Abstract
Background and Aims
Connective tissue growth factor (CTGF) expression is intimately associated with hepatic fibrotic pathophysiology. In this study, CTGF production and action was investigated in ethanol-treated mouse primary hepatic stellate cells (HSC) or human LX-2 cells.
Methods
CTGF, transforming growth factor-beta1 (TGF-β1), alpha-smooth muscle actin (α-SMA) or collagen α1(I) mRNA were quantified by real-time PCR after treatment of HSC with ethanol or acetaldehyde. CTGF protein production was assessed by immunoprecipitation or ELISA. Ethanol-stimulated CTGF transcription was investigated using CTGF promoter reporter constructs. The TGF-β1- or CTGF-dependency of ethanol-induced CTGF, α-SMA or collagen α1(I) was determined using small interfering RNA (siRNA) to TGF-β1 or CTGF.
Results
In human steatohepatitis, CTGF was produced by presumptive activated HSC. In cultured human or mouse HSC, production of CTGF, α-SMA and/or collagen was increased by ethanol treatment, an effect mimicked by acetaldehyde and blocked by 4-methylpyrazole (4-MP) or N-acetylcysteine (NAC). CTGF promoter activity was stimulated in a sustained fashion by ethanol or TGF-β1. Mutation of the Smad site or basal control element (BCE-1) in the CTGF promoter caused a 5-fold reduction in ethanol-stimulated CTGF promoter activity. Administration of TGF-β1 siRNA or CTGF siRNA significantly decreased ethanol- or acetaldehyde-stimulated mRNA or protein levels of CTGF, α-SMA or collagen I in LX-2 cells. In mouse HSC, TGF-β1- or ethanol-stimulated CTGF, α-SMA or collagen I were significantly attenuated by CTGF siRNA.
Conclusions
Ethanol-induced α-SMA or collagen α1(I) in HSC are mediated via TGF-β-dependent CTGF production, highlighting potential therapeutic benefits of targeting CTGF in alcoholic liver disease.
Introduction
Excessive chronic alcohol consumption is a leading cause of hepatic fibrosis, which is a major cause of morbidity and mortality worldwide. More than 10% of the US population abuse or are dependent on alcohol resulting in ~27,000 cases of alcoholic liver disease each year [1]. Substantial progress has been made over the last 10–20 years in understanding some of the pathophysiological mechanisms that underlie alcoholic liver fibrosis. Activation of hepatic stellate cells (HSC) is a key event by which this otherwise quiescent cell type expresses α-smooth muscle actin (α-SMA), assumes a myofibroblastic phenotype, and synthesizes fibrillar collagens [2]. In acute liver injury, this response is transient and an important component of wound repair by facilitating matrix contraction and restoration. During chronic liver injury, the activated phenotype persists resulting in an unabated deposition of fibrillar collagens into the interstitial spaces which ultimately compromises normal hepatic function [2]. The initiation and perpetuation of HSC activation is regulated by many signaling molecules including, in particular, transforming growth factor-β (TGF-β) [3]. However, since TGF-β has important immunomodulatory functions and acts a tumor suppressor for epithelial cells (hepatocytes), it is a difficult molecule to effectively target in fibrotic liver disease.
Connective tissue growth factor (CTGF) has emerged as an alternative candidate for therapeutic interventions because it is often activated as a TGF-β immediate early gene and is itself pro-fibrotic [4–7]. CTGF stimulates mitogenesis, chemotaxis, adhesion and matrigenesis in cultured HSC, is expressed by activated HSC in fibrotic liver, and is produced by HSC in vitro as a function of their activation or after exposure of the cells to TGF-β [5, 8, 9]. Here, we investigated mechanisms of ethanol-mediated CTGF production and action in the human LX-2 HSC line [10] and in cultures of primary mouse HSC.
Materials and Methods
Human Liver Samples
Samples of human alcoholic liver disease (n=6) were biopsy or surgical specimens archived at Harbor-UCLA Medical Center (Torrance, CA. USA). Tissues were fixed in buffered zinc formalin, sectioned to 4 μm, and processed for CTGF or α-SMA immunohistochemistry.
Cell Culture
LX-2 cells (from Scott L. Friedman, Mount Sinai School of Medicine, New York, NY, USA) were grown for 24 hours in DMEM / 10% FBS and then cultured in serum-free medium for the next 24 hours. Primary HSCs from male Swiss Webster mice were isolated [9] and cultured for up to 10 days before incubation in DMEM / 1% FBS for 12 hours. Triplicate wells of cells were incubated for up to 48 hours in the presence of 0 – 25 mM ethanol or 0–100 μM acetaldehyde. In some experiments, cells were also treated with 2mM 4-methylpyrazole (4-MP; an inhibitor of ethanol oxidation via inhibition of alcohol dehydrogenase I (ADHI)) or 1mM N-acetyl-l-cysteine (NAC; a reactive oxygen species scavenger). Cells were then evaluated for CTGF, α-SMA, TGF-β1 or collagen α1(I) mRNA expression or CTGF protein production.
RNA extraction and real-time polymerase chain reaction (PCR)
Total RNA was extracted using RNA STAT-60 (TEL-TEST, Friendswood, TX, USA) according to the manufacturer’s protocol. First-strand cDNA was synthesized using 2 μg of total RNA in a 20μl final volume by reverse transcription utilizing SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) with oligo-dT(18)-primers (Invitrogen, Carlsbad, CA, USA). The real-time PCR reactions were performed using SYBR Green Master Mix kit according to the manufacturer’s instructions (Applied Biosystems, Forster City, CA, USA). RNA for CTGF, TGF-β1, α-SMA, or collagen α1(I) were amplified using ABI Prism 7000 Sequence Detection System (Applied Biosystems, Forster City, CA, USA). The primers (Invitrogen, Carlsbad, CA, USA) are shown in Table 1. For all real-time PCR studies, negative controls were a non-reverse transcriptase reaction and a non-sample reaction (data not shown). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified as an internal standard.
Table 1.
Primer sequences for amplification of human or mouse CTGF, TGF-β1, α-SMA, collagen α1(I) or GAPDH
| Gene | GeneBank accession number | Primers | Product size (bp) | |
|---|---|---|---|---|
| Sense | Anti-sense | |||
| CTGF (human) | NM_001901 | 5’AATGCTGCGAGGAGTGGGT3’ | 5’CGGCTCTAATCATAGTTGGGTCT3’ | 94 |
| CTGF (mouse) | NM_010217 | 5’CACTCTGCCAGTGGAGTTCA3’ | 5’AAGATGTCATTGTCCCCAGG3’ | 111 |
| TGF-β1 (human) | NM_000660 | 5’ACCTGAACCCGTGTTGCTCT3’ | 5’CTAAGGCGAAAGCCCTCAAT3’ | 208 |
| α-SMA (human) | NM_030059 | 5’GATGGGCATCTATCAGATAC3’ | 5’AAGCATTTCTGATGGTGATG3’ | 115 |
| α-SMA (mouse) | NM_007392 | 5’GGCTCTGGGCTCTGTAAGG3’ | 5’CTCTTGCTCTGGGCTTCATC3’ | 148 |
| Collagen α1(I) (human) | NM_000088 | 5’GAACGCGTGTCATCCCTTGT3’ | 5’GAACGAGGTAGTCTTTCAGCAACA3’ | 91 |
| Collagen α1(I) (mouse) | NM_007742 | 5’GCCCGAACCCCAAGGAAAAGAAGC3’ | 5’CTGGGAGGCCTCGGTGGACATTAG3’ | 148 |
| GAPDH (human) | NM_002046 | 5’TGCACCACCAACTGCTTAGC3’ | 5’GGCATGGACTGTGGTCATGAG3’ | 66 |
CTGF Radioimmunoprecipitation Assay and ELISA
LX-2 cells were cultured in cysteine- and methionine-free DMEM (GIBCO-Invitrogen Carlsbad, CA, USA) containing 100μCi /ml [35S]methionine/cysteine (MP Biomedicals, Irvine, CA, USA) in the presence of 0 – 25mM ethanol for 48h. Cell lysates (1mg/ml) were incubated at 4°C for 12 hours with 190 μg/ml CTGF antiserum (“NH3”; an in-house IgY raised against recombinant human CTGF) and immunoprecipated for 2h at room temperature using IgY precipitating agarose beads (Millipore, Temecula, CA, USA). Immunoprecipitates were analyzed on 18% SDS-PAGE gels, dried and exposed to X-ray film (Amersham Biosciences, Piscataway, NJ, USA). Supernatants of LX-2 cell lysates were analyzed using a human CTGF ELISA (Antigenix America, Huntington Station, NY).
Transfection of LX-2 cells with CTGF promoter reporter constructs
LX-2 cells were cultivated for 24h in 12-well plates in DMEM with 10% FBS medium at 37°C and then transfected using Lipofectamine TM 2000 (Invitrogen, Carlsbad, CA, USA) at a ratio of 3μl:1μg DNA per well. Cells were transfected (3μg/well) with plasmids containing a secreted alkaline phosphatise (SEAP) reporter gene fused to the wild type CTGF promoter (nucleotides −805 to +17), deletion mutant: −805 to −166 or −805 to −86, or individual point mutants targeting the Smad site or the basal control element (BCE-1) [11]. SEAP reporter activity was calculated after adjustment for differences among samples in transfection efficiency as determined by co-transfection with a cytomegalovirus (CMV) promoter-β-galactosidase (CMV-β-gal) reporter gene. Post-transfection, cells were serum-starved for 18h prior to addition of 0–50 mM ethanol or 0–20 ng/ml TGF-β1 for 0–240 mins. CTGF promoter activity assays were performed with Phospha-Light kit (Applied Biosystems, Forster City, CA, USA) to detect SEAP reporter expression and β-galactosidase expression was determined by Galacto-star kit (Applied Biosystems). SEAP levels were measured using a LMax II 384 luminometer (Molecular Devices, Sunnyvale, CA, USA).
SiRNA suppression of TGF-β1 or CTGF in LX-2 cells or primary mouse HSC
Human TGF-β1 or CTGF GIPZ lentiviral shRNAmir plasmids or negative scramble siRNA were obtained from Open Biosystem (Huntsville, AL, USA). The human TGF-β1 siRNA target sequences were 5’-ACGAGCCCTGGACACCAACTAT-3’ (sense) and 3’-ATAGTTGGTGTCCAGGGCTCGG 5’ (antisense) and the human CTGF siRNA target sequences were 5’-CCAGCACCAGAATGTATATTAA-3’ (sense) and 3’-TTAATATACATTCTGGTGCTGT-5’ (antisense). Mouse CTGF siRNA was cloned into pRNAT-CMV3.1/Neo vector from GenScript Corporation (Piscataway, NJ, USA). Mouse CTGF siRNA target sequences were 5’-CGCAAGATCGGAGTGTGCTTCAAGAGAGCACACTCCGATCTTGCGGTT -3’ (sense) and 3’-GGCGTTCTAGCCTCACACGAAGTTCTCTCGTGTGAGGCTAGAACGCCAA-5’ (antisense). Cells were seeded in 35mm dishes (105 cells / dish) and transfected with 3.2 μg plasmid or negative scramble siRNA using either LipofectamineTM 2000 alone (LX-2 cells) or in combination with electroporation (primary mouse HSC) using a Nucleofector Kit (Lonza, Koln, Germnay). The transfected cells were incubated in medium containing 10% FBS for 12 hours and then switched to serum-free medium with or without test agents. Transfection efficiency in LX-2 cells was approximately 70% as monitored by co-transfection with 0.8μg of the green fluorescent protein (GFP) -expressing plasmid, pEGFP (Invitrogen, Carlsbad, CA, USA). Transfection efficiency in primary mouse HSC was approximately 10% as determined by co-expression of GFP contained in the pRNAT-CMV3.1/Neo vector.
Fluorescent-activated cell sorting (FACS)
Transfected cells were sorted using a Beckman Coulter Epics Altra cell sorter (HighWycombe), collected into DMEM/10%FBS, and plated into 6-well plates. At least 30,000 events were acquired for analysis. Cells were treated with 0 – 25mM ethanol for 48h and then treated with Trizol reagent (Gibco BRL) for RNA extraction or fixed with formalin for immunofluorescence assay.
Immunohistochemistry
Fixed LX-2 cells or tissue sections were incubated with NH1 anti-CTGF IgY (5μg/ml; see [12]), anti-α-SMA (1:100, Dako Cytomatio, Denmark) or anti-collagen I (1:250, Abcam, Cambridge, MA) followed by incubation with Alexa Fluor® 488 goat-anti chicken IgY (1:1000, Invitrogen, Carlsbad, CA), Alexa Fluor® 568 goat-anti rabbit IgG (1:1000, Invitrogen, Carlsbad, CA) and Alexa Fluor® 647 goat-anti mouse IgG (1:1000, Invitrogen, Carlsbad, CA) for 1 hour at room temperature. Next, the cells were mounted with Vectashield Mounting Medium with DAPI (Vector Laboratories, Burlingame, CA), and examined by confocal laser microscopy (LSM510, Carl Zeiss Co. Ltd, Jena, Germany).
Statistical Analysis
All experiments were performed at least three times with triplicate measurements, with data expressed as mean ± s.e.m. The intensity of immunoreactive bands on radioimmunoprecipitation was quantified by image analysis (Scion Corporation, Frederick, MD). These data, as well as data from real-time PCR, promoter activity assays and ELISA assay, were analyzed by student’s t-test using Sigma plot 11.0 software (SPSS Inc., Chicago, IL) and P values < 0.05 were considered statistically significant.
Results
Production of CTGF by activated HSC in human alcoholic liver disease
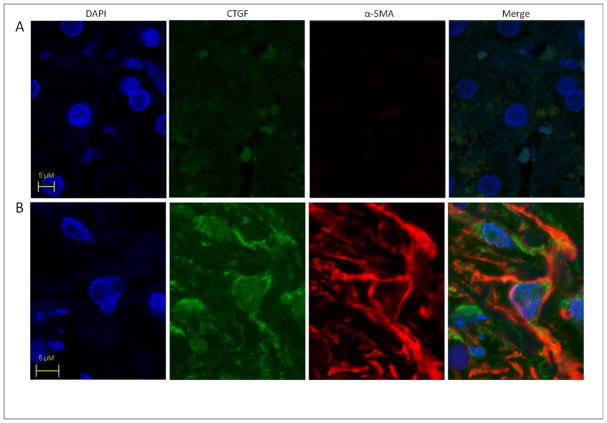
Fibrotic regions in livers from patients with alcoholic steatohepatitis contained myofiboblastic cells that strongly for both α-SMA and CTGF whereas this pattern of staining was absent in normal liver. Representative data are shown in Figure 1. Since these data showed that CTGF is produced by presumptive activated HSC in alcohol liver disease, we undertook detailed studies of ethanol-CTGF circuitry in human LX-2 cells in vitro.
Fig. 1. CTGF production by myofibroblasts in human steatohepatitis.
Sections of (A) normal human liver or (B) steatohepatitis with moderate activity and septal fibrosis stage 3 were stained with DAPI for nuclear localization and processed for CTGF and α-SMA immunohistochemistry. Images were merged to verify co-localization of CTGF and α-SMA to the same myofibroblastic cells (i.e. presumptive activated HSC) within the fibrotic regions (yellow-orange color). Data are representative of six samples of human alcoholic liver fibrosis.
Ethanol-induced CTGF, α-SMA or TGF-β1 mRNA expression and CTGF protein production in LX-2 cells
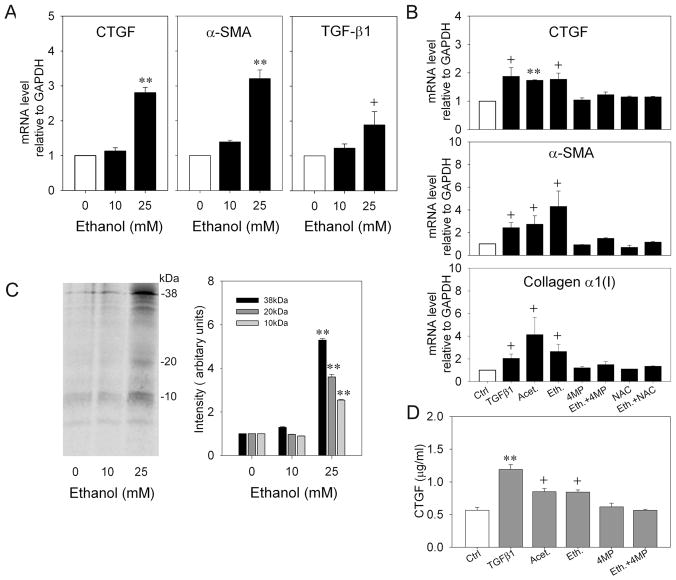
Initial studies showed that CTGF mRNA expression in LX-2 cells in response to ethanol was time-dependent, with peak stimulation at 48 hours (data not shown). As assessed by real-time PCR, CTGF mRNA levels in response to ethanol were also dose-dependent, and exposure of LX-2 cells to 25mM ethanol for 48 hours under reduced serum conditions caused a 2- to 4- fold increase (p<0.001) in expression of CTGF as compared to non-treated control cells (Figure 2A). To verify that CTGF protein production was also stimulated, LX-2 cells were labeled with [35S]methionine/cysteine during 48 hours of treatment with 0–25mM ethanol prior to immunoprecipitation of CTGF from the cell lysates. As shown in Figure 2C, CTGF-immunoreactive bands corresponding to the full length 38kDa protein and 20kDa or 10kDa cleavage products were induced 2.5- to 6-fold by ethanol (Figure 2C). These data were confirmed by CTGF ELISA of cell lysates (Figure 2D).
Fig.2. Effect of ethanol, TGF-β1 or acetaldehyde on CTGF, TGF-β1, α-SMA or collagen α 1(I) expression in LX-2 cells.
(A.B) Cultured LX-2 cells were serum-starved for 24h prior to 48-hour treatment with 0–25mM ethanol ± 2mM 4MP ± 1mM NAC, 5ng/ml TGF-β1 or 100μM acetaldehyde. RNA was subjected to real-time PCR and expression of CTGF, TGF-β1, α-SMA or collagen α 1(I) was normalized to that of GAPDH. (**p<0.001, + p<0.05 v no treatment). (C) LX-2 cells were labeled with 100μCi /ml [35S] methionine/cysteine in the presence or absence of 0–25mM ethanol for 48h. CTGF immunoprecipitates were run on 18% SDS-PAGE gels and analyzed by autoradiography. The figure shows a representative blot (left) and quantification of three independent experiments (right). (**p<0.001 v no ethanol). (D) Lysates from serum-starved LX-2 cells treated for 48h with 0–25mM ethanol ± 2mM 4MP ± 1mM NAC, 5ng/ml TGF-β1 or 100μM acetaldehyde were assayed for CTGF content by ELISA. (**p<0.001, + p<0.05 v Ctrl).
The increase in CTGF protein production or mRNA expression in ethanol-treated LX-2 cells was comparable to that elicited by 5ng/ml TGF-β or 100 μM acetaldehyde (Figure 2B, D). Since acetaldehyde is a principal product of ADH-mediated ethanol oxidation, LX-2 cells were treated with 4-MP, an inhibitor of ADHI. Ethanol-stimulated CTGF production was subsequently ablated by 4-MP whereas basal CTGF production was unaffected. Moreover, the expression of α-SMA or collagen α1(I) by LX-2 cells was also stimulated by TGF-β1, acetaldehyde or ethanol, the latter of which was also blocked by 4-MP (Figure 2B,D). Finally, since acetaldehyde can also be generated from ethanol by the action of cytochrome P450 2E1 (CYP 2E1) which also generates reactive oxygen species (ROS), LX-2 cells were treated with N-acetyl-cysteine (NAC), a broad-spectrum anti-oxidant. This treatment was effective in blocking ethanol-stimulated but not basal production of CTGF, α-SMA or collagen α1(I) (Figure 2B,D).
TGF-β1-induced CTGF expression in HSC depends on Smad and BCE elements in the CTGF promoter
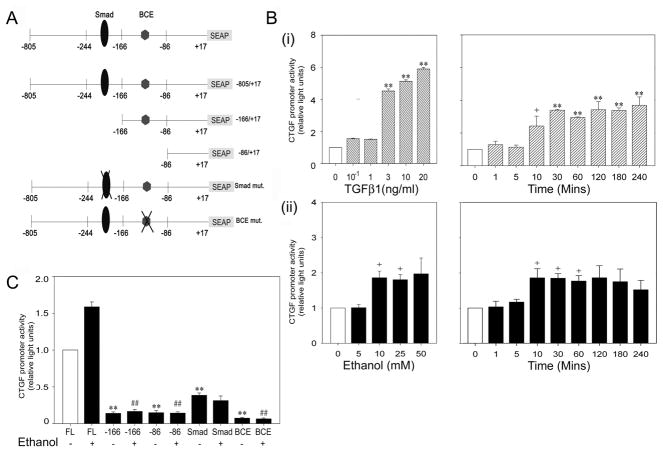
TGF-β1 mRNA in LX-2 cells was also stimulated by ethanol at similar concentrations to those required for CTGF production (Figure 2A). To verify that the CTGF promoter in LX-2 cells was TGF-β-responsive, cells were transfected with plasmids containing the CTGF promoter (nucleotides −805 to +17) fused to a SEAP reporter gene (Figure 3A) and subsequently stimulated with TGF-β1. CTGF promoter activity was stimulated up to 6-fold in a dose-dependent manner by 1–20ng/ml TGF-β1 while, in response to 10ng/ml TGF-β1, activation of the CTGF promoter by ~2-fold was evident after 10 minutes and maintained at a maximum ~3-fold increase between 30 and 240 minutes after stimulation (Figure 3B). CTGF promoter activity was stimulated 2-fold by 10–50 mM ethanol whereas 5 mM was ineffective and the increase in CTGF promoter activity by 10 mM ethanol was maximal at 10 minutes and maintained for 240 minutes (Figure 3B). Thus, either TGF-β1 or ethanol caused a rapid (within 10 minutes) and sustained (up to 240 minutes) activation of CTGF promoter activity, a temporal association that supported a possible functional link between TGF-β and ethanol in stimulating CTGF promoter activity. To address this question directly, LX-2 cells were transfected with CTGF promoter constructs that harbored deletion mutation downstream of the Smad or BCE response elements, or point mutations within these same elements (Figure 3A). As shown in Figure 3C, basal CTGF promoter activity was completely absent from the deletion mutants, showing that neither the sequence between nucleotides −166 and +17 or −86 and +17 (the latter of which contained the BCE element) were sufficient to drive the CTGF promoter under basal conditions. Also, the point mutation in either the Smad site or the BCE site resulted in significant attenuation of basal CTGF production. Somewhat higher levels of residual basal CTGF promoter activity was observed for the Smad mutants as compared to the BCE mutant, showing that this activity was Smad-independent but BCE-dependent. When cells were treated with 25mM ethanol, CTGF promoter activity was increased as expected, but this effect was absent from the deletion mutants and the BCE point mutant which exhibit the same, highly reduced levels of activity as under basal conditions. The Smad mutant exhibited slight but nonetheless substantially lower activity after ethanol treatment which was comparable to that exhibited by this mutant under basal conditions. These results showed that ethanol-stimulated activity in LX-2 cells was Smad- and BCE-dependent, while basal CTGF promoter activity was BCE-dependent and partially Smad-dependent.
Fig.3. Transcriptional response elements in the CTGF promoter that drive ethanol-stimulated reporter activity in LX-2 cells.
(A) Organization of CTGF promoter (−805 to +17), fused to a SEAP reporter gene showing the location of key transcriptional elements that were altered by point mutation or deletion mutation. (B) Cultured LX-2 cells were transfected with plasmids containing the CTGF promoter-reporter, serum-starved for 18h, and then treated with (i) 0–20 ng/ml TGF-β1 for 6h (left) or 10 ng/ml TGF-β1 for 0–240 mins (right) or (ii) 0–50 mM ethanol for 2h (left) or 25 mM ethanol for 0–240 mins (right). SEAP reporter activity was calculated after adjustment for differences among samples in transfection efficiency as determined by co-transfection with a β-gal reporter. (**p<0.001, +p<0.05 v no treatment). (C) LX-2 cells were transfected with wild type or mutant CTGF promoter reporters after which cells were treated with 0 or 25mM ethanol for 48h. SEAP reporter activity was calculated after adjustment for differences among samples in transfection efficiency as determined by co-transfection with a β-gal reporter. (**p<0.001 v FL alone; # # p<0.001 v FL + ethanol).
Knockdown of TGF-β1 reduces ethanol-mediated CTGF or α-SMA expression
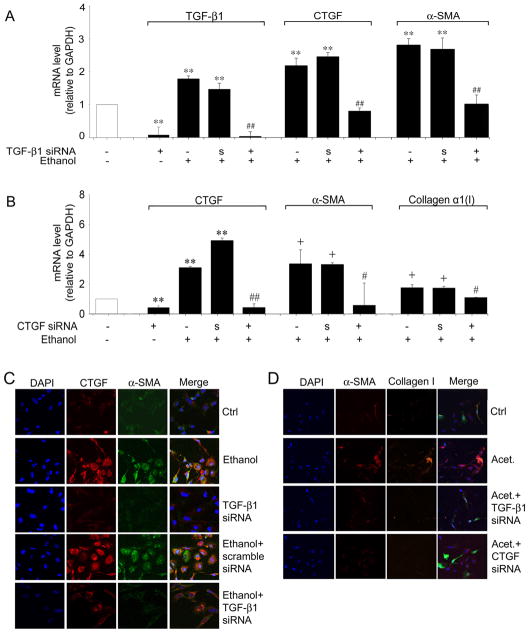
The CTGF promoter studies suggested that TGF-β response elements were involved in ethanol-mediated CTGF production. TGF-β1 knockdown experiments were thus performed by transfecting LX-2 cells with TGF-β1 siRNA and examining CTGF production by the transfected cells after stimulation with 25mM ethanol. As shown in Figure 4A, ethanol-mediated expression of either CTGF or α-SMA mRNA was completely antagonized by TGF-β1 siRNA but not by a scrambled sequence. To verify that these changes were reflected at the protein level, CTGF or α-SMA immunocytochemistry was performed on the cells under the same treatment conditions. As shown in Figure 4C, staining for CTGF or α-SMA proteins was strongly increased after ethanol treatment, a phenomenon that persisted in cells that were transfected with scrambled siRNA, but which was strongly attenuated in TGF-β1 siRNA-treated cells. Similarly, the production of α-SMA or collagen I proteins in response to acetaldehyde was highly attenuated in cells transfected with TGF-β1 siRNA (Figure 4D).
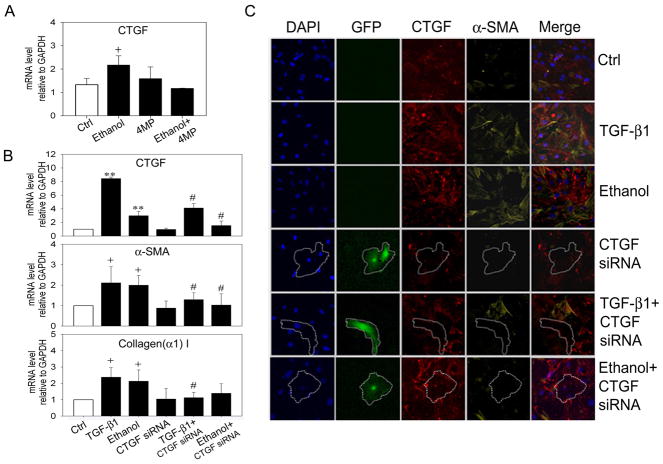
Fig. 4. Effect of TGF-β1 siRNA or CTGF siRNA on ethanol-induced CTGF, α-SMA or collagen α1(I) production in LX-2 cells.
GFP-positive LX-2 cells were collected by FACS 24 hours after co-transfection with 0.8μg pEGFP and 3.2μg TGF-β1 siRNA (A,C,D), CTGF siRNA (B,D), or scrambled siRNA (A, B, C). Sorted cells were seeded into 6-well plates (A,B) or 8-well culture slides (C,D), serum-starved for 18h, and treated with 25 mM ethanol for 48h. The figure shows real-time PCR analysis for cells treated with TGF-β siRNA (A) or CTGF siRNA (B). Data are normalized to GAPDH and are shown relative to the expression of each gene product in control non-treated cells (open bar). “S”, scrambled siRNA. (**p<0.001 v control, +p<0.05 v control, ## p<0.001 v ethanol, #p<0.05 v ethanol). Immunohistochemical detection of CTGF (C), α-SMA (C,D) or collagen I (D) by confocal microscopy is shown for LX-2 cells treated with ethanol (C) or acetaldehyde (D) after transfection with TGF-β1 siRNA (C,D) or CTGF siRNA (D). ×200.
Knockdown of CTGF attenuates ethanol-induced α-SMA or collagen I expression
We next investigated whether ethanol-or acetaldehyde-mediated α-SMA expression was directly attributable to the production of CTGF by performing CTGF knockdown experiments with CTGF siRNA. Ethanol-stimulated mRNA expression of CTGF, α-SMA mRNA or collagen α1(I) or acetaldeyhyde-stimulated production of α-SMA or collagen I proteins was blocked by CTGF siRNA but not by a scrambled sequence (Figures 4B, D, and data not shown). These results showed that LX-2 cell α-SMA or collagen I expression in response to ethanol or acetaldehyde was dependent on CTGF mRNA production.
Ethanol-CTGF axis in primary mouse HSC
Ethanol responses were also evaluated in mouse HSC that had been freshly isolated from normal livers and maintained in culture for 2–12 days. Treatment of primary mouse HSC with ethanol on Day 2 of culture resulted in a 2-fold stimulation of CTGF mRNA expression, an effect that was blocked by co-incubation of the cells with 4-MP or NAC (Figure 5A). Ethanol also stimulated increased expression of mRNA for CTGF, α-SMA or collagen α1(I) in Day 12 cells and this was blocked by transfection of the cells with CTGF siRNA (Figure 5B). Moreover, TGF-β1-stimulated mRNA for CTGF, α-SMA or collagen α1(I) was also blocked by CTGF siRNA (Figure 5B). Since GFP and CTGF siRNA were driven off the same vector, confocal immunofuorescent imaging was used to demonstrate that GFP-positive cells exhibited diminished CTGF and α-SMA co-expression after ethanol or TGF-β treatment of CTGF siRNA-transfected cells (Figure 5C). Collectively, these data showed that ethanol or TGF-β-mediated production of α-SMA or collagen α1(I) were dependent on the production of CTGF.
Fig.5. Ethanol-induced CTGF-dependent pathways in primary mouse HSC.
(A) Effect of ethanol on Day 2 mouse primary mouse HSC in the presence or absence of 4-MP. (+ p < 0.05 v control). (B,C) Day 10 HSC were transfected for 24 h with 3μg pRNAT-CMV3.1/Neo vector containing mouse CTGF siRNA or non-target siRNA, serum-starved for 12h, and then treated with 5ng/ml TGF-β1 or 25 mM ethanol for 36h. Cells were processed for (B) real-time PCR ( + p<0.05 v control; # p<0.05 v ethanol or TGF-β) or (C) immunohistochemical detection of CTGF or α-SMA. GFP-positive (i.e. CTGF siRNA-transfected) HSC are outlined and show reduced immunofluorescent staining for CTGF and α-SMA.
Discussion
During chronic alcoholic liver injury, the combined actions of alcohol-induced transcriptional activators, growth factors, pro-inflammatory cytokines, and oxidant stress contributes to the transformation of quiescent HSC into collagen-producing myofibroblasts. HSC have previously been shown to produce CTGF in vitro as a function of culture-induced activation or after their stimulation by TGF-β [5, 8, 9]. In primary mouse HSC, activation of CTGF gene transcription by TGF-β involves Smad and Ets-1 elements in the CTGF promoter [8]. Moreover, CTGF expression in HSC following TGF-β stimulation or culture-induced activation is dependent on the TGF-β type I receptor showing that TGF-β paracrine and autocrine pathways play a central role in CTGF production in activated HSC [8].
In these studies, we showed that presumptive activated HSC in human steatotic liver were CTGF-positive suggesting that CTGF is involved in alcohol-induced fibrosis. Ethanol treatment of human LX-2 cells resulted in a dose-dependent stimulation of CTGF promoter activity, CTGF mRNA levels, and CTGF protein production. These changes were dependent on the production of acetaldehyde or oxidant stress and were mirrored by concomitant increases in TGF-β mRNA expression. Treatment of LX-2 cells with acetaldehyde also resulted in enhanced expression of CTGF mRNA and CTGF protein production. Comparable responses were observed in primary mouse HSC as shown by increased CTGF mRNA expression in response to TGF-β or ethanol, the latter of which was associated with the production of acetaldehyde or oxidant stress.
A functional link between CTGF and TGF-β was established by knockdown of TGF-β mRNA in ethanol-treated LX-2 cells using TGF-β siRNA, an approach that resulted in highly diminished levels of CTGF mRNA or protein. Thus, the production of TGF-β upon exposure of HSC to ethanol appears to be required for the production of CTGF, a phenomenon that was supported by the CTGF promoter reporter studies which implicated the action Smad and BCE-1 in the CTGF response to ethanol. Previous studies in other cell types showed that TGF-β stimulation of CTGF mRNA relies on the functional Smad element in the CTGF promoter and that, while the BCE-1 site is involved with basal CTGF promoter activity, it also indirectly responsive to TGF-β since it is a response element for endothelin 1 which is induced by TGF-β and is essential for TGF-β to induce CTGF [13, 14]. Our data showed that ethanol-stimulated activity was Smad- and BCE-dependent, while basal CTGF promoter activity was BCE-dependent and partially Smad-dependent, the latter suggesting that TGF-β-independent pathways may contribute at least partially to the high constitutive production of CTGF, at least in LX-2 cells.
Ethanol-stimulated α-SMA production in human or mouse HSC was also a CTGF-dependent effect that occurred downstream of TGF-β since ethanol- or acetaldehyde-stimulated α-SMA mRNA expression and protein production were blocked by either TGF-β1 siRNA or CTGF siRNA. Importantly, the finding using CTGF siRNA that CTGF production in primary HSC is required for the stimulation of α-SMA by ethanol or TGF-β1 has important impact regarding mechanisms by which HSC activation is perpetuated or exacerbated. These data support earlier in vivo studies which have shown that the frequency of myofibroblasts (including activated HSC) in experimental models of liver fibrosis can be reduced by administration of CTGF siRNA [15]. However, since this in vivo strategy was not designed to target the HSC population exclusively, the data presented in our study are the first evidence that ablation of HSC-derived CTGF is sufficient to attenuate α-SMA production in the cells and show that CTGF plays an important role in driving the activated phenotype of these cells. Similar conclusions have been made from studies of CTGF-overexpressing transgenic mice in which a liver-specific CTGF transgene was shown to sustain and exacerbate the activated HSC phenotype as opposed to initiate it [16].
Our data further revealed that ethanol-induced production of collagen in human or mouse HSC is CTGF-dependent as shown by the ability of CTGF siRNA to reduce the levels of ethanol-dependent collagen I(α1) mRNA. While ethanol-stimulated CTGF-dependent collagen production has not been previously reported, our finding that TGF-β-dependent collagen production in primary mouse HSC is also CTGF-dependent is consistent with a growing body of evidence has shown that antagonism of basal or TGF-β-induced CTGF mRNA expression by CTGF antisense oligonucleotides, siRNA or hammerhead ribozyme, results in decreased collagen production in a diverse variety of cell culture systems [17]. Additionally, CTGF siRNA was effective at reducing collagen expression and deposition in carbon tetrachloride or N-nitrosodimethylamine models of liver fibrosis in rodents [15, 18]. While these data have convincingly demonstrated the anti-collagenic actions of antagonists of CTGF RNA, our data are the first to show that such a strategy is effective in response to ethanol and thus provide support for its use as a potential therapy of fibrosis associated with alcoholic liver disease.
Acknowledgments
Supported by NIH/NIAAA grants 5R01AA016003 (DRB) and Alcohol Center Grant PA 50-011999 Morphology Core (SWF)
Abbreviations
- HSC
hepatic stellate cell
- CTGF
connective tissue growth factor
- TGF-β
transforming growth factor beta
- α-SMA
alpha smooth muscle actin
- DMEM
Dulbecco’s modified Eagles’ medium
- FBS
fetal bovine serum
- PCR
polymerase chain reaction
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- RIPA
radioimmunoassay
- SEAP
secreted alkaline phosphatase
- BCE
basal control element
- CMV
cytomegalovirus
- GFAP
green fluorescent protein
- siRNA
small interfering ribonucleic acid
- FACS
fluorescent activated sorting
- ELISA
enzyme-linked immunosorbent assay
- ADH
alcohol dehrogenase
- 4-MP
4-methylpyrazole
- NAC
N-acetylcysteine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Crawford JM. Liver Cirrhosis. In: MacSween RNM, Burt AD, Portmann BC, Ishak KG, Scheuer PJ, Anthony PP, editors. Pathology of the Liver. Harcourt Publishers Limited; London: 2002. pp. 575–619. [Google Scholar]
- 2.Friedman SL. Hepatic fibrosis -- overview. Toxicology. 2008;254:120–129. doi: 10.1016/j.tox.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Gressner AM, Weiskirchen R, Breitkopf K, Dooley S. Roles of TGF-beta in hepatic fibrosis. Front Biosci. 2002;7:d793–807. doi: 10.2741/A812. [DOI] [PubMed] [Google Scholar]
- 4.Rachfal AW, Brigstock DR. Connective tissue growth factor (CTGF/CCN2) in hepatic fibrosis. Hepatol Res. 2003;26:1–9. doi: 10.1016/s1386-6346(03)00115-3. [DOI] [PubMed] [Google Scholar]
- 5.Williams EJ, Gaca MD, Brigstock DR, Arthur MJ, Benyon RC. Increased expression of connective tissue growth factor in fibrotic human liver and in activated hepatic stellate cells. J Hepatol. 2000;32:754–761. doi: 10.1016/s0168-8278(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 6.Paradis V, Dargere D, Vidaud M, De Gouville AC, Huet S, Martinez V, et al. Expression of connective tissue growth factor in experimental rat and human liver fibrosis. Hepatology. 1999;30:968–976. doi: 10.1002/hep.510300425. [DOI] [PubMed] [Google Scholar]
- 7.Abou-Shady M, Friess H, Zimmermann A, di Mola FF, Guo XZ, Baer HU, et al. Connective tissue growth factor in human liver cirrhosis. Liver. 2000;20:296–304. doi: 10.1034/j.1600-0676.2000.020004296.x. [DOI] [PubMed] [Google Scholar]
- 8.Leask A, Chen S, Pala D, Brigstock DR. Regulation of CCN2 mRNA expression and promoter activity in activated hepatic stellate cells. J Cell Commun Signal. 2008;2:49–56. doi: 10.1007/s12079-008-0029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang G, Brigstock DR. Integrin expression and function in the response of primary culture hepatic stellate cells to connective tissue growth factor (CCN2) J Cell Mol Med. 2010 doi: 10.1111/j.1582-4934.2010.01072.x. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu L, Hui AY, Albanis E, Arthur MJ, O'Byrne SM, Blaner WS, et al. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leask A, Holmes A, Black CM, Abraham DJ. Connective tissue growth factor gene regulation. Requirements for its induction by transforming growth factor-beta 2 in fibroblasts. J Biol Chem. 2003;278:13008–13015. doi: 10.1074/jbc.M210366200. [DOI] [PubMed] [Google Scholar]
- 12.Charrier AL, Brigstock DR. Connective tissue growth factor production by activated pancreatic stellate cells in mouse alcoholic chronic pancreatitis. Lab Invest. 2010:1–10. doi: 10.1038/labinvest.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leask A, Sa S, Holmes A, Shiwen X, Black CM, Abraham DJ. The control of ccn2 (ctgf) gene expression in normal and scleroderma fibroblasts. Mol Pathol. 2001;54:180–183. doi: 10.1136/mp.54.3.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi-wen X, Kennedy L, Renzoni EA, Bou-Gharios G, du Bois RM, Black CM, et al. Endothelin is a downstream mediator of profibrotic responses to transforming growth factor beta in human lung fibroblasts. Arthritis Rheum. 2007;56:4189–4194. doi: 10.1002/art.23134. [DOI] [PubMed] [Google Scholar]
- 15.Li G, Xie Q, Shi Y, Li D, Zhang M, Jiang S, et al. Inhibition of connective tissue growth factor by siRNA prevents liver fibrosis in rats. J Gene Med. 2006;8:889–900. doi: 10.1002/jgm.894. [DOI] [PubMed] [Google Scholar]
- 16.Tong Z, Chen R, Alt DS, Kemper S, Perbal B, Brigstock DR. Susceptibility to liver fibrosis in mice expressing a connective tissue growth factor transgene in hepatocytes. Hepatology. 2009;50:939–947. doi: 10.1002/hep.23102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brigstock DR. Strategies for blocking the fibrogenic actions of connective tissue growth factor (CCN2): From pharmacological inhibition in vitro to targeted siRNA therapy in vivo. J Cell Commun Signal. 2009;3:5–18. doi: 10.1007/s12079-009-0043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.George J, Tsutsumi M. siRNA-mediated knockdown of connective tissue growth factor prevents N-nitrosodimethylamine-induced hepatic fibrosis in rats. Gene Ther. 2007;14:790–803. doi: 10.1038/sj.gt.3302929. [DOI] [PubMed] [Google Scholar]