Stahler DR, et al. Molecular and evolutionary history of melanism in North American gray wolves. Science. 2009;323:1339–1343. doi: 10.1126/science.1165448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnara JT.1998Comparative anatomy and physiology of pigment cells in nonmammalian tissuesIn: Nordlund JJ, Boissy RE, Hearing VJ, King RA and Ortonne J (eds).The Pigmentary System Oxford University Press: New York [Google Scholar]
- Barrett RD, Rogers SM, Schluter D. Natural selection on a major armor gene in threespine stickleback. Science. 2008;322:255–257. doi: 10.1126/science.1159978. [DOI] [PubMed] [Google Scholar]
- Barrett RD, Rogers SM, Schluter D. Environment specific pleiotropy facilitates divergence at the Ectodysplasin locus in threespine stickleback. Evolution. 2009;63:2831–2837. doi: 10.1111/j.1558-5646.2009.00762.x. [DOI] [PubMed] [Google Scholar]
- Barsh GS. The genetics of pigmentation: from fancy genes to complex traits. Trends Genet. 1996;12:299–305. doi: 10.1016/0168-9525(96)10031-7. [DOI] [PubMed] [Google Scholar]
- Beavis WD.1998QTL analyses: power, precision and accuracyIn: Paterson AH (ed).Molecular Dissection of Complex Traits CRC Press: Boca Raton, RL; 145–162. [Google Scholar]
- Bell MA, Foster SA. The Evolutionary Biology of the Threespine Stickleback. Oxford University Press: Oxford; 1994. [Google Scholar]
- Boulding EG, Culling M, Glebe B, Berg PR, Lien S, Moen T. Conservation genomics of Atlantic salmon: SNPs associated with QTLs for adaptive traits in parr from four trans-Atlantic backcrosses. Heredity. 2008;101:381–391. doi: 10.1038/hdy.2008.67. [DOI] [PubMed] [Google Scholar]
- Broman KW, Sen S. A Guide to QTL Mapping with R/qtl. Springer: New York; 2009. [Google Scholar]
- Candille SI, Kaelin CB, Cattanach BM, Yu B, Thompson DA, Nix MA, et al. A beta-defensin mutation causes black coat color in domestic dogs. Science. 2007;318:1418–1423. doi: 10.1126/science.1147880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro T. The adaptive significance of coloration in mammals. Bioscience. 2005;55:125–136. [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Jr, Shapiro MD, Brady SD, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM, Routman EJ, Duarte FA, van Swinderen B, Cothran K, Perel C. Quantitative trait loci for murine growth. Genetics. 1996;142:1305–1319. doi: 10.1093/genetics/142.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Jr, Dickson M, Grimwood J, et al. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- Colosimo PF, Peichel CL, Nereng K, Blackman BK, Shapiro MD, Schluter D, et al. The genetic architecture of parallel armor plate reduction in threespine sticklebacks. PLoS Biol. 2004;2:e109. doi: 10.1371/journal.pbio.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Brooks S, Bellone R, Bailey E. Missense mutation in exon 2 of SLC36A1 responsible for champagne dilution in horses. PLoS Genet. 2008;4:e1000195. doi: 10.1371/journal.pgen.1000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresko WA, Amores A, Wilson C, Murphy J, Currey M, Phillips P, et al. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc Natl Acad Sci USA. 2004;101:6050–6055. doi: 10.1073/pnas.0308479101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerge RW. Mapping and analysis of quantitative trait loci in experimental populations. Nat Rev Genet. 2002;3:43–52. doi: 10.1038/nrg703. [DOI] [PubMed] [Google Scholar]
- Eizirik E, David VA, Buckley-Beason V, Roelke ME, Schaffer AA, Hannah SS, et al. Defining and mapping mammalian coat pattern genes: multiple genomic regions implicated in domestic cat stripes and spots. Genetics. 2010;184:267–275. doi: 10.1534/genetics.109.109629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Larson G, Ribeiro HS, Li N, Andersson L. Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 2009;5:e1000341. doi: 10.1371/journal.pgen.1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SA, Garcia VB, Town MY. Cannibalism as the cause of an ontogenetic shift in habitat use by fry of the threespine stickleback. Oecologia. 1988;74:577–585. doi: 10.1007/BF00380056. [DOI] [PubMed] [Google Scholar]
- Gratten J, Beraldi D, Lowder BV, McRae AF, Visscher PM, Pemberton JM, et al. Compelling evidence that a single nucleotide substitution in TYRP1 is responsible for coat-colour polymorphism in a free-living population of Soay sheep. Proc Biol Sci. 2007;274:619–626. doi: 10.1098/rspb.2006.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Borowsky R, Tabin CJ. A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus. PLoS Genet. 2009;5:e1000326. doi: 10.1371/journal.pgen.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Protas M, Conrad M, Scheid PE, Vidal O, Jeffery WR, et al. Synteny and candidate gene prediction using an anchored linkage map of Astyanax mexicanus. Proc Natl Acad Sci USA. 2008;105:20106–20111. doi: 10.1073/pnas.0806238105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, Odenthal J, Mullins MC, Lin S, Farrell MJ, Vogelsang E, et al. Mutations affecting pigmentation and shape of the adult zebrafish. Dev Genes Evol. 1996;206:260–276. doi: 10.1007/s004270050051. [DOI] [PubMed] [Google Scholar]
- Hagen DW. Isolating mechanisms in threespine sticklebacks (Gasterosteus) J Fish Res Board Canada. 1967;24:1637–1692. [Google Scholar]
- Hoekstra HE. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity. 2006;97:222–234. doi: 10.1038/sj.hdy.6800861. [DOI] [PubMed] [Google Scholar]
- Kelsh RN. Genetics and evolution of pigment patterns in fish. Pigment Cell Res. 2004;17:326–336. doi: 10.1111/j.1600-0749.2004.00174.x. [DOI] [PubMed] [Google Scholar]
- Kerns JA, Cargill EJ, Clark LA, Candille SI, Berryere TG, Olivier M, et al. Linkage and segregation analysis of black and brindle coat color in domestic dogs. Genetics. 2007;176:1679–1689. doi: 10.1534/genetics.107.074237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ullmann B, Walker C, Wilson C, Currey M, Phillips PC, et al. Evolution and development of facial bone morphology in threespine sticklebacks. Proc Natl Acad Sci USA. 2005;102:5791–5796. doi: 10.1073/pnas.0408533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley DM, Peichel CL.2007The molecular genetics of evolutionary change in sticklebacksIn: Ostlund-Nilsson S, Mayer I and Huntingford F (eds).Biology of the Three-Spined Stickleback CRC Press: Boca Raton [Google Scholar]
- Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YF, et al. A role for a neo-sex chromosome in stickleback speciation. Nature. 2009;461:1079–1083. doi: 10.1038/nature08441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga A, Inagaki H, Bessho Y, Hori H. Insertion of a novel transposable element in the tyrosinase gene is responsible for an albino mutation in the medaka fish, Oryzias latipes. Mol Gen Genet. 1995;249:400–405. doi: 10.1007/BF00287101. [DOI] [PubMed] [Google Scholar]
- Kondo S, Iwashita M, Yamaguchi M. How animals get their skin patterns: fish pigment pattern as a live Turing wave. Int J Dev Biol. 2009;53:851–856. doi: 10.1387/ijdb.072502sk. [DOI] [PubMed] [Google Scholar]
- Lopes SS, Yang X, Muller J, Carney TJ, McAdow AR, Rauch GJ, et al. Leukocyte tyrosine kinase functions in pigment cell development. PLoS Genet. 2008;4:e1000026. doi: 10.1371/journal.pgen.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchinko KB. Predation's role in repeated phenotypic and genetic divergence of armor in threespine stickleback. Evolution. 2009;63:127–138. doi: 10.1111/j.1558-5646.2008.00529.x. [DOI] [PubMed] [Google Scholar]
- Miller CT, Beleza S, Pollen AA, Schluter D, Kittles RA, Shriver MD, et al. cis-Regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131:1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz B, Bradl M. Mosaic expression of a tyrosinase fusion gene in albino mice yields a heritable striped coat color pattern in transgenic homozygotes. Proc Natl Acad Sci USA. 1991;88:9643–9647. doi: 10.1073/pnas.88.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro RE, Ramos-Balderas JL, Guerrero I, Pelcastre V, Maldonado E. Pigment dilution mutants from fish models with connection to lysosome-related organelles and vesicular traffic genes. Zebrafish. 2008;5:309–318. doi: 10.1089/zeb.2008.0549. [DOI] [PubMed] [Google Scholar]
- Nichols KM, Edo AF, Wheeler PA, Thorgaard GH. The genetic basis of smoltification-related traits in Oncorhynchus mykiss. Genetics. 2008;179:1559–1575. doi: 10.1534/genetics.107.084251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page-McCaw PS, Chung SC, Muto A, Roeser T, Staub W, Finger-Baier KC, et al. Retinal network adaptation to bright light requires tyrosinase. Nat Neurosci. 2004;7:1329–1336. doi: 10.1038/nn1344. [DOI] [PubMed] [Google Scholar]
- Parichy DM. Evolution of danio pigment pattern development. Heredity. 2006;97:200–210. doi: 10.1038/sj.hdy.6800867. [DOI] [PubMed] [Google Scholar]
- Peichel CL, Nereng KS, Ohgi KA, Cole BL, Colosimo PF, Buerkle CA, et al. The genetic architecture of divergence between threespine stickleback species. Nature. 2001;414:901–905. doi: 10.1038/414901a. [DOI] [PubMed] [Google Scholar]
- Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, et al. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- Protas ME, Patel NH. Evolution of coloration patterns. Annu Rev Cell Dev Biol. 2008;24:425–446. doi: 10.1146/annurev.cellbio.24.110707.175302. [DOI] [PubMed] [Google Scholar]
- Quigley IK, Manuel JL, Roberts RA, Nuckels RJ, Herrington ER, MacDonald EL, et al. Evolutionary diversification of pigment pattern in Danio fishes: differential fms dependence and stripe loss in D albolineatus. Development. 2005;132:89–104. doi: 10.1242/dev.01547. [DOI] [PubMed] [Google Scholar]
- Quinn TP. The Behavior and Ecology of Pacific Salmon and Trout. University of Washington Press: Seattle; 2005. [Google Scholar]
- Roberts RB, Ser JR, Kocher TD. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science. 2009;326:998–1001. doi: 10.1126/science.1174705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruxton GD, Sherratt TN, Speed MP. Avoiding Attack: The Evolutionary Ecology of Crypsis, Warning Signals and Mimicry. Oxford University Press: Oxford; 2004. [Google Scholar]
- Seehausen O, Maynew PJ, Van Alphen JJM. Evolution of colour patterns in East African cichlid fish. J Evol Biol. 1999;12:514–534. [Google Scholar]
- Starcevic M, Dell'Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Biol Chem. 2004;279:28393–28401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- Steiner CC, Weber JN, Hoekstra HE. Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biol. 2007;5:e219. doi: 10.1371/journal.pbio.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA. Mouse coat color mutations: from fancy mice to functional genomics. Dev Dyn. 2006;235:2401–2411. doi: 10.1002/dvdy.20840. [DOI] [PubMed] [Google Scholar]
- Stevens M, Merilaita S. Animal camouflage: current issues and new perspectives. Philos Trans R Soc Lond B Biol Sci. 2009a;364:423–427. doi: 10.1098/rstb.2008.0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M, Merilaita S. Defining disruptive coloration and distinguishing its functions. Philos Trans R Soc Lond B Biol Sci. 2009b;364:481–488. doi: 10.1098/rstb.2008.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi N, Hoffmann M, Dreyer C. Natural variation of male ornamental traits of the guppy, Poecilia reticulata. Zebrafish. 2008;5:265–278. doi: 10.1089/zeb.2008.0548. [DOI] [PubMed] [Google Scholar]
- Tripathi N, Hoffmann M, Willing EM, Lanz C, Weigel D, Dreyer C. Genetic linkage map of the guppy, Poecilia reticulata, and quantitative trait loci analysis of male size and colour variation. Proc Biol Sci. 2009;276:2195–2208. doi: 10.1098/rspb.2008.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uy JA, Moyle RG, Filardi CE, Cheviron ZA. Difference in plumage color used in species recognition between incipient species is linked to a single amino acid substitution in the melanocortin-1 receptor. Am Nat. 2009;174:244–254. doi: 10.1086/600084. [DOI] [PubMed] [Google Scholar]
- Valenzano DR, Kirschner J, Kamber RA, Zhang E, Weber D, Cellerino A, et al. Mapping loci associated with tail color and sex determination in the short-lived fish Nothobranchius furzeri. Genetics. 2009;183:1385–1395. doi: 10.1534/genetics.109.108670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen JW, Voorrips RE. JoinMap 3.0, Software for the Calculation of Genetic Linkage Maps. Plant Research International: Wageningen, The Netherlands; 2001. [Google Scholar]
- Watanabe M, Iwashita M, Ishii M, Kurachi Y, Kawakami A, Kondo S, et al. Spot pattern of leopard Danio is caused by mutation in the zebrafish connexin41.8 gene. EMBO Rep. 2006;7:893–897. doi: 10.1038/sj.embor.7400757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Koshikawa S, Williams TM, Carroll SB. Generation of a novel wing colour pattern by the Wingless morphogen. Nature. 2010;464:1143–1148. doi: 10.1038/nature08896. [DOI] [PubMed] [Google Scholar]
- Widelitz RB, Baker RE, Plikus M, Lin CM, Maini PK, Paus R, et al. Distinct mechanisms underlie pattern formation in the skin and skin appendages. Birth Defects Res C Embryo Today. 2006;78:280–291. doi: 10.1002/bdrc.20075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Beldade P. Development and evolution of insect pigmentation: genetic mechanisms and the potential consequences of pleiotropy. Semin Cell Dev Biol. 2009;20:65–71. doi: 10.1016/j.semcdb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Wu R, Lin M. Functional mapping—how to map and study the genetic architecture of dynamic complex traits. Nat Rev Genet. 2006;7:229–237. doi: 10.1038/nrg1804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.