Abstract
Tamoxifen resistance is a major problem in the treatment of Estrogen Receptor (ER) positive patients. We have previously reported that Hexamethylene bis-acetamide Inducible Protein 1 (HEXIM1) inhibits ERα activity by competing with ERα for binding to cyclin T1, a subunit of Positive Transcription Elongation b (P-TEFb). This results in the inhibition of the phosphorylation of RNA polymerase II (RNAPII) at serine 2 and the inhibition of transcription elongation of ERα target genes. Since HEXIM1 can inhibit ER activity, we examined whether it plays a critical role in the inhibitory effects of tamoxifen on ER. We observed that tamoxifen induced HEXIM1 recruitment to the promoter region of ER target genes and decreased the recruitment of cyclin T1 and serine 2 phosphorylated RNAPII to the coding regions of these genes. Conversely, in cells wherein HEXIM1 expression has been downregulated we observed attenuation of the inhibitory effects of tamoxifen on estrogen-induced cyclin T1 recruitment to coding regions of ER target genes. As a consequence, downregulation of HEXIM1 resulted in the attenuation of the repressive effects of tamoxifen on estrogen-induced gene expression and proliferation. Conferring clinical relevance to our studies is our analysis of human breast cancer tissue samples that indicated association of lower expression of HEXIM1 with tumor recurrence in patients who received tamoxifen. Our studies provide a better understanding of the mechanistic basis for the inhibitory effect of tamoxifen on ER activity and may suggest new therapeutic targets for the treatment of tamoxifen resistant breast cancer.
Keywords: Estrogen Receptor, Tamoxifen, HEXIM1, Breast Cancer
INTRODUCTION
Lifetime exposure to estrogen is a major risk factor for breast cancer since estrogen plays an important role in the initiation of tumorigenesis (Yager JD and Davidson NE 2006). Estrogen carries out its functions by activating nuclear receptors, Estrogen Receptor α (ERα) and Estrogen Receptor β (ERβ) that function as ligand-dependent nuclear transcription factors (Mangelsdorf et al 1995, Yager and Davidson 2006). When estrogen binds to ERα, the receptor changes conformation and binds to estrogen-responsive elements (EREs) located in the promoter region of ERα target genes (Gruber CJ et al 2002, Mangelsdorf et al 1995). The interaction between ERα and ERE leads to the recruitment of other transcription factors that facilitate gene transcription (Mangelsdorf et al 1995). ERα target gene transcription mediates mammary cell proliferation, increased cell division, and DNA synthesis that elevates the risk of replication errors and can initiate tumorigenesis (Deroo and Korach 2006).
Our laboratory identified an ER coregulator, Estrogen Downregulated Gene 1 (EDG1) as a protein whose expression is down-regulated by estrogens (Wittmann et al 2003). EDG1 is also known as hexamethylene-bis-acetamide (HMBA)-inducible protein 1 (HEXIM1) because it can be upregulated in vascular smooth muscle cells by the differentiating agent HMBA (Kusuhara M et al 1999). HEXIM1 was shown to inhibit the positive transcription elongation factor b (P-TEFb) by interacting with the cyclin T1 subunit (Wittmann et al 2005, Yik et al 2003). We have reported that cyclin T1 and E2-liganded-ER interact on the promoter region of ER target genes allowing for ERα to compete with HEXIM1 for cyclin T1 (Ogba et al 2008, Wittmann et al 2005). E2-liganded ER then enhances P-TEFb kinase activity and phosphorylation of RNA polymerase II (RNAPII) at serine 2, resulting in transcriptional elongation. On the contrary, increased HEXIM1 expression results in inhibition of cyclin T1 recruitment (Ogba et al 2008) and inhibition of E2-induced phosphorylation of RNAP II at serine 2 (Ogba et al 2008). As a result, HEXIM1 inhibited estrogen-induced gene expression and proliferation (Ogba et al 2008). Furthermore, levels of HEXIM1 expression were decreased in breast tumor tissue when compared to adjacent normal breast tissues in all tumor grades (Wittmann et al 2003).
Approximately two-thirds of breast tumors express ERα (Cleator et al 2009). ERα is a valuable predictive factor since ERα positive breast cancer cells depend on estrogen and can be treated with antiestrogens such as tamoxifen (Lewis and Jordan 2005). Although, tamoxifen has different agonist or antagonist effects in different tissues, tamoxifen works as an antagonist in breast tissues by binding to ERα and blocking the interaction between estrogen and the ER (Musgrove and Sutherland 1994). It has been effectively used as an adjuvant therapy for ERα positive patients to reduce mortality and prolong survival time (Osborne 1998). However, the efficacy of tamoxifen treatment is limited by the development of resistance, with approximately half of the patients developing tamoxifen resistance after five years of therapy (Anderson et al 2007, Zwart et al 2009). Proposed bases for tamoxifen resistance include loss of ER expression/function, activation of growth factor signaling pathways that circumvent tumor reliance on ERα signaling, and alterations in the expression of co-regulatory proteins involved in mediating ER target gene transcription (Hurvitz SA and Pietras RJ 2008, Lahusen et al 2009, Lavinsky et al 1998, Riggins et al 2007, Ring and Dowsett 2004, Scott et al 2007).
More than 70% of tamoxifen-resistant tumors still express functional ER (Cleator et al 2009) and because HEXIM1 inhibits ERα transcriptional activity, we hypothesized that HEXIM1 is a critical factor in the inhibition of ER activity by tamoxifen and loss of HEXIM1 may contribute to tamoxifen resistance. To investigate the hypothesis, we examined the recruitment of HEXIM1, cyclin T1 and serine 2 phosphorylated RNAP II to ER target genes in the presence of tamoxifen. Furthermore, we downregulated HEXIM1 expression to demonstrate a functional role for HEXIM1 in the inhibition of ER activity by tamoxifen. To confer clinical relevance to our in vitro studies, immunohistochemical studies were conducted to examine if there was a correlation between HEXIM1 expression and disease recurrence in patients who had been treated with tamoxifen.
RESULTS
Tamoxifen enhances the recruitment of HEXIM1 to ERα target genes
We have previously reported that endogenous HEXIM1 interacted with E2-liganded ERα in breast cells and was recruited to the promoter regions of ERα target genes (Wittmann et al 2005). Moreover, we observed that trans-hydroxytamoxifen (TOT)-liganded ERα also interacted with HEXIM1 (Wittmann et al 2005). We hypothesized that HEXIM1 DNA binding was regulated by TOT. To investigate this hypothesis, we carried out chromatin immunoprecipitation (ChIP) assays in MCF-7 cells and examined the effect of TOT on HEXIM1 occupancy on the ER-target genes, pS2 and Cyclin D1 (CCND1). In TOT-treated cells, we observed a significant increase in HEXIM1 occupancy at estrogen responsive regions within the pS2 promoter when compared to vehicle or E2-treated cells (Figure 1A). We also observed increased HEXIM1 occupancy within the CCND1 promoter in TOT-treated cells (Supplementary Figure 1A).
Figure 1. Tamoxifen treatment resulted in enhanced recruitment of HEXIM1 and inhibition of the recruitment of cyclin T1 and RNAP II to an ER responsive gene.
A. MCF-7 cells were treated with 100 nM E2 or 1 uM TOT for 90 minutes and processed for ChIP assays. ChIP assays were performed with antibodies against HEXIM1 or nonspecific rabbit IgG (as a control for immunoprecipitation). Panel on the left, DNA fragments were analyzed by PCR using primers specific for the promoter region of pS2. Panel on the right, Quantifications of HEXIM1 enrichment at the pS2 promoter. Columns represent the mean of three replicates; bars, SE. *, P < 0.05. B. ChIP assays were performed with antibodies against cyclin T1 or non-specific rabbit IgG (as a control for immunoprecipitation). Panel on the left, DNA fragments were analyzed by PCR using primers specific for the coding region of pS2. Panel on the right, Quantifications of cyclin T1 enrichment at the pS2 promoter or coding regions. Each column represent the mean of three replicates; bars, SE. *, P < 0.05. C. Samples were processed for ChIP assays using antibodies against serine 2 phosphorylated RNAP II. Panel on the left, DNA fragments were analyzed by PCR using primers specific for coding region of pS2. Panel on the right, Quantifications of RNAP II enrichment at the pS2 coding region. Columns represent the mean of three replicates; bars, SE. *, P < 0.05.
Tamoxifen inhibits the recruitment of cyclin T1 and phosphorylated RNAP II to ERα target genes
Our previous studies indicated that HEXIM1 interacted with and inhibited ERα activity by competing with ERα for binding to the cyclin T1 subunit of P-TEFb. In doing so, HEXIM1 inhibited phosphorylation of RNAP II carboxy terminal domain (CTD) at serine 2 and transcriptional elongation by RNAP II (Ogba et al 2008, Wittmann et al 2005, Yik et al 2003). We determined whether TOT, by increasing HEXIM1 recruitment could also inhibit P-TEFb recruitment and the resulting phosphorylation of RNAP II. ChIP assays were performed to study the binding of cyclin T1 and RNAP II to the promoter and coding regions of the pS2 gene. We examined both regions of ER target genes to investigate whether the effects of tamoxifen on the recruitment of cyclin T1 or phosphorylated RNAP II reflected effects on transcription initiation or elongation. We observed no significant decrease in cyclin T1 binding to the promoter region of pS2 as a result of TOT treatment (Figure 1B). As we previously reported, E2 induced recruitment of cyclin T1 to the coding region of the pS2 gene (Ogba et al 2008). However, TOT treatment resulted in attenuation of E2-induced cyclin T1 recruitment to the coding region of the pS2 and CCND1 genes (Figure 1B and Supplementary Figure 1B).
We also examined recruitment of serine 2 phosphorylated RNAP II to the coding region of pS2 gene. Treatment with E2 alone resulted in increased serine 2 phosphorylated RNAP II recruitment, but co-treatment with tamoxifen significantly decreased E2-induced serine 2 phosphorylated RNAP II recruitment to basal levels (p value < 0.05) (Figure 1C). E2-induced recruitment of serine 2 phosphorylated RNAP II to the coding region of CCND1 was also significantly decreased in the presence of TOT (Supplementary Figure 1C).
In summary, tamoxifen increased the recruitment of HEXIM1 to the promoter region of pS2 and CCND1 and attenuated E2-induced cyclin T1 and phosphorylated RNAP II recruitment to the coding region of both pS2 and CCND1, supporting a critical role of HEXIM1 in tamoxifen inhibition of ERα-mediated transcription.
Our previous studies indicated that E2 increased P-TEFb kinase activity. We also observed that HEXIM1 attenuated E2–induced P-TEFb activity (Ogba et al 2008). We hypothesized that tamoxifen would inhibit E2–induced P-TEFb activity. To investigate this, we measured the kinase activity of P-TEFb in endogenous immunoprecipitates of cyclin T1 from MCF-7 cells using a synthetic peptide substrate (CTD4) phosphorylation. We observed that tamoxifen decreased E2-induced P-TEFb activity in MCF-7 cells (Supplementary Figure 2), supporting inhibition of the transcription elongation process by tamoxifen.
Downregulation of HEXIM1 attenuates the repressive effects of tamoxifen on ERα-mediated transcriptional activity
We further tested the critical role of HEXIM1 in tamoxifen inhibition of ERα-mediated transcription by downregulating HEXIM1 expression and examining repressive effects of tamoxifen on ERα-transcriptional activity. We developed MCF-7 cells stably transfected with expression vectors for control or HEXIM1 miRNA and confirmed that HEXIM1 protein expression was downregulated in HEXIM1 miRNA-transfected cells when compared to control miRNA-transfected cells (Figure 2A). We did not observe any effects on ERα protein levels, indicating that effects of downregulating HEXIM1 levels cannot be attributed to changes in ERα levels (Supplementary Figure 3A)
Figure 2. Downregulation of HEXIM1 expression resulted in the attenuation of repressive effects of tamoxifen on ERα transcription.
A. HEXIM1 miRNA- or control miRNA-transfected MCF-7 cells were treated with 10 nM E2 ± 100 nM TOT for 3 hours. Proteins were extracted from cells and processed for western blot analyses as described in the “Materials and Methods”. Image shown is representative of three independent replicates. B. HEXIM1 miRNA- or control miRNA-transfected MCF-7 cells were treated with 100 nM E2 ± 1 uM TOT for 90 minutes and processed for ChIP assays. ChIP assays were performed with antibodies against HEXIM1 or non-specific rabbit IgG (as a control for immunoprecipitation). Panel on the left, DNA fragments were analyzed by PCR using primers specific for the promoter region of pS2. Panel on the right, Quantifications of HEXIM1 enrichment at the pS2 promoter. bars, SE. a, P < 0.05 vs. control-transfected cells with the same treatment. C. ChIP assays were performed with antibodies against cyclin T1 or non-specific rabbit IgG (as a control for immunoprecipitation). Shown are quantifications of cyclin T1 enrichment at the pS2 coding region. Columns represent mean of three replicates; bars, SE. a, P < 0.05 vs. E2-treated cells. D. ChIP assays were performed with antibodies against serine 2 phosphorylated RNAP II or non-specific rabbit IgG (as a control for immunoprecipitation). Shown are quantifications of RNAP II enrichment at the pS2 coding region. Columns represent mean of three replicates; bars, SE. a, P < 0.05 vs. E2-treated cells; b, P < 0.05 vs. control-transfected cells with the same treatment.
The recruitment of HEXIM1 to the promoter regions of pS2 and CCND1 genes was decreased in HEXIM1 miRNA-transfected cells when compared to control cells regardless of treatment (Figure 2B and Supplementary Figure 3B). Recruitment of HEXIM1 in HEXIM1 miRNA-transfected cells was influenced by hormonal treatment, compounding the relationship between HEXIM1 expression and promoter occupancy. Downregulation of HEXIM1 expression resulted in attenuation of the repressive effects of TOT on E2-induced cyclin T1 recruitment to the pS2 and CCND1 coding regions (Figure 2C and Supplementary Figure 3C). Cotreatment with TOT induced an increase (although not statistically significant) in estrogen-induced recruitment of cyclin T1 to the pS2 coding region in HEXIM1 miRNA-transfected cells, rather than the decrease observed in control-transfected cells. Downregulation of HEXIM1 expression also resulted in increased basal and TOT-induced recruitment of serine 2 phosphorylated RNAP II to the pS2 coding region (Figure 2D). These data further confirm that HEXIM1 is critical for the repressive effects of tamoxifen on E2 -mediated transcription.
Downregulation of HEXIM1 results in attenuation of the inhibitory effects of tamoxifen on estrogen-induced gene expression and cell proliferation
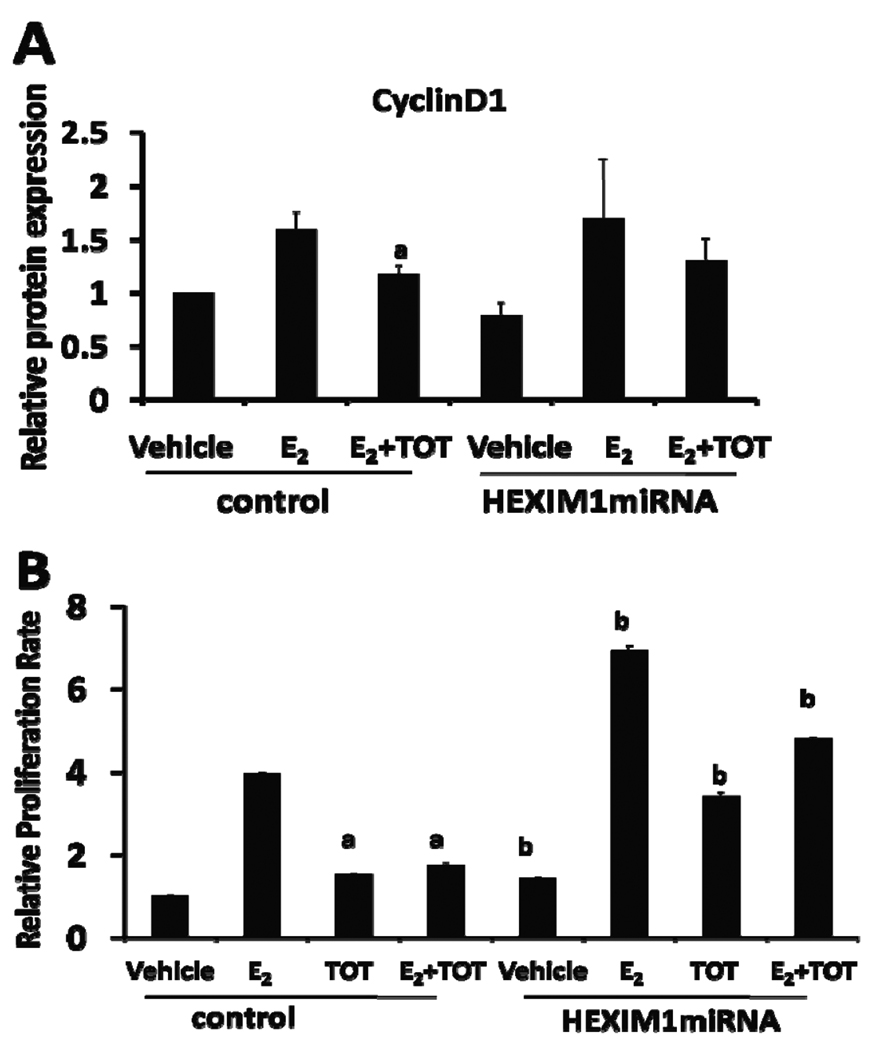
Cyclin D1, an estrogen target gene, is a D-type cyclin that regulates G1-S cell cycle progression during cell proliferation. Our previous study reported that HEXIM1 downregulated cyclin D1 expression levels in breast epithelial cells in vitro and in mouse mammary glands (Ogba et al 2008). We hypothesized that downregulation of HEXIM1 would result in attenuation of inhibitory effects of tamoxifen on E2-induced gene expression and proliferation. To investigate this, control miRNA- and HEXIM1 miRNA-transfected MCF-7 cells were treated with E2 or both E2 and TOT for 3 hours. In control cells, we observed that E2 treatment increased cyclin D1 expression while TOT treatment attenuated E2-induced cyclin D1 expression (Figure 3A). However, in HEXIM1 miRNA-transfected cells, we did not observe a significant decrease in E2-induced cyclin D1 expression as a result of TOT treatment (Figure 3A). pS2 mRNA levels were significantly increased in TOT-treated HEXIM1 miRNA-transfected MCF-7 cells when compared to TOT-treated control-transfected cells (Supplementary Figure 4A). We observed attenuation of TOT repressive affect as a result of downregulation of HEXIM1 expression in another breast epithelial cell line, T47D (Supplementary Figure 4B).
Figure 3. Downregulation of HEXIM1 results in attenuation of the inhibitory effects of tamoxifen.
A. HEXIM1 miRNA- or control miRNA-transfected MCF-7 cells were treated with 10 nM E2 ± 100 nM TOT for 3 hours. Proteins were extracted from cells and processed for western blot analyses as described in the “Materials and Methods”. Levels of cyclin D1 protein were quantitated and normalized to GAPDH. Columns represent the mean of three independent replicates; bars, SE. a, P < 0.05 vs. E2-treated cells B. HEXIM1 miRNA- or control miRNA-transfected MCF-7 cells were plated onto 96 well plates and treated with 10 nM E2, 100 nM TOT, or both 10 nM E2 and 100 nM TOT for 7 days. MTT assays were performed as described in Materials and Methods. Columns, mean of three independent replicates with 5 wells for each treatment group; bars, SE. a, P < 0.05. vs. E2-treated cells; b, P < 0.05 vs. control-transfected cells with the same treatment.
Because TOT had reduced capacity for inhibiting E2-ER regulated transcription as a result of downregulation of HEXIM1 expression, we examined if downregulation of HEXIM1 expression will also attenuate the inhibition of breast cancer cell proliferation by tamoxifen. In control cells, tamoxifen significantly reduced E2-induced cell proliferation from 4-fold to 1.7-fold (Figure 3B). On the contrary, inhibitory effects of tamoxifen were significantly attenuated in HEXIM1 miRNA-transfected cells. We also observed enhanced E2-induced proliferation in HEXIM1 miRNA-transfected cells (Figure 3B).
HEXIM1 expression is associated with decreased probability of cancer recurrence in breast cancer patients who received tamoxifen treatment
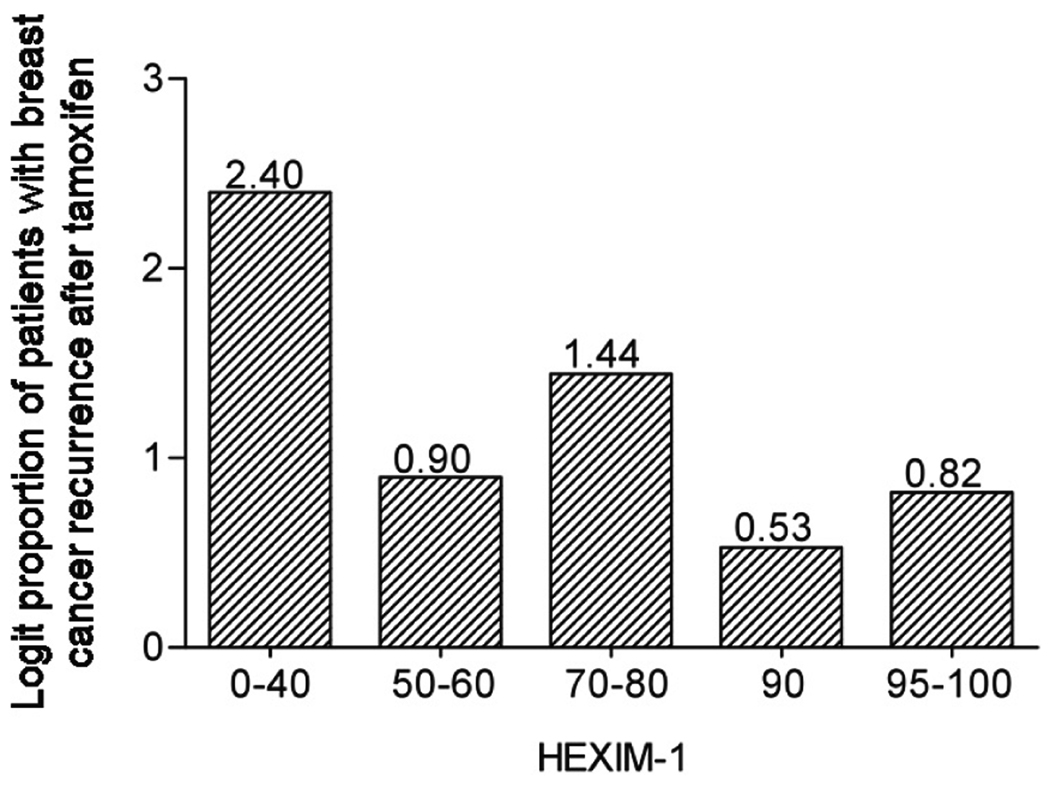
Our previous studies indicated reduced levels of HEXIM1 expression in breast cancer tissues when compared to normal adjacent breast tissue (Wittmann et al 2003). We also observed that breast tumor cells expressing HEXIM1 were more likely to express ERα than the proliferation marker Ki67. Since HEXIM1 plays critical roles in the inhibitory effects of tamoxifen, we examined the expression levels of HEXIM1 in a case control study of breast cancer patients treated with tamoxifen. This case control study consisted of 58 matched pairs of ER positive tumors specimens (116 patients), from patients matched with regard to 5 criteria; year of diagnosis, patient age at diagnosis, TNM stage (tumor size, lymph nodes, metastasis; I, II, or III), and radiation therapy (yes or no). This group was limited to women with at least 5 years of follow-up. Cases were defined as having recurred earlier than their matched controls. HEXIM1 expression was assessed by immunohistochemical staining (Supplementary Figure 5). The proportion of cases decreased with increased HEXIM1 expression (Figure 4). The estimated odds ratio of HEXIM1 expression for the cases was less than 1, indicating that higher HEXIM1 expression was associated with lower probabilities of recurrence. The odds of recurrence (case versus control) decreased by an estimated factor of 0.84 for each 10 percent increase in HEXIM1 expression (Table 1). These data support our in vitro studies revealing an important role for HEXIM1 in the inhibitory effects of tamoxifen and that loss of HEXIM1 may be a contributing factor to the development of tamoxifen resistance.
Figure 4. Expression levels of HEXIM1 in breast tissue from case control study of breast cancer patients treated with tamoxifen.
Sections from tumor specimens obtained from matched “control” and “case” patients were stained for endogenous HEXIM1 as described in the "Materials and Methods" section. “Cases” were defined as having recurred earlier than their matched controls. Shown is the graph of logit (log(p/(1-p)) proportion of patient (p) with breast cancer recurrence after tamoxifen within each of five approximately equal size groups based on HEXIM1 expression.
Table 1.
Estimates (95% CIs) of odds ratios for the association between marker percent (10% increase) and earlier recurrence (case versus control). Maximum number of matched pairs is 56.
| Marker status |
Est. Odd Ratio= (95%CI) |
p-value |
|---|---|---|
| HEXIM1 | 0.84 (0.68,0.99) | 0.039 |
DISCUSSION
The present study provides novel evidence that HEXIM1 is critical for tamoxifen inhibition of ERα activity. Our studies also suggest that tamoxifen-liganded ER has activities on its own and a more proactive role beyond competitively inhibiting the interaction of ERα with estrogens. Tamoxifen-liganded ERα induced recruitment of HEXIM1 to ER target genes, resulting in the inhibition of recruitment of cyclin T1 and serine 2 phosphorylated RNAP II to the coding regions of these ER target genes. We also demonstrated a functional role for HEXIM1 in the inhibition of ER-mediated gene expression and proliferation by tamoxifen. Most importantly, our immunohistochemical studies indicate a correlation between HEXIM1 expression and disease recurrence in patients who have been treated with tamoxifen.
Coactivator and corepressor proteins play critical roles in mediating transcriptional activation by the ER, and enhancement of coactivator or inhibition of corepressor expression may contribute to the tamoxifen resistant phenotype (Ring and Dowsett 2004, Scott et al 2007). Overexpression of the coactivators Amplified in Breast Cancer 1 (AIB1) or Nuclear Coactivator-3 (NCoA3) in breast cancer has been correlated with tamoxifen resistance and decreased overall survival (Lahusen et al 2009). Corepressors have been reported to interact with tamoxifen-liganded ER (Lavinsky et al 1998, Webb et al 2003). In vitro studies have indicated that reduced levels of corepressors N-COR and SMRT resulted in attenuation of the inhibitory effects of tamoxifen (Lavinsky et al 1998). However modulation of the recruitment of AIB1, NCoA3, N-COR, or SMRT to ER target genes by tamoxifen has not been reported. Correlations between the levels of known corepressors and tamoxifen resistance in human breast tissue samples have also not been reported. We have previously reported that HEXIM1 interacts with tamoxifen-liganded ER (Wittmann et al 2005). We now report on the consequences of this interaction and show that tamoxifen enhances the recruitment of HEXIM1 to the promoters of ER target genes that, in turn, inhibits the recruitment of cyclin T1 and phosphorylated RNAP II.
To demonstrate the functional relevance of tamoxifen-induced recruitment of HEXIM1 in the inhibitory effects of tamoxifen, we stably downregulated HEXIM1 expression and showed that decreased HEXIM1 recruitment to the promoter region of pS2 and CCND1 resulted in attenuation of tamoxifen inhibition of E2-induced recruitment of cyclin T1. Moreover, downregulation of HEXIM1 resulted in the enhancement of E2-induced cell proliferation, and attenuated the inhibitory effects of tamoxifen on cyclin D1 expression and cell proliferation. Therefore, our studies provide evidence for a critical role of HEXIM1 in the inhibitory effects of tamoxifen.
E2 binds to ER at the hydrophobic pocket of ligand binding domain (LBD) that is sealed by helix 12 providing a platform for recruiting coactivator proteins. Tamoxifen also binds to ER at LBD and repositions helix 12. A key difference between E2-liganded ER and SERM- or tamoxifen- ER complex is that helix 12 is prevented from sealing the ligand binding pocket (Shiau et al 1998). HEXIM1 has been shown to interact with the E/F domain of ERα in vitro (Wittmann et al 2005). The binding of tamoxifen to ER resulted in several residues exposing different side chains than those observed in E2-liganded ER (Shiau et al 1998), perhaps allowing for an interaction with HEXIM1 to be more energetically favored. Based on our current studies, we speculate that tamoxifen can enhance HEXIM1 recruitment to ER target genes because of conformational changes that occur after tamoxifen binds to ER that allows for higher affinity binding to HEXIM1 than E2-liganded ER. Further studies should be done to gain a better understanding of structural basis for the interaction of HEXIM1 with tamoxifen-liganded ER.
The clinical relevance of our findings was supported by our immunohistochemical analysis of HEXIM1 expression in human breast cancer specimens. Our studies indicated that lower expression of HEXIM1 was significantly associated with increased risk of tumor recurrence in patients who received tamoxifen treatment. Thus we speculate that loss of HEXIM1 may be involved in the development of tamoxifen resistance. Therefore, therapies aimed at increasing HEXIM1 expression can be developed for tamoxifen-resistant patients to improve breast cancer survival. We are currently testing this possibility using mouse models generated in our laboratory.
MATERIALS AND METHODS
Cell culture and transfection
Construction of expression vectors for control or HEXIM1 miRNA were described previously (Ogba et al 2008). A Pol II promoter-driven miRNA expression vector system (Invitrogen, CA) was used. To make pcDNA-HEXIM1 miRNA, miRNA oligos (Ogba et al 2008) were annealed and cloned into the pcDNA 6.2 GW/EmGFP vector (Invitrogen) according to the manufacturer’s instructions. MCF-7 cells were transfected with pcDNA 6.2-GW/EmGFP-miR expression vectors containing either the HEXIM1 miRNA insert or a control miRNA insert. Following blasticidin selection, cells expressing the highest level of GFP were flow-sorted and expanded
Western Blot
Cells were treated with 10 nM E2 or both 10 nM E2 and 100 nM trans-hydroxy-tamoxifen (TOT) for 4 hours. Western blot analyses have been described previously (Ogba et al 2008).
Chromatin Immunoprecipitation
Control- and HEXIM1 miRNA-transfected MCF-7 cells were plated onto 150-mm plates. Before harvesting, cells were treated with 100 nM 17β estradiol (E2) and/or 1 uM trans-hydroxytamoxifen (TOT) for 45 or 90 minutes. ChIP assays were carried out as previously described (Wittmann et al 2005).
CTD Kinase assay
Kinase assays were performed as previously described protocols (Ogba et al 2008).
Proliferation assay
MCF-7 cells transfected with expression vector for control miRNA or HEXIM1 miRNA were plated onto 96 well plates. Cells were treated with 10 nM E2, 100 nM TOT, or both 10 nM E2 and 100 nM TOT for 7 days. Cell proliferation was assessed using the MTT based Cell Growth Determination Kit from Sigma-Aldrich according to the manufacturer’s protocol.
Immunohistochemistry
Human breast tissue samples were obtained from the Cooperative Breast Cancer Tissue Resource (CBCTR). All samples were confirmed to be ER positive by IHC, and pairs were matched by 5 criteria including: year of diagnosis by 5 year categories, patient age at diagnosis by 5 year categories, TNM stage, and radiation therapy. Controls were defined as having verified recurrence free survival greater than that for their matched case. Sixty-four matched pairs were initially identified, and 6 pairs were eventually eliminated due to technical problems with at least one of the pairs slides.
We carried out immunohistochemical staining to detect HEXIM1 levels. Breast tissue samples were fixed in formalin, embedded in paraffin, and sectioned at 5 micron thickness. Paraffin sections were deparaffinized and rehydrated. To unmask epitopes we used heat –induced antigen retrieval technique using 10 mM Citrate buffer (pH 6.0). After peroxidase block, samples were blocked with PBS containing 10% goat serum and 10% Triton X-100. Sections were incubated with HEXIM1 rabbit polyclonal antibody (1:100 dilution) and biotinylated goat anti-rabbit IgG secondary antibody (1:200 dilution). Duplicate sections were immunostained with nonspecific rabbit IgG. Staining was classified according to intensity of HEXIM1 expression in tumor cells in 4 categories: negative (0), weak (1), moderate (2), strong (3), and very strong (4).
Statistics
The odds ratio (for the association between HEXIM1 levels and recurrence) and corresponding 95% confidence interval were estimated using conditional logistic regression in the SAS Logist Procedure (SAS System Version 9.2, SAS Institute Inc.). The p-value was obtained using an exact test of the null hypothesis that the odds ratio equals 1. Statistical significance was accepted at p < 0.05.
Supplementary Material
Acknowledgments
We thank Wei Wang (CWRU Department of Epidemiology and Biostatistics) for help with construction of graphs representing the relationship between disease recurrence and expressions levels of HEXIM1 in breast tissue. This work was supported by National Institute of Health grant CA92440 to M.M.M and American Cancer Society grant RSG CCE-110689 to J.J.P.
Abbreviations
- ERα
Estrogen receptor alpha
- E2
17-beta estradiol
- HEXIM1
Hexamethylene inducible gene-1
- TOT
trans-hydroxy- tamoxifen
- P-TEFb
Positive transcription elongation factor b
- CTD
Carboxy-repeat terminal domain
- RNAP II
RNA polymerase II
- CCND1
cyclin D1
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
REFERENCES
- Anderson H, Bulun S, Smith I, Dowsett M. Predictors of response to aromatase inhibitors. J Steroid Biochem Mol Biol. 2007;106:49–54. doi: 10.1016/j.jsbmb.2007.05.024. [DOI] [PubMed] [Google Scholar]
- Cleator SJ, Ahamed E, Coombes RC, Palmieri C. A 2009 update on the treatment of patients with hormone receptor-positive breast cancer. Clin Breast Cancer. 2009;9(Suppl 1):S6–S17. doi: 10.3816/CBC.2009.s.001. [DOI] [PubMed] [Google Scholar]
- Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116:561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber CJ, Tschugguel W, Schneeberger C, Huber JC. Production and actions of estrogens. N Engl J Med. 2002;346:340 –352. doi: 10.1056/NEJMra000471. [DOI] [PubMed] [Google Scholar]
- Hurvitz SA, Pietras RJ. Rational management of endocrine resistance in breast cancer. Cancer. 2008;9:2385–2397. doi: 10.1002/cncr.23875. [DOI] [PubMed] [Google Scholar]
- Kusuhara M, Nagasaki K, Kimura K, Maass N, Manabe T, Ishikawa S, et al. Cloning of hexamethylene-bis-acetamide-inducible transcript, HEXIM1, in human vascular smooth muscle cells. Biomed Res. 1999;20:273–279. [Google Scholar]
- Lahusen T, Henke RT, Kagan BL, Wellstein A, Riegel AT. The role and regulation of the nuclear receptor co-activator AIB1 in breast cancer. Breast Cancer Res Treat. 2009;116:225–237. doi: 10.1007/s10549-009-0405-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavinsky RM, Jepsen K, Heinzel T, Torchia J, Mullen TM, Schiff R, et al. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc Natl Acad Sci U S A. 1998;95:2920–2925. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JS, Jordan VC. Selective estrogen receptor modulators (SERMs): mechanisms of anticarcinogenesis and drug resistance. Mutat Res. 2005;591:247–263. doi: 10.1016/j.mrfmmm.2005.02.028. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrove EA, Sutherland RL. Cell cycle control by steroid hormones. Semin Cancer Biol. 1994;5:381–389. [PubMed] [Google Scholar]
- Ogba N, Chaplin LJ, Doughman YQ, Fujinaga K, Montano MM. HEXIM1 regulates 17beta-estradiol/estrogen receptor-alpha-mediated expression of cyclin D1 in mammary cells via modulation of P-TEFb. Cancer Res. 2008;68:7015–7024. doi: 10.1158/0008-5472.CAN-08-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339:1609–1618. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- Riggins RB, Schrecengost RS, Guerrero MS, Bouton AH. Pathways to tamoxifen resistance. Cancer Lett. 2007;256:1–24. doi: 10.1016/j.canlet.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring A, Dowsett M. Mechanisms of tamoxifen resistance. Endocr Relat Cancer. 2004;11:643–658. doi: 10.1677/erc.1.00776. [DOI] [PubMed] [Google Scholar]
- Scott DJ, Parkes AT, Ponchel F, Cummings M, Poola I, Speirs V. Changes in expression of steroid receptors, their downstream target genes and their associated co-regulators during the sequential acquisition of tamoxifen resistance in vitro. Int J Oncol. 2007;31:557–565. doi: 10.3892/ijo.31.3.557. [DOI] [PubMed] [Google Scholar]
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- Webb P, Nguyen P, Kushner PJ. Differential SERM effects on corepressor binding dictate ERalpha activity in vivo. J Biol Chem. 2003;278:6912–6920. doi: 10.1074/jbc.M208501200. [DOI] [PubMed] [Google Scholar]
- Wittmann BM, Wang N, Montano MM. Identification of a novel inhibitor of breast cell growth that is down-regulated by estrogens and decreased in breast tumors. Cancer Res. 2003;63:5151–5158. [PubMed] [Google Scholar]
- Wittmann BM, Fujinaga K, Deng H, Ogba N, Montano MM. The breast cell growth inhibitor, estrogen down regulated gene 1, modulates a novel functional interaction between estrogen receptor alpha and transcriptional elongation factor cyclin T1. Oncogene. 2005;24:5576–5588. doi: 10.1038/sj.onc.1208728. [DOI] [PubMed] [Google Scholar]
- Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- Zwart W, Rondaij M, Jalink K, Sharp ZD, Mancini MA, Neefjes J, et al. Resistance to antiestrogen arzoxifene is mediated by overexpression of cyclin D1. Mol Endocrinol. 2009;23:1335–1345. doi: 10.1210/me.2008-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.