Abstract
Although surgery and radiotherapy have been the standard treatment modalities for head and neck squamous cell carcinoma (HNSCC), the integration of cisplatin (CDDP)-based therapy has led to improvements in local and regional control of disease for patients. However, many trials show that only 10–20% of patients benefit from this treatment intensification, which can result in profound treatment-associated morbidity and mortality. Moreover, the marginal survival improvement suggests that CDDP resistance is an innate characteristic of HNSCC. To elucidate the biological mechanisms underpinning CDDP resistance in HNSCC, we utilized an experimental model of CDDP resistance in this disease. We first observed significant enhancements in local tumor growth and metastasis, as well as adverse survival, in CDDP-resistant (CR) tumors compared with sensitive tumors. To elucidate the molecular mechanisms of this phenotype, we undertook a systems biology-based approach utilizing high-throughput PCR arrays, and we identified a significant suppression of KiSS1 mRNA and protein expression in the CR cells, but no significant regions of genomic loss with array comparative genomic hybridization. Genetic suppression of KiSS1 in CDDP-sensitive cell lines rendered them CR, an observation that was mechanistically linked to alterations in glutathione S-transferase-π expression and function. We next confirmed that, in human HNSCC tumors, loss of KiSS1 expression was associated with metastatic human HNSCC tumors compared with non-metastatic tumors. Genetic reconstitution of KiSS1 in CR cells abrogated cellular migration and induced CDDP sensitivity. To confirm these findings in a murine model, either CR or KiSS1-transfected CR cells were studied in an orthotopic model of HNSCC, or survival studies revealed significant improvement in survival of the mice bearing CR-KiSS1 tumors. Mechanistically, alterations in apoptotic pathways and CDDP metabolism contributed to KiSS1-associated chemotherapy sensitization. These studies provided further direct evidence for the role of KiSS1 loss in biologically aggressive HNSCC and suggest potential targets for therapy in CR cancers.
Keywords: head and neck squamous cell carcinoma, KiSS1, chemotherapy resistance, cisplatin, metastasis
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer diagnosis worldwide, and affects over 44 000 individuals in the United States each year. The integration of systemic therapy in the definitive management of HNSCC has evolved over the past 15 years. Landmark prospective, randomized, multi-institutional trials have clearly shown that cisplatin (CDDP)-based chemotherapeutic strategies are effective for local and regional control at multiple tumor sites in the head and neck when used adjuvantly or concomitantly with radiotherapy (Cooper et al., 2004; Forastiere et al., 2003). However, the results of many of these trials show that only 10–20% of patients benefit from this treatment intensification, which can result in profound treatment-associated morbidity and mortality. Moreover, survival is only marginally improved, suggesting that resistance to CDDP is an innate component of HNSCC. The etiology of this CDDP resistance in HNSCC has unclear biological mechanisms and has received little attention to date.
Multiple mechanisms of CDDP resistance have been proposed in previous studies and include: AKT1 amplification (Liu et al., 2007); metabolic dysregulation; glutathione-induced cellular trapping of CDDP (Hamaguchi et al., 1993); enhanced cellular export; secondary mutations in BRCA2; among many others (reviewed by Kelland (2007)). However, for HNSCC, mechanisms of CDDP resistance have received little attention. Further, overcoming this resistance for other epithelial malignancies has yielded only few relevant targets, and translation of these into the clinical realm has not been forthcoming owing to the dependency on in vitro cell line data.
Cogent data support a role for KiSS1 in the progression and metastasis of various tumors (Welch et al., 1994; Lee and Welch, 1997; Ohtaki et al., 2001). Several studies have shown that loss of KiSS1 expression has been associated with increased metastasis and cancer progression in a number of human cancers, including esophageal, bladder, brain, breast, ovarian, and pancreatic and melanoma (reviewed by Nash and Welch (2006)). The KiSS1 gene encodes the KiSS peptides of various molecular weights that have diverse functions, and was initially identified as a metastasis suppressor in melanoma and breast cancer systems. KiSS peptides have been implicated in numerous physiological processes including: puberty, fertility and vascular homeostasis, mediated through the G-protein-coupled receptor-54. Although correlative studies have shown a downregulation of KiSS1 mRNA and KiSS peptide protein expression in advanced tumors, the mechanisms for this anti-metastatic capability is only slowly emerging, and may be related in part to effects on suppressing angiogenesis (Cho et al., 2009) and inducing tumor dormancy (Nash et al., 2007). Molecular insights into KiSS1-directed effects on migration and invasion have been gleaned and may be due in part through interactions with protein kinase C (Jiang et al., 2005) and nuclear factor (NF)-κB (Sung-Gook et al., 2009). However, the clinical relevance of KiSS1 is now emerging and has been linked to disease progression in only selected cancers (Nash and Welch, 2006).
Collectively, our understanding of anti-metastatic role of KiSS1 from extensive studies in other tumor types, in conjunction with our studies, have led us to hypothesize that KiSS1 mechanistically contributes to CDDP sensitivity in HNSCC. In this study, we identify KiSS1 as an important mediator of CDDP resistance in HNSCC and as a potential target for therapeutic strategies for this disease.
Results
CDDP-resistant HNSCC is associated with an aggressive phenotype
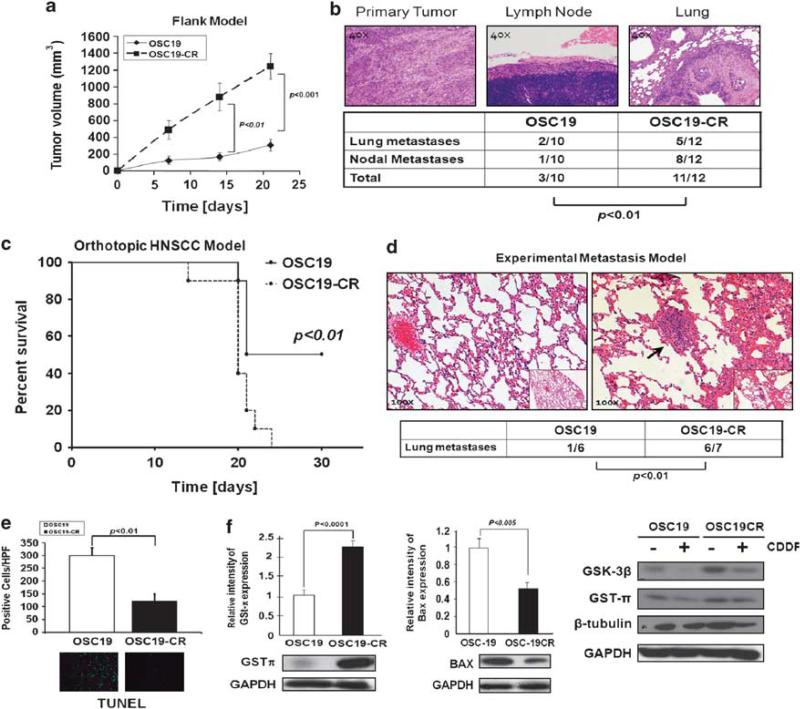
To elucidate the biological mechanisms underpinning CDDP resistance in HNSCC, we have developed a cell-based model of drug resistance by long-term exposure of tumor cell lines to escalating doses of CDDP over a 6-month period. The characteristics of these cells have been reported previously (Yilmaz et al., 2010). We next utilized a mouse model of HNSCC to assess the in vivo biological behavior of the CDDP-resistant (CR) cells, and observed a profound enhancement of both local tumor growth (Figure 1a), lymphatic and distant metastasis in the CR cells, compared with the parental cells (Figure 1b). Utilizing micro-computed tomography imaging, we identified radiographically visible pulmonary metastasis in the mice harboring the CR tumors, but not in the parental mice (data not shown). Further, in an orthotopic model of HNSCC, CR tumors were associated with adverse survival compared with the parental tumors (Figure 1c). Moreover, an in vivo experimental metastasis model showed a higher incidence of lung metastasis in the CR-inoculated mice compared with the parental cell lines (Figure 1d). We further detected a marked decrease in apoptosis (terminal deoxynucleotidyl transferase dUTP nick-end labeling) in the CR-derived tumors, suggesting a more aggressive phenotype in these tumors (Figure 1e).
Figure 1.
Cisplatin-resistant HNSCC is associated with aggressive biological behavior in mouse models of HNSCC. (a) Flank tumor model identifies significant tumor growth for CDDP-resistant (OSC19CR) cell lines compared with CDDP-sensitive cells (OSC19). Results are representative of three independent experiments. Differences between tumor volumes were evaluated by the non-parametric Mann–Whitney test. Results are reported as mean±s.e.m. A P-value <0.05 was considered significant. (b). Representative hematoxylin and eosin of tumor, lymph node and lung metastasis showing increased metastatic efficiency to both regional and distant sites for CR cell lines. Differences between the number of metastases per group were evaluated by the non-parametric Mann–Whitney test. Results are reported as mean±s.e.m. (c). Decreased survival of mice harboring orthotopically implanted CR lines (n = 6) compared with parental lines (n = 6), in an orthotopic model of HNSCC, as determined by the Kaplan–Meier method. Results are representative of three independent experiments. (d) After injection into the tail vein, mice were monitored for the development of pulmonary metastasis with micro-computed tomography imaging (parental, n = 6; CR, n = 7). all After 6 weeks, mice were killed and the lungs of mice were examined both grossly and microscopically for pulmonary metastasis. Differences between the number of metastases per group were evaluated by the non-parametric Mann–Whitney test. Results are reported as mean±s.e.m. Results are representative of three independent experiments. (e) Significant suppression of terminal deoxynucleotidyl transferase dUTP nick-end labeling staining is noted in the CR tumors compared with parental tumors (P<0.001). (f) CR and parental cell lines were profiled for known CDDP-resistant proteins under basal (left, middle panels) and CDDP treatment conditions (right panel) by SDS–PAGE. Protein lysates from subconfluent HNSCC cell lines were separated by SDS–PAGE and assessed with the indicated antibodies.
To determine the underlying pathway dysregulation in the chemo-resistant phenotype, known mediators of CDDP resistance were analyzed by western blot. Although levels of β-tubulin were unchanged in the CR cells, glycogen synthase kinase-3β and glutathione S-transferase (GST)-π levels were upregulated compared with parental cell lines, whereas the antiapoptotic Bax protein was downregulated, showing an association of the CR phenotype with previously reported mediators of CDDP resistance (Figure 1f) (Bai et al., 1996; Arai et al., 2000). Alterations in proteins associated with epithelial–mesenchymal transition were also seen in the CR cells (Supplementary Figure 1), suggesting that CDDP resistance is associated with a biologically aggressive behavior and tumor metastasis, a phenotype similar to that observed in human cancers.
KiSS1 loss is associated with CDDP resistance in HNSCC
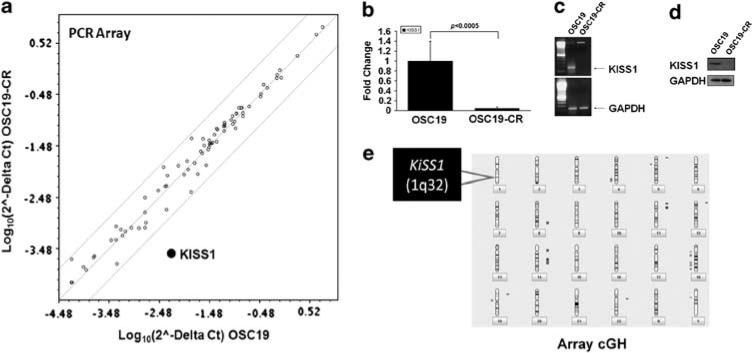
We next undertook a systems biology-based approach to elucidate the molecular mechanisms associated with the observed biological phenotype in CR cells. Utilizing high-throughput PCR-based microarrays, we identified a profound decrease in KiSS1 mRNA expression in the CR cell lines compared with the parental cells (Figures 2a and b). We further confirmed these observations utilizing reverse transcription (RT)–PCR (Figure 2c), and also observed complete loss of KiSS1 at the protein level (Figure 2d), although no significant differences were noted in the levels of G-protein-coupled receptor-54, the receptor for KiSS1 (Supplementary Figure 2). Array comparative genomic hybridization (CGH) analysis identified no significant genomic loss at 1q32, the region of the genome that encodes for KiSS1, suggesting that transcriptional suppression may explain KiSS1 loss in CR cells (Figure 2e).
Figure 2.
Differential expression of KiSS1 in CDDP-resistant HNSCC. (a) High-throughput PCR-based microarray analysis revealed that the expression of KiSS1 was significantly downregulated in CDDP-resistant cell line. Individual genes are plotted as a function of fold-expression change for the parental (x axis) and CR (y axis) cell lines. KiSS1 is highlighted in bold. (b) Graphical representation of mRNA expression levels of KiSS1 in parental and CR cell lines from the PCR array experiments. Results are representative of three independent experiments. Quantitative RT–PCR (c) and western blot (d) were performed to determine the mRNA and protein expression of KiSS1 in the indicated cell lines. (e) Ideogram representation of the array CGH experimental data, with areas of gain (blue) and loss (red) notated adjacent to the respective regions of the genome. A full colour version of this figure is available at the Oncogene journal online.
Genetic suppression of KiSS1 induces CDDP resistance in HNSCC
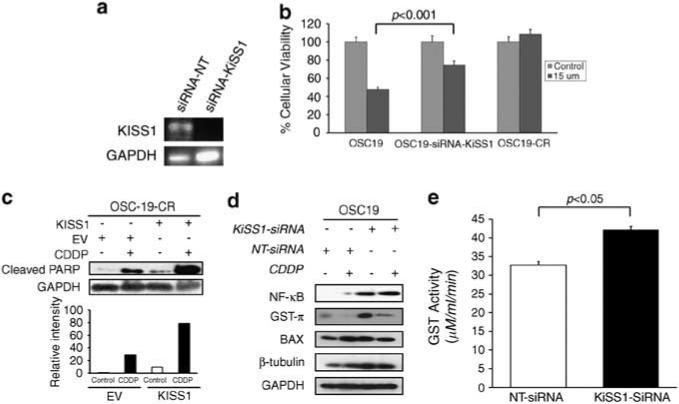
To further explore the direct role of KiSS1 in CDDP resistance, we inhibited its expression in CDDP-sensitive parental cells and analyzed the in vitro therapeutic impact. HNSCC cell lines were transiently transfected with small interfering RNA (siRNA) constructs targeting either KiSS1 or a non-targeting construct. After showing effective inhibition of KiSS1 mRNA expression (Figure 3a), we then evaluated the impact of KiSS1 downmodulation on CDDP response. When KiSS1 expression was abrogated through siRNA in CDDP-sensitive cells, a relative induction of CDDP resistance ensued, but this effect did not result in complete chemotherapy resistance (Figure 3b). These results suggested that the loss of KiSS1 expression induces partial resistance to platinum-based chemotherapy, providing further support for the importance of KiSS1 as a mediator of a chemo-resistant phenotype in HNSCC.
Figure 3.
Genetic knockdown of KiSS1 induces CDDP resistance in HNSCC. (a) Cells were transfected with either an siRNA sequence targeting KiSS1 (siRNA-KiSS1) or a non-targeting siRNA (siRNA-NT). Efficiency of genetic inhibition was determined by PCR. (b) Genetic knockdown of KiSS1 in CDDP-sensitive cells induces CDDP resistance. Depicted cell lines were transfected with the denoted siRNA constructs, exposed to CDDP (15 μm) and then analyzed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. Differences between groups were analyzed by utilizing ANOVA, followed by post hoc comparisons based on Bonferroni's Multiple Comparison Test. Results are reported as mean±s.e.m. A P-value <0.05 was considered significant and all are two-tailed. Experiments were performed in octuplicate and repeated three times. (c) Genetic knockdown of KiSS1 in CDDP-sensitive cells suppresses PARP cleavage. SDS–PAGE was utilized to determine the induction of PARP cleavage in non-targeted siRNA (siRNA-NT)- or KiSS1-targeted siRNA (siRNA-KiSS1)-transfected cells under CDDP treatment conditions (upper panel). Cells were exposed to CDDP for 24 h, lysed and proteins were separated by 10% SDS–PAGE and analyzed for the indicated antibodies. Intensity data depicts suppressed PARP cleavage in siRNA-KiSS1-transfected cells that were exposed to CDDP. Bar graph represents densitometry results. (d) Levels of proteins associated with CDDP sensitivity were analyzed by SDS–PAGE in the depicted cell lines after exposure to CDDP. Increased GST-π expression in the KiSS1-inhibited cells was consistent with the induction of CDDP resistance in the sensitive cell lines. (e) The alteration in GST function was assessed to establish the impact of KiSS1 knockdown in CDDP-sensitive cells. Specific GST activity is expressed on the y axis and was calculated dividing the absorbance change per minute with the extinction coefficient for CDNB (A = 9.6 per mm) and the total protein content of the cell homogenate. Results are reported as mean±s.e.m. A P-value <0.05 was considered significant, and all are two-tailed. Experiments were performed in triplicate and repeated three times. A full colour version of this figure is available at the Oncogene journal online.
KiSS1 modulates GST-π function to regulate chemo-resistance in HNSCC
To further define the mechanisms by which KiSS1 regulates CDDP resistance in HNSCC, we evaluated the impact of KiSS1 knockdown on pathways associated with CDDP resistance. The loss of KiSS1 was associated with a marked inhibition of poly(ADP-ribose) polymerase (PARP) cleavage when cells were exposed to CDDP, which revealed another level of regulation of KiSS1 in apoptotic and drug resistance (Figure 3c). Moreover, cells with suppressed KiSS1 had a significant upregulation of GST-π and NF-κb expression, but minimal alterations in Bax and β-tubulin (Figure 3d). To further refine the implication of differential GST-π expression when KiSS1 expression was reduced, we next assessed the functional impact of KiSS1 on GST-π activity. When KiSS1 expression was abrogated in CDDP-sensitive cells, GST-π activity was significantly elevated, an effect that was accompanied by a concomitant induction of CDDP resistance (Figure 3e). These data suggested that KiSS1 impacts the CDDP resistance phenotype, and is associated with an alteration of GST-π levels and function, providing a potential target for therapy-resistant HNSCC.
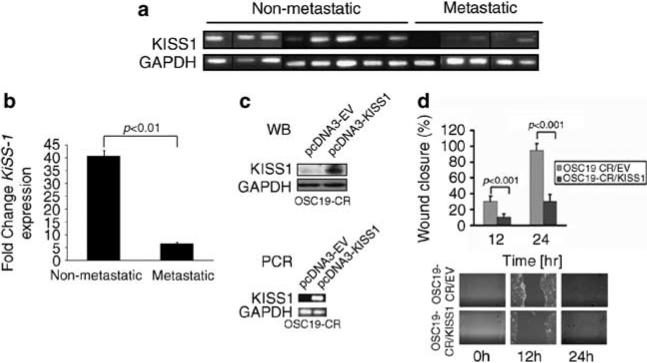
KiSS1 expression is downregulated in metastatic HNSCC KiSS1, a suppressor of metastasis, has been associated with hematogenous metastasis among patients with various cancer types. However, the clinical relevance of KiSS1 in HNSCC has not been explored to date. Further, the association of KiSS1 with regional lymphatic metastasis has received little attention, as KiSS1 loss is primarily associated with hematogenous metastasis in other disease models. We investigated whether KiSS1 was associated with clinical metastasis, and analyzed the relative expression of KiSS1 among patients with HNSCC who underwent surgical resection for their disease (Supplementary Table 1). In nearly every case of non-metastatic HNSCC, KiSS1 mRNA was detected in the tumor samples (Figures 4a and b). However, KiSS1 mRNA was minimally detectable in tumors from patients with lymphatic and hematogenous metastases. These findings suggested that KiSS1 loss may be involved not only in hematogenous metastasis, but may also contribute to the development of lymphatic metastasis in HNSCC. These data provided further support for the clinical relevance of KiSS1 in this disease phenotype.
Figure 4.
KiSS1 is lost in metastatic HNSCC tumors. (a) Real-time (RT)–PCR analysis of KiSS1 and GAPDH in metastatic and nonmetastatic human HNSCC tumors, showing decreased expression or loss of KiSS1 in the metastatic tumors. Specimens run on different gels are separated by a black vertical line. (b) KiSS1 densitometric expression in human tumors from (a) were normalized to GAPDH levels, and data were averaged and analyzed with Student's t-test. (c) CDDP-resistant cell lines were transfected with either an empty vector (pcDNA3-EV) or a vector containing the full-length KiSS1 coding sequence. The expression of KiSS1 was analyzed by SDS–PAGE (western blot, top panel) and real-time RT–PCR (PCR, bottom panel). (d) Mock- or KiSS1-tranfected cells were seeded onto six-well plates, grown to confluency and assessed for haptotaxis after a scratch wound was made. Images were taken at 0, 12 and 24 h, and the degree of wound closure was determined with ImagePro. Experiments were performed in triplicate and repeated three times. Columns represent percentage of wound closure from 0 h; bars represent s.e.m.; magnification × 100. A full colour version of this figure is available at the Oncogene journal online.
Restoration of KiSS1 suppresses motility and restores chemo-sensitivity in HNSCC
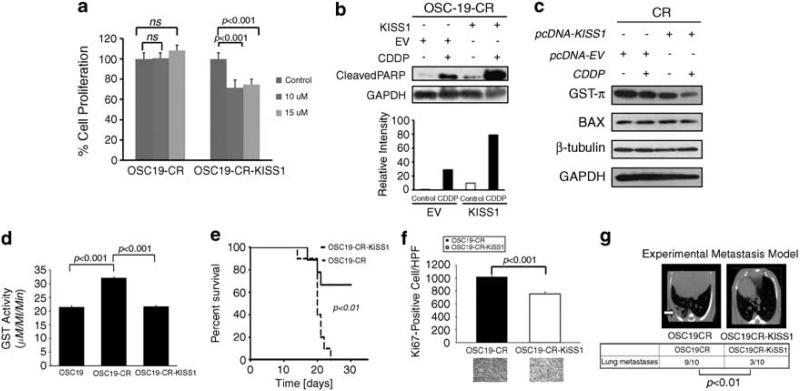
To assess directly the role of KiSS1 in the aggressive HNSCC phenotype and CDDP resistance, CR cells were stably transfected with a KiSS1 expression vector (CR-KiSS1), or an empty vector (CR-EV) (Figure 4c). Wound healing assays revealed that cellular migration was inhibited in the KiSS1-transfected cells compared with control vector, but not completely abrogated, which suggested that the aggressive in vivo motile behavior may be due in part to the loss of KiSS1 in the CR cells (Figure 4d). We next assessed the chemotherapeutic sensitivity of the KiSS1-overexpressing CR cells and found a profound reversal of CDDP resistance in these cells compared with the CR cells, suggesting a direct role for KiSS1 in mediating CDDP therapy resistance in HNSCC (Figure 5a, Supplementary Figure 3). To explore a potential mechanism as to how KiSS1 may modulate the response to CDDP, we evaluated the function of GST-π, a known mediator of CDDP inactivation in cancers. CR-KiSS1 cells had a relative downregulation of GST-π compared with the CR-EV cells (Figure 5b), and CR-KiSS1 cells also showed an enhanced cleavage of PARP, a marker of apoptotic induction, although there was minimal change in other mediators of CDDP resistance in these cells (Figure 5c). Moreover, functional assays showed enhanced GST-π activation in the CR cells, an effect that was abrogated when KiSS1 was genetically overexpressed in these cells (Figure 5d). These data suggested that KiSS1 impacts the CDDP resistance phenotype, and is associated with altered GST-π levels and function, providing a potential pathway for targeting approaches in therapy-resistant HNSCC.
Figure 5.
Re-expression of KiSS1 reverses the CR phenotype in vitro and in vivo. (a) Empty-vector- or KiSS1-tranfected cells were treated with depicted concentrations of CDDP and assessed for cellular proliferation after 24 h of exposure. Differences between groups were analyzed by utilizing ANOVA, followed by post hoc comparisons based on Bonferroni's Multiple Comparison Test. Results are reported as mean±s.e.m. A P-value <0.05 was considered significant, and all are two-tailed. Experiments were performed in octuplicate and repeated three times. (b) SDS–PAGE was utilized to determine the induction of PARP cleavage in mock-transfected or KiSS1-tranfected CR cells under CDDP treatment conditions (upper panel). Cells were exposed to CDDP for 24 h, lysed and proteins were separated by 10% SDS–PAGE and analyzed for the indicated antibodies. Intensity data depicts relative increase in PARP cleavage in CDDP-treated KiSS-overexpressing cells. (c) Levels of proteins associated with CDDP-resistant were analyzed by SDS–PAGE in the depicted cell lines after exposure to CDDP. Reduced GST-π expression in the KiSS1-expressing cells was consistent with the re-establishment of CDDP sensitivity in CR cells. (d) The alteration in GST function was assessed to establish the impact of KiSS1 overexpression in the CR cells. Specific GST activity is expressed on the y axis and was calculated dividing the absorbance change per minute with the extinction coefficient for CDNB (A = 9.6 per mm) and the total protein content of the cell homogenate. Results are reported as mean±s.e.m. A P-value <0.05 was considered significant, and all are two-tailed. Experiments were performed in triplicate and repeated three times. (e) OSC19-CR cells transfected with either an empty vector (OSC19CR) or a KiSS1 overexpression construct (OSC19CR-KiSS1) were injected into the tongues of nude mice. Mice were then observed and killed when moribund. (f) Mice harboring orthotopically implanted OSC19CR-KiSS1 tumors (n = 7) had improved survival compared to those with OSC10CR tumors (n = 6), as determined by the Kaplan–Meier method. Results are representative of three independent experiments. Immunohistochemical analysis of Ki67 revealed statistically significant decrease in cellular proliferation in the KiSS1-tranfected CR tumors compared with the CR tumors (P<0.001). (g) After injection into the tail vein, mice were monitored for the development of pulmonary metastasis with micro-computed tomography imaging (OSC19CR, n = 10; OSC19CR-KiSS1, n = 10). After 6 weeks, mice were killed and the lungs of all mice were examined both grossly and microscopically for pulmonary metastasis. Differences between the number of lung metastases per group were evaluated by the non-parametric Mann–Whitney test. Representative micro-computed tomography images are shown. Results are reported as mean±s.e.m. Results are representative of three independent experiments. A full colour version of this figure is available at the Oncogene journal online.
Re-expression of KiSS1 suppresses tumor growth and metastasis in animal models of HNSCC
Previous studies have shown the anti-metastatic role of KiSS1 in other models of cancer, but the impact of KiSS1 on local tumor growth has been debated. To determine whether KiSS1 may inhibit metastasis on HNSCC cells in vivo, we compared CR and CR-KiSS1 cells for their metastatic potential in relevant mouse models of HNSCC. Either CR or CR-KiSS1 cells were inoculated into the tongues of nude mice to establish orthotopic tongue tumors. Survival studies revealed significant improvement in mouse viability in the CR-KiSS1 mice (Figure 5e) and decreased cellular proliferation, as evidenced by lower Ki67 values (Figure 5f). These results paralleled findings from an experimental metastasis model that revealed a significant inhibition of metastatic foci in the lungs of the KiSS1-overexpressing CR cells (Figure 5g). We next determined whether anchorage-independent growth, an in vitro metric of oncogenic potential, was altered by KiSS1 in CR cells. In soft agar experiments, anchorage-independent growth was significantly abrogated in the KiSS1-transfected CR cells, as compared to empty-vector-transfected CR cells (Supplementary Figure 4). In addition, proliferation was only marginally increased in the chemo-resistant cells compared with the KiSS1-transfected cells (Supplementary Figure 5). These experiments suggested that, although cellular proliferation was increased in the chemo-resistant cells, this alone did not explain the increased metastatic rate seen in these cells compared with the KiSS1-transfected cells.
Discussion
In this study, we report a novel mechanism of chemotherapy resistance in HNSCC, regulated by KiSS1, a mediator of aggressive biological behavior human cancer. First, we developed chemotherapy-resistant cell lines that faithfully recapitulated the inherent phenotypic characteristics in multiple in vivo models of HNSCC. High-throughput genomic analysis revealed distinct molecular alteration in KiSS1 levels between CDDP-sensitive and CR cells, findings that were confirmed through a number of approaches. We further identified loss of KiSS1 in patients with metastatic HNSCC, but not in non-metastatic tumors, establishing the clinical importance of KiSS1 in this disease. Using genetic and pharmacological manipulation, we showed that loss of KiSS1 induced CDDP resistance, and, conversely, re-introduction of KiSS1 reversed these biological properties in vitro and in vivo. Finally, a mechanistic link between KiSS1 and GST-π, a known regulator of CDDP sensitivity, was revealed through expression and functional assays. These findings support a new biological role for KiSS1 in the chemotherapeutic response, and highlight a potential treatment avenue for recalcitrant HNSCC.
Most cancers have defects in some part of the DNA damage response, culminating in apoptotic resistance, whereas normal cells have a full complement of DNA repair response. Mutational status of TP53 has also been extensively studied for its correlation with tumor grade, patient survival and treatment outcome in many cancers (Soussi, 2007; Soussi and Wiman, 2007). The results of these studies have, however, been inconsistent with various groups reporting both positive and negative correlations of p53 mutations (Kigawa and Terakawa, 2000; Yazlovitskaya et al., 2001; Ganjavi et al., 2005). Another mechanism for CDDP resistance in squamous cell carcinoma is dysregulation of Bcl-2 (Chanvorachote et al., 2006), which impairs the mitochondrial apoptotic function by neutralizing the proapoptotic Bcl-2 family members such as Bax and Bak. In HNSCC, high endogenous tumor-associated Bcl-2 expression promoted CDDP resistance. In contrast, endogenous Bcl-XL showed no correlation with CDDP sensitivity and tumor recurrence (Michaud et al., 2009). In a similar vein, utilizing KiSS1 expression in patients with HNSCC as a biomarker may be useful for predicting poor outcomes or resistance to CDDP therapy, warranting an alternative treatment paradigm. Although apoptotic resistance has been established as a mechanism of CDDP resistance in various cancers (Niedner et al., 2001; Stewart, 2007), a link between KiSS1, apoptosis resistance and therapy resistance have not been identified to date. Extensive data support the role of KiSS1 as a metastasis suppressor, although recent data suggest that high levels of KiSS1 are associated with adverse outcomes among patients with estrogen-positive breast cancers (Marot et al., 2007). These seemingly contradictory findings perhaps may be resolved by evaluating these studies through the lens of the results shown here, wherein we showed that therapy-resistant tumors have suppressed levels of KiSS1. As biochemical and genetic modification of KiSS1 expression directly impacts the response of tumors to platinum-based chemotherapy through alterations in both apoptotic and drug metabolism pathways, these intracellular systems may also impart KiSS1-mediated anti-metastatic phenotype as well.
Previous work has shown that the decreased expression of KiSS-1 and the increased metastatic potential of melanoma were associated with the loss of heterozygosity at chromosome 6q16.3–q23, a locus mapped to the location of KiSS1 transcriptional co-activators, including CRSP3, DRIP130 and SP1 (Goldberg et al., 2003; Mitchell et al., 2007). Although in the specific melanoma model used by Goldberg et al. (2003), loss of heterozygosity at the 6q16.3–q23 locus and the resulting loss of the regulatory elements on chromosome 6 led to the loss of KiSS-1, in other models and metastatic tumors, loss of KiSS-1 is not associated with loss of heterozygosity. In this study, we have conducted CGH analysis on parental and CR cell lines and did not see any alteration of the chromosome, suggesting that alternate mechanisms may exist that impair the transcription and expression of KiSS-1. Although we have not yet studied the relative expression of the regulatory elements, we strongly believe that long-term exposure of cells to CDDP alter the molecular interactions of these regulatory elements, thereby inhibiting the transcription of KiSS-1. It is conceivable that other mechanisms, including DNA methylation and miRNA regulation, result in the same phenotypic changes associated with the loss of the regulatory elements, as has already been shown for other chemotherapy-resistant tumors (Kim et al., 2010). Future studies on the mechanism for KiSS1 loss will elucidate this issue.
CDDP detoxification is commonly associated with the development of drug resistance through increased activity of GST-π, which is a member of the superfamily of multifunctional enzymes that have key roles in cellular detoxification (Di Pietro et al., 2010). GST-π has been associated with the development of CDDP resistance in several cancer types, including ovarian cancer (Masanek et al., 1997), head and neck cancer (Cullen et al., 2003) and lung cancer (Bai et al., 1996). In agreement with previous studies, this study also showed that CR HNSCC cells have an elevated level of GST-π. Several transcriptional mechanisms, including p53 and NF-κB, have been shown to regulate GST-π expression in chemo-resistant cell lines under different experimental models (Moffat et al., 1996; Antoun et al., 2000; Lo et al., 2008). Although a p53–GST-π interaction have a significant role in the protection of cellular genome against CDDP, our experimental models lack p53, which suggests that an alternative transcription factor act in the regulation of GST-π. Morceau et al. (2004) investigated the regulation of GST-π, and their study revealed a tumor necrosis factor-α-inducible NF-κB binding motif within the promoter region of GST-π that is responsible for the activation of the GST-π expression. (Moffat et al., 1996). Here we show that the loss of KiSS1 induced by long-term exposure of cancer cells to CDDP was associated with decreased transcription of GST-π, potentially through NF-κB. Further, we show for the first time that the enzymatic activity of total GST-π is negatively regulated by the level of KISS-1 expression in cells selected for CDDP resistance. This result further confirms that GST-π have an important role in the protection of chemo-resistant cell lines and promotion of tumor proliferation.
The cardinal process of apoptosis involves a cascade of reactions that culminate in the activation of effector caspases (caspase-3, -6 and -7), which ultimately mediate the enzymatic cleavage of a wide range of substrates including PARP1 (Rouleau et al., 2010), a nuclear enzyme that is induced in response to DNA damage to facilitate base excision repair of DNA single-strand breaks (Hu et al., 2005). A number of studies have suggested that NF-κB have a central role in cell survival by inhibiting apoptosis and by regulating multitude of critical genes responsible for cell proliferation, metastasis and chemotherapy resistance (Karin and Lin, 2002; Li and Stark, 2002). In this study, we show that genetic alterations in KiSS1 levels can profoundly impact the expression of NF-κB and potentiate the ability of cells to terminally effect apoptosis by PARP1 cleavage. Potentially through its activation of downstream mediators, KiSS1 can modulate distinct members of the intrinsic pathway, although a putative mechanism has not been established. As shown previously, G-protein-coupled receptor-54 can activate downstream extracellular signal-regulated kinase (Navenot et al., 2009b) and Rho kinase (Navenot et al., 2009a) pathways, thus inducing apoptosis, findings that are in line with our current data. We extend these findings to show that this may be an autocrine effect, one in which the presence of KiSS1 enhances CDDP-induced apoptosis. Further, the ability of dormant tumor cells to survive in the metastatic niche may be due in part to their ability to upregulate antiapoptotic and drug metabolism genes as a result of KiSS1 loss. These observations become even more significant in light of recent trends to use PARP1 inhibitors, often in combination with platinum compounds, for various cancers (Fong et al., 2009; Khan et al., 2010). A putative therapeutic approach may involve indirectly upregulating KiSS1 expression to induce drug sensitivity, or directly introducing recombinant KiSS1 to re-establish CDDP sensitivity.
Conclusions
In summary, our data suggest that KiSS1 may be an important mechanistic regulator of the phenotypic behavior of treatment resistance in HNSCC. These include: (1) the acquisition of enhanced metastatic potential and differential KiSS1 expression in CR HNSCC; (2) the loss of KiSS1 in CDDP-sensitive cells induces chemoresistance; (3) restoration of KiSS1 abrogates the metastatic phenotype and sensitizes cells to CDDP; (4) the induction of an epithelial–mesenchymal transition-like transcriptional program in CR tumor cells; and (5) the loss of KISS1 in CR cell lines leads to impairment of the apoptotic machinery and enhanced CDDP metabolism. Collectively, these data suggest that KiSS1 is required for CDDP sensitivity and may regulate metastatic potential, which may be mediated in part by altered apoptotic response and CDDP detoxification.
Materials and methods
Cell lines and reagents
The HNSCC cell lines were maintained in Dulbecco's modied Eagle's medium supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 1 mm sodium pyruvate, 0.1 mm minimum essential medium non-essential amino acids, minimum essential medium vitamin solution and penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA). pCDNA/3.1-KiSS1 was obtained from Dr D Boyd (Yan et al., 2001) and empty vector of pCDNA/3.1 was purchased from Invitrogen. Validated siRNA against KiSS1 (no. 4392420) was purchased from Ambion (Applied Biosystems, Austin, TX, USA). For endpoint PCR analysis, PCR primer sets for KiSS1 (VHPS-1609) and GAPDH (VHPS-3541) were purchased from Real Time Primers (Elkins Park, PA, USA). For PCR rray analysis, First Strand Synthesis kit (C03), Cyber green master mix (PA-011) and RT2 Profiler PCR Array plates (PAHS-028) were purchased from SB Biosciences (Frederick, MD, USA). The following antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA) goat anti-rabbit antibody (Santa Cruz; Sc-2004); and goat anti-mouse antibody (Sc-2005) KiSS-1 (Sc-15400). GST-π (3369), cleaved PARP (9541) and Bax (2772) were purchased from Cell signaling (Danvers, MA, USA). GAPDH (AM4300) was purchased from Ambion (Applied Biosystems).
siRNA and plasmid transfections
Cells were seeded in 10-cm dishes and grown overnight to 70% confluency, trypsinized and transfected with siRNA targeting KiSS1, GAPDH or a non-targeting construct, using Amaxa nucleofector (Lonza, Portsmouth, NH, USA). Total RNA and proteins were isolated at 24 and 72 h post-transfection, respectively. For stable transfection of KiSS1, cells were seeded overnight and transfected with either pCDNA/3.1 or pCDNA/3.1 containing KiSS1, using Lipofectamine according to the procedures of the manufacturer (Invitrogen). Selection of transfectants was started after 24 h with G418 (Invitrogen). Selection medium was changed every 3 days for 2 weeks, and the transfected cell lines were maintained in G418 (0.5 mg/ml). KiSS1 overexpression was confirmed by sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE).
Western blotting
All western blotting analysis was performed as described previously (Kupferman et al., 2009). Densitometry data were analyzed by using the Fujifilm Multigauge Software (Stamford, CT, USA), and statistical analysis was performed by utilizing either conventional Student's t-test or analysis of variance (ANOVA), followed by post hoc comparisons based on modified Newman–Keuls–Student procedure, where appropriate. Results are reported as mean±s.e.m. A P-value <0.05 was considered significant and all are two-tailed.
Experimental animals and orthotopic implantation of tumor cells
Male athymic nude mice (NCI-nu), aged 8–12 weeks, were purchased from the Animal Production Area of the National Cancer Institute-Frederick Cancer Research and Development Center (Frederick, MD, USA). The mice were used in accordance with Animal Care and Use Guidelines of The University of Texas MD Anderson Cancer Center under a protocol approved by the Institutional Animal Care and Use Committee. For establishment of HNSCC tumors, cells were injected into either the tongue or thigh of athymic nude mice with 5 × 104 cells, as described previously (Kupferman et al., 2010). Tumor volume was measured weekly, and the differences between tumor volumes were evaluated by the non-parametric Mann–Whitney test. Results are reported as mean±s.e.m. A P-value <0.05 was considered significant. Survival was calculated by the method of Kaplan and Meier. For the experimental model of metastasis, 1 × 105 cells were injected into the tail vein of mice. At the end of all experiments, mice were killed and tissues were collected for hematoxylin and eosin staining and immunohistochemistry analysis. Terminal deoxynucleotidyl transferase dUTP nick-end labeling and Ki67 immunohistochemistry staining and analysis were performed as described previously (Kupferman et al., 2010).
Wound healing assay
Wound healing assay was performed using IBIDI (Troy, New York, USA) culture insert method in a six-well plate, as described previously (Liang et al., 2007). Cells (3 × 104) were seeded in each of the reservoirs, grown overnight and wound closure measurements were taken 24 h following removal of the inserts. Experiments were performed in triplicate and repeated three times. Measurements were taken at indicated time points and were quantified with ImagePro. Differences between groups were analyzed by utilizing either conventional Student's t-test or ANOVA followed by post hoc comparisons based on Bonferroni's Multiple Comparison Test, where appropriate. Results are reported as mean±s.e.m. A P-value <0.05 was considered significant, and all are two-tailed.
In vitro GST activity assay
Total (cytosolic and microsomal) GST activity was measured by the conjugation of 1-chloro-2,4-dinitrobenzene (CDNB) with reduced glutathione following the procedures of the manufacturer (Sigma-Aldrich, St Louis, MO, USA). Briefly, cells were homogenized in phosphate-buffered saline by ultrasonic disruption for three pulses of 5 s each using Fisher's Sonic Dismembrator (Fisher, Pennsylvania, PA, USA). Total protein concentration was determined by using RC DC Protein Assay kit (BioRad, Hercules, CA, USA). Aliquots of cell homogenates (200 μG) were mixed with 1× sample buffer and added to a GST reaction substrate master mix (Dulbecco's phosphate-buffered saline, 2 mm reduced glutathione and 1 mm CDNB). Conjugation of substrate was measured by calorimetric reading at 340 nm at intervals of 1 min for 7 min using DU 640B spectrophotometer (Beckman, Brea, CA, USA). Measurements were recorded, and the change in absorbance was determined by plotting the absorbance values against time. Specific GST activity (μm/ml/min) was calculated dividing the absorbance change per minute with the extinction coefficient for CDNB (A = 9.6 per mm) and the total protein content of the cell homogenate. Differences between groups were analyzed by utilizing ANOVA, followed by post hoc comparisons based on Bonferroni's Multiple Comparison Test, where appropriate. Results are reported as mean±s.e.m. A P-value <0.05 was considered significant, and all are two-tailed.
RNA isolation, RT–PCR and PCR array
RNA was isolated from cells by using Qiagen RNeasy Mini Kit (Valencia, CA, USA) following the manufacturer's protocol. The quality and quantity of RNA was analyzed with the Nanodrop (ND-1000) spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA) at the absorbance ratio of 260 and 280 nm. The single-strand cDNA was synthesized from 2 μg of total RNA by using RT2 First Strand synthesis Kit following the manufacturer's instructions (SA Biosciences, Frederick, MD, USA; C-03). For gene expression array analysis, quantitative RT–PCR was carried out with Bio-Rad CFX96 RT–PCR Detection system (Hercules, CA, USA). Comparison of the relative expression of genes was characterized by using Human Tumor Metastasis RT2 Profiler PCR Array (SABiosciences, Frederick, MD, USA; catalog no. PHAS-028). Experiments were repeated three times, and the data was analyzed using Excel-based PCR Array Data Analysis Templates (SABiosciences). Confirmatory RT–PCR was performed by using 0.5 μg of total RNA. Human tumors were collected from patients undergoing surgical resection under an institutional-approved protocol. RNA was extracted and RT–PCR was performed with the following primers (Real Time Primers, Elkins Park, PA, USA) as described previously (O'Donnell et al., 2005): KiSS1 (F): CTT GGC AGC TAC TGC TTT TC, (R) GTA GCA GCT GGC TTC CTC TC; GAPDH (F) GAG TCA ACG GAT TTG GTC GT, (R) TTG ATT TTG GAG GGA TCT CG.
Densitometry data were analyzed by using the Fujifilm Multigauge Software, and expression of KiSS1 was normalized to GAPDH levels. Statistical analysis was performed by utilizing conventional Student's t-test. Results are reported as mean±s.e.m. A P-value <0.05 was considered significant and all are two-tailed.
Array CGH experiments
Genomic DNA was extracted from HNSCC cell lines (Qiagen) and analyzed as described previously. DNA (1 mg) from the parental and CR cell lines were used as the sample and reference, respectively, and were differentially labeled with either cyanine 3-deoxycytidine triphosphate or cyanine 5-deoxycytidine triphosphate (Perkin-Elmer, Boston, MA, USA) by random prime labeling. DNA samples were then aliquoted onto SpectralChip 2600 arrays (Perkin-Elmer) and hybridized. Images of the arrays were captured using a two-laser ScanArray scanner (Perkin-Elmer) and quantitated using GenePix software (Molecular Devices, Sunnyvale, CA, USA). The data were analyzed and reports were generated using SpectralWare analysis software (Perkin-Elmer).
Cell proliferation assay
OSC19, OSC19-CR and OSC19-CR/KISS1 cells were seeded at 1 × 105 cells per well in six-well plates and aliquots were counted every 24 h by Coulter counter for five consecutive days. Results are reported as mean±s.e.m. A P-value <0.05 was considered significant.
Soft agar assay
Six-well culture plates were coated with 1 ml bottom agar mixture (Dulbecco's modied Eagle's medium with 20% fetal bovine serum, 0.6% agar) and allowed to solidify at room temperature. Cells (3 × 103) were suspended in Dulbecco's modied Eagle's medium agar mixture (20% fetal bovine serum, 0.3% agar) and plated over the bottom layer of the 0.6% agarmedium mixture. After 12 days, plates were stained with 0.005% crystal violet and colonies larger than 100 μm were counted.
Supplementary Material
Acknowledgements
We thank Yongxiang Li BS for technical assistance and Terri Astin for administrative assistance. We also thank the members of the Small Animal Imaging Facility for their assistance with the mouse studies, Taylor Appleberry for assistance with the array CGH studies and Dr Douglas Boyd (MDACC) for providing the KiSS1 construct. This work is supported by the following funding sources: NIH Grant K08-DE019185 (MEK), Triological Society Career Development Award (MEK), MD Anderson Cancer Center Physician-Scientist Program (MEK) and by the RNR Cross Foundation (TJ, EH). This research is also supported in part by the National Institutes of Health through MD Anderson's Cancer Center Support Grant CA016672.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Antoun G, Baylin SB, Ali-Osman F. DNA methyltransferase levels and altered CpG methylation in the total genome and in the GSTP1 gene in human glioma cells transfected with sense and antisense DNA methyltransferase cDNA. J Cell Biochem. 2000;77:372–381. [PubMed] [Google Scholar]
- Arai T, Yasuda Y, Takaya T, Hayakawa K, Toshima S, Shibuya C, et al. Immunohistochemical expression of glutathione transferase-pi in untreated primary non-small-cell lung cancer. Cancer Detect Prev. 2000;24:252–257. [PubMed] [Google Scholar]
- Bai F, Nakanishi Y, Kawasaki M, Takayama K, Yatsunami J, Pei XH, et al. Immunohistochemical expression of glutathione S-transferase-Pi can predict chemotherapy response in patients with nonsmall cell lung carcinoma. Cancer. 1996;78:416–421. doi: 10.1002/(SICI)1097-0142(19960801)78:3<416::AID-CNCR6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Chanvorachote P, Nimmannit U, Stehlik C, Wang L, Jiang BH, Ongpipatanakul B, et al. Nitric oxide regulates cell sensitivity to cisplatin-induced apoptosis through S-nitrosylation and inhibition of Bcl-2 ubiquitination. Cancer Res. 2006;66:6353–6360. doi: 10.1158/0008-5472.CAN-05-4533. [DOI] [PubMed] [Google Scholar]
- Cho S-G, Yi Z, Pang X, Yi T, Wang Y, Luo J, et al. Kisspeptin-10, a KISS1-derived decapeptide, inhibits tumor angiogenesis by suppressing Sp1-mediated VEGF expression and FAK/Rho GTPase activation. Cancer Res. 2009;69:7062–7070. doi: 10.1158/0008-5472.CAN-09-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JS, Pajak TF, Forastiere AA, Jacobs J, Campbell BH, Saxman SB, et al. Postoperative concurrent radiotherapy and chemotherapy for high-risk squamous-cell carcinoma of the head and neck. N Engl J Med. 2004;350:1937–1944. doi: 10.1056/NEJMoa032646. [DOI] [PubMed] [Google Scholar]
- Cullen KJ, Newkirk KA, Schumaker LM, Aldosari N, Rone JD, Haddad BR. Glutathione S-transferase pi amplification is associated with cisplatin resistance in head and neck squamous cell carcinoma cell lines and primary tumors. Cancer Res. 2003;63:8097–8102. [PubMed] [Google Scholar]
- Di Pietro G, Magno LA, Rios-Santos F. Glutathione S-transferases: an overview in cancer research. Expert Opin Drug Metab Toxicol. 2010;6:153–170. doi: 10.1517/17425250903427980. [DOI] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Forastiere AA, Goepfert H, Maor M, Pajak TF, Weber R, Morrison W, et al. Concurrent chemotherapy and radiotherapy for organ preservation in advanced laryngeal cancer. N Engl J Med. 2003;349:2091–2098. doi: 10.1056/NEJMoa031317. [DOI] [PubMed] [Google Scholar]
- Ganjavi H, Gee M, Narendran A, Freedman MH, Malkin D. Adenovirus-mediated p53 gene therapy in pediatric soft-tissue sarcoma cell lines: sensitization to cisplatin and doxorubicin. Cancer Gene Ther. 2005;12:397–406. doi: 10.1038/sj.cgt.7700798. [DOI] [PubMed] [Google Scholar]
- Goldberg SF, Miele ME, Hatta N, Takata M, Paquette-Straub C, Freedman LP, et al. Melanoma metastasis suppression by chromosome 6: evidence for a pathway regulated by CRSP3 and TXNIP. Cancer Res. 2003;63:432–440. [PubMed] [Google Scholar]
- Hamaguchi K, Godwin AK, Yakushiji M, O'Dwyer PJ, Ozols RF, Hamilton TC. Cross-resistance to diverse drugs is associated with primary cisplatin resistance in ovarian cancer cell lines. Cancer Res. 1993;53:5225–5232. [PubMed] [Google Scholar]
- Hu H, Jiang C, Ip C, Rustum YM, La J. Methylseleninic acid potentiates apoptosis induced by chemotherapeutic drugs in androgen-independent prostate cancer cells. Clin Cancer Res. 2005;11:2379–2388. doi: 10.1158/1078-0432.CCR-04-2084. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Berk M, Singh LS, Tan H, Yin L, Powell C, et al. KiSS1 suppresses metastasis in human ovarian cancer via inhibition of protein kinase C alpha. Clin Exp Metast. 2005;22:369–376. doi: 10.1007/s10585-005-8186-4. [DOI] [PubMed] [Google Scholar]
- Karin M, Lin A. NF-[kappa]B at the crossroads of life and death. Nat Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- Khan K, Araki K, Wang D, Li G, Li X, Zhang J, et al. Head and neck cancer radiosensitization by the novel poly(ADP-ribose) polymerase inhibitor GPI-15427. Head Neck. 2010;32:381–391. doi: 10.1002/hed.21195. [DOI] [PubMed] [Google Scholar]
- Kigawa J, Terakawa N. Adenovirus-mediated transfer of a p53 gene in ovarian cancer. Adv Exp Med Biol. 2000;465:207–214. doi: 10.1007/0-306-46817-4_19. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Kang HS, Jung SY, Min SY, Lee S, Kim SW, et al. Methylation patterns of genes coding for drug-metabolizing enzymes in tamoxifen-resistant breast cancer tissues. J Mol Med. 2010;88:1123–1131. doi: 10.1007/s00109-010-0652-z. [DOI] [PubMed] [Google Scholar]
- Kupferman ME, Jayakumar A, Zhou G, Xie T, Dakak-Yazici Y, Zhao M, et al. Therapeutic suppression of constitutive and inducible JAK\STAT activation in head and neck squamous cell carcinoma. J Exp Ther Oncol. 2009;8:117–127. [PubMed] [Google Scholar]
- Kupferman ME, Jiffar T, El-Naggar A, Yilmaz T, Zhou G, Xie T, et al. TrkB induces EMT and has a key role in invasion of head and neck squamous cell carcinoma. Oncogene. 2010;29:2047–2059. doi: 10.1038/onc.2009.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Welch DR. Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene, KiSS-1. Cancer Res. 1997;57:2384–2387. [PubMed] [Google Scholar]
- Li X, Stark GR. NF[kappa]B-dependent signaling pathways. Exp Hematol. 2002;30:285–296. doi: 10.1016/s0301-472x(02)00777-4. [DOI] [PubMed] [Google Scholar]
- Liang C-C, Park AY, Guan J-L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protocols. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- Liu L-Z, Zhou X-D, Qian G, Shi X, Fang J, Jiang B-H. AKT1 amplification regulates cisplatin resistance in human lung cancer cells through the mammalian target of rapamycin/p70s6k1pathway. Cancer Res. 2007;67:6325–6332. doi: 10.1158/0008-5472.CAN-06-4261. [DOI] [PubMed] [Google Scholar]
- Lo HW, Stephenson L, Cao X, Milas M, Pollock R, Ali-Osman F. Identification and functional characterization of the human glutathione S-transferase P1 gene as a novel transcriptional target of the p53 tumor suppressor gene. Mol Cancer Res. 2008;6:843–850. doi: 10.1158/1541-7786.MCR-07-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marot D, Bieche I, Aumas C, Esselin S, Bouquet C, Vacher S, et al. High tumoral levels of Kiss1 and G-protein-coupled receptor 54 expression are correlated with poor prognosis of estrogen receptor-positive breast tumors. Endocr Relat Cancer. 2007;14:691–702. doi: 10.1677/ERC-07-0012. [DOI] [PubMed] [Google Scholar]
- Masanek U, Stammler G, Volm M. Messenger RNA expression of resistance proteins and related factors in human ovarian carcinoma cell lines resistant to doxorubicin, taxol and cisplatin. Anticancer Drugs. 1997;8:189–198. doi: 10.1097/00001813-199702000-00010. [DOI] [PubMed] [Google Scholar]
- Michaud WA, Nichols AC, Mroz EA, Faquin WC, Clark JR, Begum S, et al. Bcl-2 blocks cisplatin-induced apoptosis and predicts poor outcome following chemoradiation treatment in advanced oropharyngeal squamous cell carcinoma. Clin Cancer Res. 2009;15:1645–1654. doi: 10.1158/1078-0432.CCR-08-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DC, Stafford LJ, Li D, Bar-Eli M, Liu M. Transcriptional regulation of KiSS-1 gene expression in metastatic melanoma by specificity protein-1 and its coactivator DRIP-130. Oncogene. 2007;26:1739–1747. doi: 10.1038/sj.onc.1209963. [DOI] [PubMed] [Google Scholar]
- Moffat GJ, McLaren AW, Wolf CR. Functional characterization of the transcription silencer element located within the human Pi class glutathione S-transferase promoter. J Biol Chem. 1996;271:20740–20747. doi: 10.1074/jbc.271.34.20740. [DOI] [PubMed] [Google Scholar]
- Morceau F, Duvoix A, Delhalle S, Schnekenburger M, Dicato M, Diederich M. Regulation of glutathione S-transferase P1-1 gene expression by NF-kappaB in tumor necrosis factor alpha-treated K562 leukemia cells. Biochem Pharmacol. 2004;67:1227–1238. doi: 10.1016/j.bcp.2003.10.036. [DOI] [PubMed] [Google Scholar]
- Nash KT, Phadke PA, Navenot J-M, Hurst DR, Accavitti-Loper MA, Sztul E, et al. Requirement of KISS1 secretion for multiple organ metastasis suppression and maintenance of tumor dormancy. J Natl Cancer Inst. 2007;99:309–321. doi: 10.1093/jnci/djk053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash KT, Welch DR. The KISS1 metastasis suppressor: mechanistic insights and clinical utility. Front Biosci. 2006;11:647–659. doi: 10.2741/1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navenot J-M, Fujii N, Peiper SC. Activation of Rho and Rho-associated kinase by GPR54 and KiSS1 metastasis suppressor gene product induces changes of cell morphology and contributes to apoptosis. Mol Pharmacol. 2009a;75:1300–1306. doi: 10.1124/mol.109.055095. [DOI] [PubMed] [Google Scholar]
- Navenot J-M, Fujii N, Peiper SC. KiSS1 metastasis suppressor gene product induces suppression of tyrosine kinase receptor signaling to akt, tumor necrosis factor family ligand expression, and apoptosis. Mol Pharmacol. 2009b;75:1074–1083. doi: 10.1124/mol.108.054270. [DOI] [PubMed] [Google Scholar]
- Niedner H, Christen R, Lin X, Kondo A, Howell SB. Identification of genes that mediate sensitivity to cisplatin. Mol Pharmacol. 2001;60:1153–1160. [PubMed] [Google Scholar]
- O'Donnell RK, Kupferman M, Wei SJ, Singhal S, Weber R, O'Malley B, et al. Gene expression signature predicts lymphatic metastasis in squamous cell carcinoma of the oral cavity. Oncogene. 2005;24:1244–1251. doi: 10.1038/sj.onc.1208285. [DOI] [PubMed] [Google Scholar]
- Ohtaki T, Shintani Y, Honda S, Matsumoto H, Hori A, Kanehashi K, et al. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411:613–617. doi: 10.1038/35079135. [DOI] [PubMed] [Google Scholar]
- Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi T. p53 alterations in human cancer: more questions than answers. Oncogene. 2007;26:2145–2156. doi: 10.1038/sj.onc.1210280. [DOI] [PubMed] [Google Scholar]
- Soussi T, Wiman KG. Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell. 2007;12:303–312. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Stewart DJ. Mechanisms of resistance to cisplatin and carboplatin. Crit Rev Oncol Hematol. 2007;63:12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Sung-Gook C, Dali L, Lewis JS, Jian L, Melissa R-V, Ying W, et al. KiSS1 suppresses TNFalpha-induced breast cancer cell invasion via an inhibition of RhoA-mediated NF-kappaB activation. J Cell Biochem. 2009;107:1139–1149. doi: 10.1002/jcb.22216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch DR, Chen P, Miele ME, McGary CT, Bower JM, Stanbridge EJ, et al. Microcell-mediated transfer of chromosome 6 into metastatic human C8161 melanoma cells suppresses metastasis but does not inhibit tumorigenicity. Oncogene. 1994;9:255–262. [PubMed] [Google Scholar]
- Yan C, Wang H, Boyd DD. KiSS-1 represses 92-kDa type IV collagenase expression by down-regulating NF-kappa B binding to the promoter as a consequence of Ikappa Balpha-induced block of p65/p50 nuclear translocation. J Biol Chem. 2001;276:1164–1172. doi: 10.1074/jbc.M008681200. [DOI] [PubMed] [Google Scholar]
- Yazlovitskaya EM, DeHaan RD, Persons DL. Prolonged wild-type p53 protein accumulation and cisplatin resistance. Biochem Biophys Res Commun. 2001;283:732–737. doi: 10.1006/bbrc.2001.4849. [DOI] [PubMed] [Google Scholar]
- Yilmaz T, Jiffar T, de la Garza G, Lin H, MacIntyre T, Brown JL, et al. Therapeutic targeting of Trk suppresses tumor proliferation and enhances cisplatin activity in HNSCC. Cancer Biol Ther. 2010;10:644–653. doi: 10.4161/cbt.10.6.12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.