Abstract
The need to develop and improve sustainable energy resources is of eminent importance due to the finite nature of our fossil fuels. This review paper deals with a third generation renewable energy resource which does not compete with our food resources, cyanobacteria. We discuss the current state of the art in developing different types of bioenergy (ethanol, biodiesel, hydrogen, etc.) from cyanobacteria. The major important biochemical pathways in cyanobacteria are highlighted, and the possibility to influence these pathways to improve the production of specific types of energy forms the major part of this review.
Keywords: Bioenergy, Biofuel, Cyanobacteria, Renewable energy, Metabolic engineering
Introduction
Fossil fuels, including oil, coal and natural gas, are providing about 85% of our energy need worldwide. The effective use of this energy resource in a productive and economic way still remains to be a major challenge. The main drawback of fossil fuels is that it is a finite resource and will be depleted in the near future. The term “peak oil” is commonly used to describe when peak oil production will be reached. Peak oil will be followed by a rapid decline in our oil reserves. Nashawi et al. (2010) predicted that peak oil will be reached as early as 2014. This finite nature of our fossil fuels and the dangers associated with nuclear energy, as evident by the recent nuclear disaster in Japan, emphasizes the importance of finding economically viable alternative energies. Alternative energy refers to renewable energy sources not derived from fossil fuels or nuclear power. Nowadays, there is a renewed interest in the development of sustainable energies promoted by the global concern that fossil fuels are finite, the rapid increase of energy consumption by industrialized countries, and the environmental problems caused by the burning of fossil fuels and from the management and storage of nuclear waste. Unlike fossil and nuclear fuels, alternative energy comes from natural resources (wind, sunlight, geothermal power and biomass) which are constantly replaced. Using these resources to supply our energy needs further supports sustainable development by lowering greenhouse gas emissions. The development and use of renewable energies provide a considerable number of benefits to nations around the world including an increment of the energy production, environmental protection, reduction in pollution and job creation. Solar (thermal or photovoltaic), wind, hydroelectric, biomass and geothermal energy currently constitute the most common sustainable sources of energy. Each one of these sources has particular properties that determine their usefulness and application in our society. The different characteristics of a specific energy resource can be evaluated in terms of sustainability indicators (Afgan and Carvalho 2002). In 2006, sustainable energies represented about 18% of the global total energy consumption (REN21 2007) and are able to substitute traditional fuels at different levels in our society including power generation, heating, transport fuel and rural energy. Because of its common use in developing countries for local energy supply, biomass represents the major source of renewable energy (constituting up to a 75% of the renewable energy sources) (Hall and Moss 1983). Bioenergy is fuel derived from biological sources (biomass) and is also referred to as biofuel. Biomass is defined as any organic material coming from any form of life or its derived metabolic products. Biofuel (either biodiesel or bioethanol) is currently the only alternative energy source able to replace transport fuel in today’s vehicles without involving major modifications to vehicle engines (Kaygusuz 2009). Biofuel is, however, not yet economically competitive with conventional energies. Additional input in order to collect, harvest and store the material is involved, resulting in higher manufacturing costs. Furthermore, biofuel possesses lower energy content than fossil fuels. Table 1 compares the calorific values for the different types of fuels. Biomass possesses important advantages if compared to other sustainable sources, for instance, it is available throughout the world, its processing is relatively simple without involving expensive equipment and it can be stored over long periods of time. In addition, bioenergy can be generated from organic waste material which might otherwise be discarded thus contributing to the waste management. One of the main controversial issues related to the production of biofuel is the competition between energy crops and edible crops for arable land and water. There is a scarcity of productive land available and areas occupied for bioenergy production may therefore serve for other more elemental uses, such as food production or conservation. Intensive cultivation of energy crops may also cause negative effects in the ecosystem biodiversity due to the substitution of local species and utilization of areas with some ecological value (RFA 2008).
Table 1.
Comparison of calorific values between conventional and alternative fuels and the corresponding references
| Fuel type | Cal value | Reference |
|---|---|---|
| Gasoline | 47.00 kJ/g | www.engineeringtoolbox.com |
| Diesel | 45.00 kJ/g | (Hanumantha Rao 2009) |
| Biodiesel | 37.27 kJ/g | http://www.berr.gov.uk |
| Methane | 35.60 kJ/L | (Sialve et al. 2009) |
| Biogas | 43.00 kJ/g | www.engineeringtoolbox.com |
| Hydrogen | 150.00 kJ/g | www.engineeringtoolbox.com |
| Coal | 27.00 kJ/g | (Matsunaga et al. 2009) |
| Ethanol | 30.00 kJ/g | www.engineeringtoolbox.com |
| Bioethanol | 26.72 kJ/g | http://bioenergy.ornl.gov |
| Rapeseed | 39.70 kJ/g | www.biofuelsb2b.com. |
| Sunflower | 39.60 kJ/g | www.biofuelsb2b.com. |
| Switchgrass | 16.70 kJ/g | www.ecn.nl |
| Wheat | 15.00 kJ/g | www.biofuelsb2b.com. |
| Peanut | 39.80 kJ/g | www.biofuelsb2b.com. |
| Sesame | 39.30 kJ/g | www.biofuelsb2b.com. |
| Soybean | 39.60 kJ/g | www.biofuelsb2b.com. |
| Jatropha | 39.07 kJ/g | (Hanumantha Rao 2009) |
| Chlorella | 21.00–28.00 kJ/g | (Scragg et al. 2002) |
| Microalgae | 25.80 kJ/g | (Matsunaga et al. 2009) |
Although biofuels are currently more expensive than fossil fuel, their production is exponentially increasing worldwide. Ethanol production experienced a twofold rise in the last 4 years reaching 67 billion litres in 2008. The increase in biodiesel production has even been more extraordinary, increasing sixfold up to 12 billion litres, in the same period of time (REN21 2009). Biodiesel and bioethanol derived from edible crops, using today’s technology, do not represent an effective alternative to substitute conventional fuel due to high costs of production and the land use competition with edible crops. Therefore, transition from the first (edible crops) and second generation (lignocellulosic biomass from dedicated non-edible crops like switchgrass and agricultural waste) to a third generation of biofuel, such as microalgae, is a promising option of sustainable biofuel production. For a description of all the different generations of biofuels, Gressel (2008) should be consulted. In addition to their higher yield per hectare, microalgae cultures do not compete with agriculture, requiring neither bio-productive lands nor freshwater (Chisti 2007, 2008; Griffiths and Harrison 2009; Mata et al. 2010, Rittmann 2008).
In this review, we will discuss the potential of a third generation of feedstock (focusing on cyanobacteria) as a viable biofuel source for energy production and compare it to first generation biofuel crops. We will also discuss the current state of the art for the production of H2, ethanol, diesel, methane, electricity and photanol from these organisms. Additionally, we will focus on the carbohydrate, lipid and amino acid metabolism and discuss the possibilities of influencing these biochemical pathways in order to improve the production of a specific biofuel and to decrease the production costs.
Cyanobacteria as a producer of third generation biofuels
The most common feedstocks used in the first and second generations of biofuel include rapeseed, sunflower, switchgrass, wheat, peanuts, sesame seeds and soybean. These sources are used to generate different liquid forms of energy including alcohols (ethanol, propanol and butanol) and vegetable oil. As mentioned previously, the major constraint of these energy crops is based on the competition with our food sources for farmland and water. To overcome this limitation, the third generation envisions a non-food biomass source for energy supply. Cyanobacteria possess certain properties which have entitled them to be one of the most promising feedstocks for bioenergy generation:
They can contain considerable amounts of lipids, which are mainly present in the thylakoid membranes.
They possess higher photosynthetic levels and growth rates compared to other algae and higher plants.
Cyanobacteria grow easily with basic nutritional requirements; they are able to survive if supplied with air [N2 (nitrogen-fixing strains) and CO2], water and mineral salts (especially phosphorous-containing salts) with light as the only energy source.
Cultivation is therefore relatively simple and inexpensive.
The accumulation of lipids in algae occurs when the organism is under stress (e.g. nutrient deprivation) and in the stationary growth phase. Another secondary advantage is that cyanobacteria, being prokaryotes, can much more readily be genetically engineered in order to enhance the production of biofuels as opposed to eukaryotic algae. Cyanobacteria possess a relatively small genome and many of them have already been completely sequenced, thus it is also less complicated to perform system biology approaches in these organisms when compared to eukaryotic algae (Rittmann 2008). The genomes of 41 strains of cyanobacteria have already been sequenced including strains that are amenable to genetic manipulation (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). Attempts to increase the biofuel content in cyanobacteria by genetic engineering has been mainly focused on Synechocystis sp. PCC 6803, which was the first cyanobacterium whose genome was sequenced (Kaneko et al. 1996) and Synechococcus sp. strain PCC 7942 whose genome has recently been sequenced (DOE Joint Genome Institute: http://genome.ornl.gov/microbiol/syn_PCC7942).
Therefore, cyanobacteria are a potential candidate for the production of biofuels and H2 (Schütz et al. 2004, Rittmann 2008). They should, however, not only be viewed as a biocatalyst of sunlight, they also possess other additional properties which allow them to become ideal candidates for the development of bio-friendly systems for energy generation. Due to their ability to thrive in elevated CO2 conditions, cyanobacteria have lately received considerable attention as a promising system for biological CO2 mitigation, driving down CO2 emissions from industrial activities. In addition, some cyanobacterial species are also able to generate NH+4, relieving the dependency on chemical fertilizers. Moreover, cyanobacteria have been applied as bioremediation agents to remove heavy metals from aquatic ecosystems and reduce the excess of phosphate and nitrate in farmlands (Hall et al. 1995; Ono and Cuello 2007). In conclusion, it can be stated that the use of cyanobacteria to harness solar energy for the production of different types of bioenergy might represent a simpler and cleaner system for the production of sustainable energy.
Cyanobacteria
Cyanobacteria as a source of renewable energy
Cyanobacteria, being photosynthetic organisms, use the sun’s energy, H2O and CO2 to synthesize their energy storage components, i.e. carbohydrates, lipids and proteins. These energy storage components form a potential feedstock which can be converted into bioenergy (Table 2) (SERI 1984). Of these three biochemical fractions, lipids have the highest energy content. To extract the energy from the lipid fraction, it has to be transesterified with a chemical process and the resulting hydrocarbons subsequently extracted. The hydrocarbons can then be used as transport fuel in the form of biodiesel. The carbohydrates may be transformed to ethanol by fermentation under dark, anoxic conditions (Stal and Moezelaar 1997) while alternatively, with the use of anaerobic digestion all three fractions can be converted to CH4 gas (SERI 1984; Sialve et al. 2009). Cyanobacteria possess unique properties which make them a promising model to transform all these C sources into valuable fuels. The following sections discuss the wide range of fuels which can be potentially obtained from cyanobacterial biomass.
Table 2.
Chemical composition of cyanobacteria (SERI 1984)
| Fuel | Energy storage component | Fuel production | Total energy (MJ/kg) | |
|---|---|---|---|---|
| Ethanol | Carbohydrate | 0.329 L/kg | 7.74 | |
| Oil | Lipids | Hydrocarbons | 1.150 L/kg | 44.96 |
| Fatty acids | 1.250 L/kg | 43.80 | ||
| Biogas | Carbohydrates | 0.370 (m3/kg) | 11.01 | |
| Lipids | 1.040 (m3/kg) | 30.95 | ||
| Proteins | 0.490 (m3/kg) | 14.58 | ||
Hydrogen
Hydrogen can be produced by many strains of cyanobacteria by the reversible activity of hydrogenase. When cyanobacteria are grown under N2-limiting conditions, H2 is formed as a byproduct of N2 fixation by nitrogenase (EC 1.7.99.2). It was also shown that non-heterocystous cyanobacteria are less efficient in H2 production than the heterocystous organisms. Several reports have reviewed cyanobacterial species capable of producing H2 (Abed et al. 2009; Das and Veziroglu 2001; Dutta et al. 2005) including at least 14 genera cultivated under different growth conditions. These genera include: Anabaena, Oscillatoria, Calothrix, Cyanothece, Anabaenopsis, Nostoc, Synechococcus, Mycrocystis, Gloebacter, Synechocystis, Aphanocapsa, Gleocapsa, Microcoelus and Chroococcidiopsis (Dutta et al. 2005). Among these genera it was shown that Anabaena spp. were able to produce the highest amount of H2 (68 μmol mg−1 chl a h−1). A comparison of the advantages which cyanobacteria have above other H2-producing microorganisms has been described elsewhere (Hall et al. 1995). Research into the production of H2 in cyanobacteria is at the moment focusing on the identification of new strains with specific H2 metabolism, optimising cultivation conditions in bioreactors and genetically modifying specific strains to enhance H2 production (McKinlay and Harwood 2010; Schütz et al. 2004; McNeely et al. 2010). The main constraint for H2 production in cyanobacteria is that hydrogenases are highly intolerant to the O2 produced during photosynthesis. In addition, the availability of the reducing agents such as ferredoxin and NADPH is another bottleneck as these are also involved in other routes like respiration. In order to enhance H2 production, it will be important to redirect part of the electron flow towards the H2-producing enzymes and to engineer oxygen-tolerant hydrogenases (Angermayr et al. 2009; Weyman 2010). Recently, an attempt to eliminate pathways that consume reducing agents has been carried out by Dismukes’ group. The mutants of Synechococcus 7002 lacking lactate dehydrogenase have resulted in a fivefold increment of the total H2 production compared to the wild type (McNeely et al. 2010). Moreover, the emergence of synthetic biology approaches will facilitate the future development of specialised strains for biofuel production (Huang et al. 2010).
Biological H2 production has been lately receiving considerably attention as a potential renewable energy source. Recently, the EC funded under the Framework Programme (FP7, 2008–2012) the SOLAR-H2 project with almost 4 million € (http://cordis.europa.eu) which aims to improve the photobiological H2 production in Cyanobacteria.
Ethanol
Ethanol produced from renewable resources is an appealing energy source due to the fact that it can be mixed with existing diesel and used without any modification of existing diesel engines (Kaygusuz 2009). Currently, bioethanol is produced by fermentation of agricultural crops, mainly sugarcane in Brazil (Goldemberg 2007) and/or corn in the US (Hill et al. 2006). Due to its large-scale production from agricultural crops (sugarcane and corn) it remains to be a controversial alternative to fossil fuel due to its negative impact on food supply and food price sustainability (Rittmann 2008). The advantage that cyanobacteria have over the traditional energy crops in producing ethanol is that they ferment naturally without the need to add yeast cultures as is the case with fermentation of traditional energy crops. This characteristic makes cyanobacteria a promising candidate for the production of ethanol. In order to study the fermentation ability of cyanobacteria, Heyer and collaborators (Heyer and Krumbein 1991) screened 37 strains and analyzed their ability of fermentation and the secretion of the fermentation products. Of the 37 strains studied, it was found that 16 strains were able to produce ethanol as one of the fermentation products while significant quantities of ethanol were produced in two Oscillatoria strains (>10 μmol/sample). Fermentation took place under dark conditions when no photosynthetical oxygen was produced, thus excluding respiration for energy production. Normally, fermentation does not represent a primary energy source for most algae and cyanobacteria. In these organisms, fermentation works at a minimum level which allows them to survive. To overcome this problem and to increase the ethanol production, genetic modification might be a possible solution. The first cyanobacterial species to be genetically modified in order to produce ethanol was Synechococcus sp. PCC 7942. The strain was transformed by inclusion of coding sequences for pyruvate decarboxylase and alcohol dehydrogenase II from Zymomonas mobilis, an obligately fermentative prokaryote. These genes were expressed under the control of the cyanobacterial rbcLS operon promoter, alone and in combination with the Escherichia coli lac promoter. The reported yields of ethanol produced by the transformed strain reached 54 nmol OD730 unit−1. liter−1. day−1 (Deng and Coleman 1999). The same genes have also been recently expressed in Synchocystis sp. PCC 6803 under the control of a different promoter, the strong light driven psbAII. This strain showed an increase in ethanol production reaching 5.2 mmol OD730 unit−1. liter−1. day−1) (Dexter and Fu 2009).
An alternative method for the production of ethanol is to produce it from cellulosic material. It has been observed that cyanobacteria deposits cellulose extracellulary at a yield of up to 25% of the cell dry weight (Dewinder et al. 1990). Synchococcus sp. PCC 7942 was modified by Nobles and Brown with the cellulose synthase genes from Gluconobacter xylinus and this transformed strain was able to produce extracellular non-crystalline cellulose. The non-crystalline nature of cellulose makes it ideal as a feedstock for ethanol production facilitating its hydrolysis (Nobles and Brown 2008).
The focus of optimising bioethanol-producing strains should start by screening several strains of cyanobacteria and using various promoters, afterwards, to establish the best combination. Studying different growth conditions as well as optimising ethanol retrieval systems could lead an increase of ethanol production to levels where it will become economically feasible (Deng and Coleman 1999). The effect of salt stress conditions on the fermentation rate has been recently evaluated in Cyanobacteria. Ethanol production in a high salt concentration medium (1.24 M NaCl) was over 100-fold higher compared with the low salt conditions (0.24 M NaCl), resulting in a production of 0.75 mmol/g (Carrieri et al. 2010). In addition, Luo and co-workers have analysed the energy consumed and greenhouse gas emissions in different ethanol-producing systems employing cyanobacteria. This study showed that these two parameters are highly influenced by the concentrations of EtOH secreted by cyanobacteria. Their modelling results reveal that initial EtOH concentrations from 0.5 to 5 wt% would be enough to develop an environmentally friendly biofuel production system with reduced energy consumption and air pollution (Luo et al. 2010).
Different countries are currently funding projects in order to improve the bioethanol production efficiency in cyanobacteria. The US Department of Energy is sponsoring a project to the value of US $1.6 million for the DNA sequencing of six strains of Cyanothece which shows promising ethanol production levels (http://news-info.wustl.edu/tips/page/normal/7719.html). The Federal Ministry of Education and Research in Germany will invest substantial funding for research on the production of bioethanol from cyanobacteria led by the Institute of Biology at Humboldt University (http://www.drivehomesafe.com/news/the_bmbf_supports_research_on_producing_ethanol_cyanobacteria-6.html).
Ethanol is today the most common biofuel worldwide although longer chain alcohols have lately attracted some attention. The longer chain alcohols possess higher energy content and can be stored and transported easier than ethanol (Atsumi et al. 2008). Recently, Radakovits et al. (2010) pointed out that influencing the keto acid pathway and thereby producing isobutanol might be a promising source of biofuel in eukaryotic microalgae. The production of longer chain alcohols (C5–C–8) in E. coli bacteria has been achieved by the overproduction of 2-keto acids, intermediates of the amino acid biosynthesis. These intermediates were later converted to butanol derivatives by the heterologous expression of 2-keto acid decarboxylase and alcohol dehydrogenase (Atsumi et al. 2008; Zhang et al. 2008). A very recent discovery that citramalate synthase occurs in cyanobacteria generates the possibility to produce propanol and butanol from 2-ketobutyrate, as this compound is an intermediate in the biosynthesis of citramalate (Wu et al. 2010). Engineering isobutanol biosynthetic pathway and overexpressing Rubisco have resulted in an enhanced production of isobutyraldehayde and isobutanol (6,230 and 3,000 μg L−1 h−1, respectively) in Synechococcus elongatus PCC 7942 (Atsumi et al. 2009).
In addition, compared to isoprene, ethanol has lower energy content and is miscible in water which requires a time-consuming and expensive distillation process before it can be used. Thus, isoprene besides being useful in the industry as the basic unit of synthetic rubber could also be suitable as biofuel. In a recent study, Synechocystis PCC 6803 was genetically modified with the Pueraria montana ispS gene to enable the production of isoprene in this microorganism (Lindberg et al. 2010). Alkanes represent another appealing chemical feedstock fuel due the high energy content they possess. Recently, two-step alkane biosynthesis has been reported in cyanobacteria. This new finding opens new possibilities for alkane production by engineered mircroorganisms (Schirmer et al. 2010).
Photanol
In order to improve biofuel production, Hellingwerf and Mattos have recently developed a new technology called the photanol approach (Hellingwerf and de Mattos 2009). Photanol has been one of the projects funded by the Dutch Ministry of Agriculture under the Biorefinery Energy Innovation Agenda (www.senternovem.nl).
In this approach, the abilities of photosynthetic and fermenting bacteria are combined in a single organism (Synechocystis sp. PCC 6803). In photoautotrophic microorganisms, CO2 is transformed into C3 sugars like glyceraldehyde-3-phosphate (G3P), which are indispensable intermediates in the biosynthesis of complex molecules involved in the basic functions and structure of the organism. In chemotrophic organisms, however, different carbohydrates are first degraded to C3 sugars to obtain energy (ATP) and converted afterwards into a variety of alcohols such as ethanol, butanol, propanediol and many others. In the photanol strategy, the properties of a chemotrophic organism have been included by means of genetic engineering into a photosynthetic organism (Synechocystis sp. PCC 6803). The C3 sugar, G3P represents in this transformed organism the central linking compound between photosynthesis and fermentation. This modified organism uses solar energy to convert CO2 into biofuel with the advantage that the number of steps to do so has been minimised. This has lead to an increase in biofuel production efficiency compared to the current biofuel production processes reaching theoretical levels of 105 L ha−1 year−1. Table 3 shows the energy productivity from the different bioenergy sources discussed above using mainly cyanobacteria as feedstock.
Table 3.
Bioenergy productivity of various energy sources using cyanobacteria as feedstocks
| Fuel | Organism | Productivity | kJ/yeara | References |
|---|---|---|---|---|
| Ethanol | Synechoccocus PCC 7942 | 54.0 nmol/L/day | <0.03 | Deng and Coleman 1999 |
| Ethanol | Synechocystis PCC 6803 | 5.2 mmol/L/day | 2593.048 | Dexter and Fu 2009 |
| Ethanol by algenol biofuels | Cyanobacteria | 56,000.0 L/ha/year | 1.31 × 109/ha | Luo et al. 2010 |
| Fatty acids | E. coli | 4.5 g/L/day | 67,671.00 | Liu et al. 2010c |
| Fatty acids | Synechocystis PCC 6803 | 6.4 nmol/L/day | <0.03 | Kaczmarzyk and Fulda, 2010 |
| Fatty acids | Synechoccocus PCC 7942 | 8.4 nmol/L/day | 0.03 | Kaczmarzyk and Fulda, 2010 |
| Isobutyraldehyde | Synechoccocus PCC 7942 | 6,230.0 μg/L/h | 136.44 | Atsumi et al. 2009 |
| Isobutanol | Synechoccocus PCC 7942 | 3,000.0 μg/L/h | 864.61 | Atsumi et al. 2009 |
| Methane | S. maxima | 0.4 L/day | 8,030.00 | Varel et al. 1988 |
| Hydrogen | S. maxima | 400.0 μmol/L/h | 994,435,200.00 | Ananyev et al. 2008 |
aCalculated from previous published data
Diesel
For the production of lipid-based biofuels, cyanobacteria have received less attention than other feedstocks such as microalgae (Miao and Wu 2006; Rodolfi et al. 2009) or crops. As an energy source, cyanobacterial biomass has traditionally been associated with the production of ethanol (Deng and Coleman 1999; Dexter and Fu 2009) or H2 (Hall et al. 1995). In 1998, 3,000 species of microalgae were screened in the Aquatic Species Program with the aim to identify species with high lipid content. In this program, little information regarding cyanobacteria was provided since they do not accumulate high amounts of lipids. It was, however, shown that cyanobacteria have the fastest growth rates and that the lipid productivity was amongst the highest in exponentially growing cultures (Sheehan et al. 1998). Spirulina also showed the highest overall utilization efficiencies in integrated liquid–gaseous fuel-processing options (SERI 1984). On the other hand, a recent comparison of different strains of microalgae revealed that although cyanobacteria possessed the highest biomass productivity, it showed a low lipid content reflecting the high metabolic cost of lipid synthesis (Francisco et al. 2010).
Around 2,000 species of cyanobacteria have been identified (Sheehan et al. 1998), but information regarding the production of biodiesel from these species or related parameters such as the biochemical profile, growth rate and energy content of the different species are scarce (Miao and Wu 2006). The implication of this is that the selection of adequate cyanobacteria strains for the production of biodiesel will not be an easy task. Table 4 summarizes the available information pertaining to the chemical composition of cyanobacteria. This information might assist in the evaluation of cyanobacteria species for industrial bioenergy production. To choose species for the large-scale production, a wide range of variables are important of which (Griffiths and Harrison 2009; Grobbelaar 2000) lipid content (percent dry weight), productivity (milligrams per litre per day) and growth rates (doubling time) are keys for the production of biodiesel. Griffiths and Harrison (2009) collected data on the biodiesel production of 55 microalgae species. Synechococcus with a production of 75 mg/L of lipids per day was among the highest yielding strains. Liu et al. (2010a) reported high secretion levels (133 mg L−1 day−1) of FFA by an engineered Synechocystis sp. However, this paper was retracted last July after some of its coauthors decided to remove their name and date from it (Liu et al. 2010b).
Table 4.
Different feedstock constituents from microalga (SERI 1984)
| Species | Protein (% dw) | Lipid content (% dw) | Carbohydrate (% dw) | Cal. value (kJ/10 g dw) | References |
|---|---|---|---|---|---|
| Phormidium sp. (F) | 62 | 11 | 16 | d.n.a. | Cañizares-Villanueva et al. 1995 |
| Calothrix crustacea (HF) | 21 | 71a | 8 | 25 | Nagarkar et al. 2004 |
| Calothrix contanerii (HF) | 27 | 64a | 8 | 29 | Nagarkar et al. 2004 |
| Gloeocapsa crepidinum (U) | 56 | 36a | 8 | 21 | Nagarkar et al. 2004 |
| Limicolaria martensiana (F) | 19 | 76a | 5 | 22 | Nagarkar et al. 2004 |
| Lyngbya semiplena (F) | 27 | 64a | 9 | 23 | Nagarkar et al. 2004 |
| Phormidium corium (F) | 50 | 34a | 16 | 33 | Nagarkar et al. 2004 |
| Phormidium tenue (F) | 63 | 22a | 15 | 31 | Nagarkar et al. 2004 |
| Spirulina subsalsa (F) | 71 | 13a | 17 | 35 | Nagarkar et al. 2004 |
| Spirulina labyrinthiformis (F) | 68 | 17a | 15 | 34 | Nagarkar et al. 2004 |
| Spirulina obliquus (F) | 50–56 | 12–14 | 10–17 | d.n.a. | Sialve et al. 2009 |
| Spirulina platensis (F) | 56–77 | 9–14 | 10–18 | d.n.a. | Ciferri 1983 |
| Spirulina maxima (F) | 60–71 | 4 | 8–13 | d.n.a. | Ciferri 1983 |
| Oscillatoria formosa (F) | 51 | 32a | 9 | 15 | Nagarkar et al. 2004 |
| Oscillatoria salina (F) | 42 | 47a | 11 | 19 | Nagarkar et al. 2004 |
| Oscillatoria subbrevis (F) | 45 | 57a | 12 | 21 | Nagarkar et al. 2004 |
| Oscillatoria spp. | d.n.a. | 13 | d.n.a. | d.n.a. | Griffiths and Harrison 2009 |
| Synechocystis spp. (U) | d.n.a. | 50 | d.n.a. | d.n.a. | Rittmann 2008 |
| Synechococcus spp. (U) | 64 | 28 | 9 | 28 | Nagarkar et al. 2004 |
| Anabaena cylindrica (HF) | d.n.a. | 5 | d.n.a. | d.n.a. | Griffiths and Harrison 2009 |
| Anabaena sp. ATCC 33047 (HF) | 45 | 10 | 28 | d.n.a. | Vargas et al. 1998 |
| Anabaena variabilis (HF) | 47 | 11 | 22 | d.n.a. | Vargas et al. 1998 |
| Anabaenopsis sp. | 52 | 11.4 | 16 | d.n.a. | Vargas et al. 1998 |
| Nodularia sp. (Chucula) (HF) | 43 | 12.6 | 17 | d.n.a. | Vargas et al. 1998 |
| Nostoc commune (HF) | 40 | 8.4 | 38 | d.n.a. | Vargas et al. 1998 |
| Nostoc paludosum (HF) | 40 | 10.4 | 27 | d.n.a. | Vargas et al. 1998 |
| Several Nostoc sp. not identified | 37–47 | 7.9–11 | 16–32 | d.n.a. | Vargas et al. 1998 |
Note that fuel production and energy are expressed in dry biomass of the class of compound (carbohydrate, lipid, protein)
HF heterocystous filamentous cyanobacteria, F non-heterocystous filamentous cyanobacteria, U unicellular cyanobacteria, d.n.a. data not available
aThe lipid content was calculated as difference between 100 and the sum of proteins and carbohydrates
Vargas and co-workers (Vargas et al. 1998) analyzed the biochemical composition of 12 N2-fixing cyanobacteria. The content of the lipids in these strains ranged between 8–12% dw, of which Nodularia and Nostoc contained the highest amount of lipids. Nagarkar et al. (2004) reported on the chemical composition and their respective calorific values of 13 cyanobacteria species (they did not report on the lipid content). They found that the calorific values varied between 15–33 kJ 10 g−1 dw with Spirulina and Phormidium responsible for the higher values. They could also correlate the high calorific values with high protein content. Vermaas and colleagues have genetically engineered a single gene mutant of Synechocystis able to accumulate up to 50% of dry weight in lipids (Rittmann 2008). A bioscience firm in the USA, Targeted Growth, has recently claimed to have developed a new technology to increase the lipid content of cyanobacteria by approximately 400% (Timmerman 2009). It should be pointed out here that absolute values rather than percentages should be provided when discussing the improvement of lipid production. Percentages offer no possibility to have an idea about the lipid content reached and therefore to appreciate the real success of the new technique developed. Another company, Synthetic Genomics Inc., announced an agreement with ExxonMobil in 2009, to develop the next generation biofuels using photosynthetic algae including microalgae and cyanobacteria. ExxonMobil will invest US $600 million to develop more efficient means to harvest the oils which the photosynthetic algae produce (Howell 2009).
Biodiesel quality is also an important factor as it should meet various specifications before commercialization according to the European or American standards (UNE-EN 14214 and US ASTM D6751, respectively). Important parameters including oxidation stability, cetane number, iodine value and cold-flow properties are closely correlated to the fatty acid composition and are determined by the degree of saturation and the chain length of the fatty acids. Low cetane numbers are associated with shorter chain lengths and an increase in the level of unsaturation in the fatty acid. Moreover, a high content in unsaturated fatty acids is responsible for decreasing the oil oxidation stability while biodiesel consisting of saturated long chain fatty acids shows poor cold-flow properties (Knothe 2008; Pinzi et al. 2009). The fatty acids C10:0, C16:1 and C18:1, have the best combination of properties to produce high quality biodiesel. Investigation of the fatty acid profile of the raw material is thus important when selecting cyanobacteria species for biodiesel production. Several reports of the fatty acid composition of different cyanobacteria species grown under different conditions are available in the scientific literature. The fatty acid composition is often governed by the growth temperature where the degree of unsaturation increases at lower temperatures and the biosynthesis of shorter acyl chains occur (Liu et al. 2005). An increase of cyanobacterial biomass and lipid content is observed in strains grown in wastewater from the swine industry (Cañizares-Villanueva et al. 1995). The amount of polyunsaturated fatty acids (PUFA) was also decreased and those of monounsaturated fatty acids (MUFA) were increased at high light intensities (Walsh et al. 1997). The N2 level is also known to influence the biochemical composition in cyanobacteria. The presence of a combined N2 source drove up the protein content and drove down the amounts of lipids and carbohydrates in cyanobacteria, although biomass productivity was only slightly affected. Finally, biochemical composition can also vary according to the growth phase, with the highest lipids occurring as the culture entered in the stationary phase (Vargas et al. 1998).
Generally, unicellular types of cyanobacteria lack PUFA, while most of the filamentous species contain high levels of di- and trienoic fatty acids (Kenyon 1972; Kenyon et al. 1972). Thus, unicellular strains represent the most suitable choice for high quality biodiesel production because they have a larger MUFA amount.
Methane
Cellular biomass can be transformed to CH4, under anoxic conditions, through a process known as anaerobic digestion (AD). After lipid extraction from cyanobacterial biomass, the remaining material can be converted into CH4 by this process raising the total energy recovery. This will lead to a more favourable or positive energetic balance of the overall biofuels production by cyanobacteria, which could also decrease the total costs of the process for bioenergy production. Furthermore, when the algae accumulate less than 40% of lipids, the oil recovery results in a 21% of the energetic value but the energetic costs of lipid recovery involved are higher than 30% (Sialve et al. 2009). In this case, AD represents an ideal choice for the total energetic recovery of biomass.
The production of CH4 by an organism is highly correlated with its biochemical composition (C, N2, phosphorous and oligonutrients). The methanogenic activity of cyanobacterial biomass in general is less desirable (0.31–0.47 L CH4 L−1 day−1) compared to cattle/swine waste or from mixed slurry (in the range of 3–6 L CH4 L−1 day−1) (Chellapandim et al. 2010; Varel et al. 1988). Therefore, not much research has been performed on the CH4 production by cyanobacteria biomass leading to insufficient information in literature in order to compare the yield of different cyanobacteria strains. However, there are some examples that deserve to be mentioned. Spirulina maxima is one of the species studied in terms of the CH4 production by AD (Samson and Leduy 1982, 1986; Varel et al. 1988). Mesophilic temperatures at 35°C seem to be optimum for a maximum yield (around 0.40 L CH4/g VS fed) with an energy conversion efficiency of 59%. Compared to other microalgae, the CH4 production of S. maxima is comparable to the yields obtained from Scenedesmus spp. and Chlorella spp. (0.4–0.8 L CH4 L−1 day−1). Further studies should analyze the effect of different parameters on the production efficiency such as lack of nutrients, poor C/N ratio and accumulation of toxic compounds in the organisms which could reduce the gas productivity. The main constraints in the CH4 production and the key factors influencing the AD yield in microalgae have recently been comprehensively analyzed by Sialve (Sialve et al. 2009).
Several studies have illustrated that the CH4 production from cyanobacteria can be successfully combined with the natural ability of certain species to mitigate contaminants from the environment. The N2-fixing cyanobacterium, Anabaena sp., was shown to be able to biodegrade cyanides and thereby producing CH4 in batch reactors. This study carried out by Gantzer (Gantzer and Maier 1990) showed that Anabaena reduced cyanides by nitrogenase to CH4 and NH3, an enzyme normally responsible for the reduction of N2. It was found that the rate for CH4 production was ten times faster than expected based on literature values. Other examples in the literature describe cyanobacteria participating in a two-step system for the production of CH4 either by producing nutrients (used by methanogenic bacteria) or by removing CO2 from biogas and thereby improving its quality. Sustainable CH4 production has been achieved in Synechococcus PCC 7942 from atmospheric CO2 and solar energy, where the photosynthetic products including glucose or acetic acid were used as nutrients by a methanogenic bacterium in the CH4 generation (Koshland 2009). Another species, Arthrospira platensis has also been involved in biogas production by removing CO2 from the biogas formed by AD of sewage sludge (Converti et al. 2009).
Currently, “The Baltic EcoEnergy Cluster” is starting a project to produce biogas with high CH4 and H2 contents from algae and cyanobacterial biomass. The algae will be harvested from the Gulf of Gdansk and the Vistula submersion. The project is expected to be completed in 2013 (BiofuelDigest 2009).
From an economic point of view, the cost of CH4 from cyanobacteria is still far more expensive when compared to the CH4 derived from fossil fuels. In the coming years, this situation might change with efforts to improve the current technology resulting in economically reasonable prices of the gas. The process of CH4 production alone or coupled to other bioenergy producing processes, therefore needs further investment in research (Rittmann 2008).
Electricity and fuel cells
It has recently been reported that microorganisms can convert light into electric energy with the use of photoelectrochemical cells. In these cells, high-energy electrons produced by the light excitation in the photosystems are transferred to an electron mediator, which in turn transfers them to an electrode and thereby producing electricity. Examples in the literature have studied strains like Anabaena (Yagishita et al. 1996) and Synechococcus or Synechocystis (Moriuchi et al. 2008; Tsujimura et al. 2001; Yagishita et al. 1996) for this purpose. Dr. Ilia Baskakov’s research at the University of Maryland (Baltimore, USA) focused on the study of the electricity production in cyanobacteria has been funded by the prestigious Elkins Professorship Award (www.umbi.umd.edu).
Cyanobacterial species can also act as natural resources of H2 in fuel cells (Dawar et al. 1998). Behera and co-workers employed Spirulina in fuel cells for H2 production. This species possess a known high nutritional value that could reduce the production costs of the energy production by this mean (Behera et al. 2007).
Central carbon metabolism in cyanobacteria
Cyanobacteria are aerobic photototrophic organisms generating ATP and NADPH during the light phase of photosynthesis. In the dark phase of photosynthesis, commonly known as the Calvin cycle, these molecules are used to produce sugars and other organic compounds from CO2 and water. In short, during the first step in the Calvin cycle, CO2 is assimilated by Rubisco (rbcL, EC 4.1.1.39) through the carboxylation of ribulose-1,5-bisphosphate (RuBP) to form 3-phosphoglycerate (3PG). Glucose-6-phosphate (G6P) is in turn formed from 3PG via gluconeogenesis. Finally, RuBP is recovered from fructose 6-phosphate, G3P and dihydroxyacetone phosphate in a sequence of reactions similar to the non-oxidative branch of the pentose phosphate pathway.
Enzymes involved in the Calvin cycle are encoded by genes known as cbb genes. Gibson and Tabita showed that these genes are regulated by a common promoter activated by a LysR-type transcription factor, CbbR. Knockout strains with mutations affecting CbbR were impaired in the expression of cbb genes (Gibson and Tabita 1996).
It is well known that cyanobacterial Rubisco possesses a relative low affinity for CO2 when compared to other algae or higher plants. In order to overcome this problem, they have developed a CO2-concentrating mechanism (CCM). The CCM contains two carbon-fixing enzymes, Rubisco and carbonic anhydrases, stored in carboxysomes. These storage microcompartments are thought to increase the CO2 level surrounding Rubisco away from the competing O2. A detailed understanding about the regulation of the main CCM constituents may enable the manipulation of this system to optimize the CO2 fixation. A recent study has shown that in cyanobacteria, carboxysomes possess a specific organization through the cell, not found in other prokaryotes, and this distribution is closely linked to the CO2 fixation efficiency. It seems that two cytoskeletal proteins, par A (involved in chromosome and plasmid segregation) and merB (involved in cell morphology), are involved in the organization of this specific carboxysomal pattern (Savage et al. 2010).
Inorganic carbon (Ci) transporters are also important CCM components and they are responsible for the intracellular delivery of CO2 and HCO3 (Badger et al. 2006). The overexpression of carbonic anhydrase, insertion of a more efficient Rubisco or multiple copies of Ci transporters could in principle increase CO2 fixation levels in cyanobacteria. Although Rubisco is the main enzyme responsible for the C fixation, cyanobacteria possess an additional assimilation mechanism operated by phosphoenolpyruvate carboxylase (PEPC) and malic acid to assist them in the fixation of large amounts of CO2 which is similar to the C4 pathway in plants (Yang et al. 2002).
The 2-phosphoglycolate (P-glycolate) is a noxious by-product of the Rubisco oxygenase activity which inhibits important enzymes in the Calvin cycle (phosphofructokinase and triosephosphate isomerase). Besides photorespiration, in cyanobacteria, this molecule is degraded via the glycerate and decarboxylation pathway. Certain compounds derived from these pathways, including glycolate and glycine, seem to be potential candidates to control the Ci level in cyanobacteria (Eisenhut et al. 2008).
Some cyanobacteria strains are capable of assimilating some sugars and growth in dark conditions as facultative heterotrophs. Nevertheless, cyanobacteria grown in the dark have shown lower growth rates than when grown under light conditions (Stanier and Cohenbazire 1977).
Unlike most other phototrophs, in cyanobacteria, photosynthesis and respiration co-occur in a single compartment within the cell, the thylakoid membrane. In addition, constituents from both electron transfer chains such as the redox carriers cytochrome bf complex, plastoquinone, cytochrome c6 and plastocyanin are shared as well. Although they have common elements, some of them are still specifically associated to one of the pathways. Photosystem I (PSI) and PSII are photosynthesis specific, whereas NADP dehydrogenase (ndh, EC 1.6.99.3), succinate dehydrogenase (sdh, EC 1.3.99.1) and terminal oxidases occur only in the respiratory chain. In contrast to higher plants, cyanobacteria often possess PSI/PSII ratios larger than 1. Under light conditions, PSI competes with terminal oxidase for electrons to maximize the amount of NADPH required for CO2 fixation (Vermaas 2001). Recently, Nelson (2010) has shown how this PSI could be used as a small battery charger (Nelson 2010).
Carbohydrate metabolism
Sugars are the main and most common source of metabolic energy among living organisms. Sugar catabolic pathways [glycolysis, the oxidative pentose phosphate pathway (OPP) and tricarboxylic acid cycle (TCA)] are active mainly during the dark phase of the light–dark cycle. These pathways are responsible for producing NAD(P)H and other biosynthetic metabolites involved in the normal cellular functions. The major route of glc degradation is the OPP cycle and is considered as the main CO2 fixation mechanism in cyanobacteria. The key enzymes in the oxidation of G6P through the OPP cycle are glucose-6-phosphate dehydrogenase (zwf, EC 1.1.1.49) and 6-phosphogluconate dehydrogenase. Glucose-6-phosphate dehydrogenase controlled at the level of gene expression is especially interesting from a regulation point of view. In addition, low RuBP levels significatively reduce this enzyme activity (Kaplan et al. 2008; Stanier and Cohenbazire 1977; Vanderoost et al. 1989).
Modulation of sugar catabolic pathways in cyanobacteria during the light–dark transition has been reviewed by Osanai et al. (2007). In Synechocystis sp. PCC 6803, enzymes that participate in the sugar catabolism are stimulated by the light–dark shift and by the circadian rhythm. Regulatory proteins including histidine kinase, Hik8 and the RNA polymerase sigma factor, Sig E are involved in this activation. In addition, reduced N2 concentrations also trigger the transcription of sugar catabolic genes via NtcA, the major N2 mediator. A detailed analysis of the transcriptional network in central metabolism during light periods has been provided by microarray data from Cyanothece 51142 (Stockel et al. 2008). This study revealed that the glycogen accumulated in diurnal periods is later degraded via glycolysis, OPP and the TCA cycle during dark or C depletion conditions. However, cyanobacteria possess an incomplete TCA cycle unable to work properly as a respiratory chain (discussed in the next section) (Stanier and Cohenbazire 1977). Synechocystis sp. PCC 6803 was shown to be able to grow under dark conditions with periodic light pulses at glc expenses, a phenomenon known as light-activated heterotrophic growth (LAHG). The genes hik8 and sigE seem to be involved in LAHG (Osanai et al. 2005; Singh and Sherman 2005). During LAHG conditions, the glycolytic enzyme fructose 1,6-bisphosphate aldolase (fbaA, EC 4.1.2.13) is induced by sll1330 (a putative helix-turn-helix DNA-binding protein) in Synechocystis sp. PCC 6803 (Tabei et al. 2009). Central sugar metabolism differs among photoautotrophic, heterotrophic and photomixotrophic growth conditions (reviewed by Kaplan et al. 2008). Previous studies revealed that genomic and metabolomic data provide enough information to model the central metabolism and estimate the flux balance during different conditions in Synechocystis PCC 6803 (Hong and Lee 2007). Modelling results were correlated with experimental data and may be extrapolatable to the whole cell metabolism in this organism.
Sugars and other organic compounds from the central metabolism participate in the biosynthesis of diverse cellular metabolites. Here, only the biosynthesis of major classes of carbohydrates will be discussed: glycogen, sucrose and the carbohydrates involved in salt stress response, trehalose and glucosylglycerol and cell wall polysaccharides.
Glycogen
Glycogen is the main carbon and energy storage polysaccharide in cyanobacteria. In the cell, glycogen is synthesised during light periods from assimilated CO2 (Ball and Morell 2003). The enzyme ADP-glucose diphosphorylase (AGPase; EC 2.7.7.27, encoded by the agp gene, also known as glgC) controls glycogen synthesis in bacteria in an ATP-dependent reaction and seems to be regulated by 3PG (activator) and Pi (inhibitor) (Ballicora et al. 2003; Gomez-Casati et al. 2003).
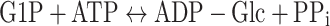 |
As discussed below, sucrose synthase can also contribute to ADP-glucose (ADP-Glc) synthesis in filamentous cyanobacteria. The formation of glycogen involves the elongation of α-1,4-linked glucan by glycogen synthase (glgA, EC 2.4.1.21) which transfers ADP-Glc to the growing chain.
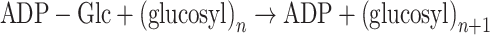 |
The last step of glycogen synthesis is the α-1,6-glycosidic bond formation catalysed by a glycogen-branching enzyme (glgB, EC 2.4.1.18). The enzyme glycogen phosphorylase (glgP, EC 2.4.1.1) is responsible for the glycogen degradation, removing glucose units to form monomers of glucose-1-phosphate (G1P). The enzyme phosphoglucomutase (pgm, EC 5.4.2.2) catalyses the conversion of G1P to G6P, which can then be used in glycolysis or in the OPP pathway. Different conditions such as N2 depletion (Yoo et al. 2007) and salt stress (Page-Sharp et al. 1998) are known to influence the glycogen accumulation in cells.
Sucrose
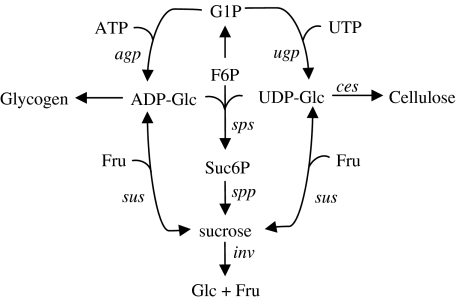
Sucrose (α-D-glucopyranosyl β-D-fructofuranoside) accumulation in cyanobacteria during conditions of salt stress or desiccation plays a fundamental role as an osmotic protective agent (Billi et al. 2000; Miao et al. 2003a). In addition, a role as storage and signalling molecule has also been associated to this disaccharide (Desplats et al. 2005).
A schematic representation of sucrose metabolism is shown in Fig. 1 [adapted from (Curatti et al. 2008)]. The main enzymes involved in the sucrose synthesis are sucrose-phosphate synthase (sps, EC 2.4.1.14) and sucrose-phosphate phosphatase (spp, EC 3.1.3.24) (Salerno and Curatti 2003). The synthesis of sucrose-6-phosphate (Suc6P) is catalysed by SPS and followed by a dephosphorylation reaction by SPP leading to the sucrose formation. Sucrose is later hydrolyzed into glc and fru in an irreversible reaction catalyzed by invertases (EC 3.2.1.26). Genomic analysis revealed the importance of these enzymes under N2-fixing conditions in filamentous cyanobacteria (Vargas et al. 2003).
Fig. 1.
Sucrose metabolic pathway in cyanobacteria (adapted from Curatti et al. 2008). Sucrose synthase has only been found in heterocyst-forming cyanobacteria. Genes encoding metabolic enzymes: agp, ugp, sps, spp, sus, inv, ces. Metabolites: ADP-Glc, G1P, UDP-Glc, F6Pm, Fru, Suc6P, Glc
Sucrose may be produced as well from activated glucose (UDP/ADP-glucose) and fructose in a reversible reaction controlled by the enzyme sucrose synthase (sus, EC 2.4.1.13). This enzyme seems to be present only in filamentous N2-fixing cyanobacteria (Curatti et al. 2000; Salerno and Curatti 2003) where it is involved in the accumulation of storage (glycogen and sucrose) and structural (such as cellulose, see below) polysaccharides (Curatti et al. 2008).
Glucosylglycerol and trehalose
Other osmoprotectants like glucosylglycerol (GG, composed of a sugar and a polyol) and trehalose also occur in cyanobacteria and they are related to the salt tolerance of the strains. GG biosynthesis involves two successive reactions similar to sucrose synthesis catalysed by the enzymes GPP-S and GPP-P, respectively. In Synechocystis sp. PCC 6803, the role in salt resistance of ggpS and ggpP (stpA) has been supported by previous studies (Hagemann et al. 1997; Marin et al. 1998).
Trehalose (α-D-glucopyranosyl-[1,1]-α-D-glucopyranoside) is known to be produced through several biosynthetic routes in the different organisms (Avonce et al. 2006). TreY-TreZ pathway is involved in the trehalose formation in Nostoc and Anabaena. TreY encodes maltooligosyltrehalose synthase (mts, EC 5.4.99.15) and treZ encodes maltooligosyl trehalose trehalohydrolase (mth, EC 3.2.1.141). Gene disruption experiments indicated that trehalose, like sucrose, plays a crucial role in dehydration (Asthana et al. 2008; Higo et al. 2006) and salt stress tolerance (Salerno et al. 2004) in different organisms.
Cell wall polysaccharides
The cyanobacterial cell wall combines features from gram-positive and gram-negative bacteria. From inside to outside a cytoplasmic membrane, a highly cross-linked peptidoglycan layer and an outer membrane with lipopolysaccharides (LPSs) are the main constituents of this cell wall (Hoiczyk and Hansel 2000). Peptidoglycan strands in cyanobacteria consist of repeating subunits of the aminosugars N-acetylmuramic acid and N-acetylglucosamine. In addition, this strand contains cross-linked peptides and it is complexed with specific polysaccharides in its structure. Peptidoglycan biosynthesis, in bacteria, is mediated by the genes murA-murG, mraY and pbp and a schematic representation can be found in Garcia et al. 2008.
The LPSs in cyanobacteria have not been extensively documented. Previous studies indicated the different composition of the LPS in marine Synechococcus sp. and proteobacteria (Snyder et al. 2009). In addition, it has been suggested that a gene homologue of lpxC, alr2270 participates in the LPS lipid A biosynthesis (Nicolaisen et al. 2009). Many cyanobacteria are also able to secrete diverse extracellular polymeric substances (EPS) into their immediate surroundings of the cell (reviewed by (Pereira et al. 2009). Cellulose has been found as the main constituent of the EPS in several cyanobacteria (Nobles et al. 2001) and the genes involved in the synthesis of this polysaccharide (cesA, EC 2.4.1.12) possess sequence resembling the plant cesA genes. As mentioned above, the sus gene seems to be involved in the cellulose biosynthesis (Curatti et al. 2000). Recently, Synechococcus sp. PCC 7942 was genetically modified to secrete non-crystalline cellulose into the growth media, which is a promising candidate for ethanol production (Nobles and Brown 2008), as discussed in “Ethanol” section.
A possible strategy to enhance the lipid content for biofuel purposes could include knocking out the genes involved in the biosynthesis of storage or osmotic protectant substances. In addition, the overexpression of genes involved in the degradation of these compounds could also increase the lipid production in the cell. Previous studies on eukaryotic algae indicate that certain starch-impaired strains accumulate higher amounts of PUFA or TAG under N2 starvation (for a review see Radakovits et al. 2010).
Redirecting the C flux to the cellulose synthesis would be another approach in order to increase the fuel content in cyanobacteria. Currently, ethanol production is derived mainly from the fermentation of cellulose. The overexpression of cesA plus a knockout of the agp, spp and sps genes could hypothetically cause an increase of the cellulose content in the cell.
Besides respiration, other polysaccharides could also in principle be catabolised by fermentation. However, as mentioned before (“Ethanol” section) in wild type cyanobacteria, fermentation does not seem to supply a significant amount of energy to the cell. Previous studies have already attempted to enhance the ethanol levels by developing genetically engineered organisms (Deng and Coleman 1999; Dexter and Fu 2009).
Lipid metabolism
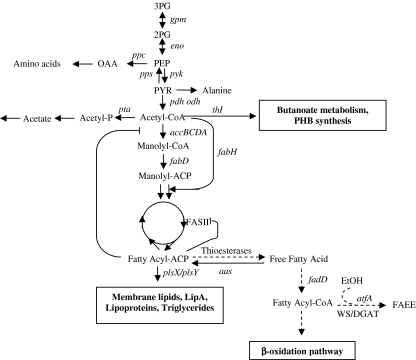
Fatty acid and protein biosynthetic pathways possess phosphoenolpyruvate (PEP) as common substrate (Fig. 2). Thus, when PEP is converted to oxaloacetate (OAA) by phosphoenolpyruvate carboxylase (ppc, EC 4.1.1.31), it enters into protein synthesis and is directed to fatty acid synthesis when transformed to malonyl-CoA. PEP is converted to pyruvate by pyruvate kinase (pyk, EC 2.7.1.40) and then by pyruvate dehydrogenase (pdhB, EC 1.2.4.1) in a second reaction to form acetyl-coenzyme A (acetyl-CoA). In addition, pyruvic acid can be converted to alanine and thus participates in protein metabolism. On the other hand, acetyl-CoA can be converted to malonyl-CoA in a rate-limiting reaction catalysed by acetyl-CoA carboxylase (accC, EC 6.4.1.2), which is the first step towards the fatty acid synthesis. Whereas high concentrations of acetyl-CoA or free fatty acids stimulated PEPC activity in E. coli, (Izui et al. 1970) in certain cyanobacterial strains, the increased levels of acetyl-CoA did not influence PEPC activity (Chen et al. 2002; Luinenburg and Coleman 1993; Owttrim and Colman 1986). PEPC from Synechococcus vulcanus was strongly activated by fructose-1,6-diphosphate while aspartate acted as a strong suppressor. This compound has been reported to reduce the PEPC activity from Coccochloris peniocystis. PEPC seems to divert the carbon flux away from fatty acid biosynthesis. Thus, the antisense expression of PEPC-coding gene (ppc), in Synechococcus sp. PCC 7002, has led to a lipid content increase in this organism (Song et al. 2008).
Fig. 2.
Simplified overview of the fatty acid biosynthesis and some of the competing pathways in cyanobacteria (adapted from Liu et al. 2010a). Pathways not present in cyanobacteria or those which are unknown are indicated with dashed lines. Genes encoding metabolic enzymes: aas, accBCDA, atfA, eno, fabD, fabH, fadD, FASII, gpm, odh, pdh, plsX/plsY, ppc, pps, pta, pyk, thI. Metabolites: 2PG, 3PG, FAEE, OAA, PEP, PYR
As pointed out before, the first committed reaction in the fatty acid biosynthesis is an enzymatic reaction catalysed by ACCase. Whereas in eukaryotes, this enzyme is constituted by a single multifunctional polypeptide, bacterial ACCase contains four proteins, a biotin carboxyl carrier protein, biotin carboxylase and the α and β subunits of carboxyltransferase (Cronan and Waldrop 2002). Previous studies have already supported the important role that ACCase possesses directing the C flow towards fatty acid synthesis (Lykidis and Ivanova 2008; Song et al. 2008). In plants, fatty acyl-ACP synthesised in the plastids is transformed to free fatty acids by acyl-ACP thioesterases and then transported from the chloroplast to the cytoplasm, thus the possibilities of influencing the control of fatty acid biosynthesis are remote.
Acyl-ACP synthesis from malonyl-CoA involves five different reactions catalysed in most bacteria by the type II or dissociate fatty acid synthase (FAS II). In the FAS II system, each reaction is catalyzed by an individual enzyme, while its eukaryotic counterpart is composed of a single multifunctional enzymatic entity (FAS I). First of all, the malonyl subunit from malonyl-CoA is transferred to ACP by the malonyl-CoA:ACP transacylase (fabD, EC 2.3.1.39). The resulting malonyl-ACP is then condensed to acetyl-CoA with the help of 3-ketoacyl-ACP synthase (fabH, EC 2.3.1.41). In E. coli, fabH is involved in the initial condensation reaction while further malonyl-ACP additions to the growing fatty acyl-ACP are carried out by fabB and fabF. Then, 3-ketoacyl-ACP reductase (fabG, EC 1.1.1.100) catalyses a reduction step yielding 3-hydroxyacyl-ACP which is dehydrated by 3-hydroxyacyl-ACP dehydrase (fabZ, EC 4.2.1.-) to produce trans-2-enoyl-ACP. The final reaction of the pathway is a reduction of the trans-2-enoyl-ACP by enoyl-ACP reductase (fabI, EC 1.3.1.9) resulting in fatty acyl-ACP used afterward in successive condensation steps. As mentioned earlier, ACCase activity is inhibited by this acyl-ACP and thereby this end product plays a fundamental role in the fatty acid synthesis control. In addition, Heath and coworkers (Heath and Rock 1995, 1996a, b) found that acyl-ACPs are also able to inhibit the 3-ketoacyl-ACP synthase and enoyl-ACP reductase activities. These findings might offer a possible explanation for the fact that the overexpression of ACCase causes a ∼100-fold rise in malonyl-CoA levels, but only a sixfold rise of fatty acid synthesis (Davis et al. 2000).
The formed fatty acyl-ACPs are later directed to the synthesis of the membrane glycerolipids including monogalactosyldiacylglycerol, digalactosyldiacylglycerol, phosphatidyl glycerol (PG) and sulfoquinovosyldiacylglycerol (Weier et al. 2005). The first step in lipid biosynthesis is the formation of a 1-acyl-sn-glycerol-3-phosphate (lysophosphatic acid). In microorganisms, this compound is known to be produced by two different mechanisms. Either the fatty acyl-ACP is directly added to a 3PG (backbone for the glycerolipid synthesis) by a sn-glycerol-3-phosphate acyltransferase (GPAT or PlsB; EC 2.3.1.15) or by a newly discovered two-reaction system catalyzed by the enzymes, PlsX and PlsY (Zhang and Rock 2008). In this system, a molecule of phosphate is added to the fatty acyl group derived from a fatty acyl-ACP chain by PlsX and then transferred to G-3-P in a reaction catalyzed by PlsY (Lu et al. 2006). Further acylation by 1-acyl-sn-glycerol-3-phosphate acyltransferase (AGPAT or PlsC; EC 2.3.1.51) forms 1,2-diacyl-sn-glycerol-3-phosphate (phosphatidic acid). The overexpression of GPAT or AGPAT led to higher lipid levels in Arabidopsis thaliana seeds (Ranalli 2007). It may be of interest to develop a similar strategy in cyanobacteria-overexpressing pls XYC genes.
Nutrient-limited conditions in E. coli trigger a series of reactions known as the stringent response. This response is modulated by guanosine tetra- (ppGpp) and pentaphosphate (pppGpp) which are included in a group of compounds called alarmones. These compounds mediate a wide spectrum of metabolic reactions. Previous results indicated that GPAT is negatively regulated by ppGpp leading to a decrease in the lipid synthesis and consequently a decrease in the fatty acid production through the accumulation of fatty acyl-ACPs, responsible of the inhibition of several steps (previously mentioned) in fatty acid synthesis. It appears that an increase in the activity levels of GPAT, PlsX and a fatty acyl-ACP thioesterase (tesA, EC 3.1.2.14) overcomes this inhibition by detaching the fatty acid from the lipid biosynthesis (Davis et al. 2000; Heath et al. 1994; Paoletti et al. 2007).
TAGs are neutral storage lipids generally synthesised from diacylglycerol in an acyl-CoA-dependent reaction catalysed by diacylglycerol acyltransferase (DGAT) (Yen et al. 2008). Additionally, in plants and yeasts an acyl-CoA independent reaction catalysed by a phospholipid-diacylglycerol acyltransferase leads also to the TAG synthesis (Dahlqvist et al. 2000). In E. coli, the heterologous expression of bifunctional DGAT from Acinetobacter baylii resulted in fatty acid ethyl ester (FAEE) synthesis in this organism. This enzyme possesses both DGAT and wax ester synthase activities being able to use acyl-CoA molecules and esterify them to ethanol to create FAEEs in vivo (Kalscheuer et al. 2006). This in vivo production of fuel molecules circumvents the necessity of industrial transesterification of TAG. The accumulation of FAEEs by the transgenic organism accounted only for a 26% of the cellular dry weight, which needs further optimization to be profitable. These levels were lower than initially expected. Steen and co-workers have also used a similar approach to produce FAEE in E. coli. Fatty acid production was improved by overexpressing a native thioesterase and fadD, together with a heterologous acyl-CoA ligase and an ester synthase (atfA). The same authors used genetic engineering to over produce fatty alcohols in E. coli with the acr1 from Acinetobacter calcoaceticus (Steen et al. 2010). Recently, similar strategies have been followed by Lu in Synechocystis sp. PCC 6803 (Lu 2010), although no yields have been reported. Radakovits engineered eukaryotic algae with specific thioesterases for the production of C12–C14 fatty acids. In this study, a high amount (around 80%) of the fatty acid produced was incorporated into triacylglycerols (Radakovits et al. 2011). In cyanobacteria, thioesterases overexpression results on the enhancement of fatty acid secretion (Roessler et al. 2009).
As previously mentioned, fatty acyl-ACP thioesterase has the capacity to uncouple fatty acid from lipid synthesis. This enzyme hydrolyses the ACP moieties from fatty acyl-ACP molecules to produce free fatty acids. In bacteria, CoA units are added to free fatty acids through an acyl-CoA synthetase (fadD, EC 6.2.1.3) and then the resulting acyl-CoA enters into the β-oxidation pathway serving as a C and energy source (Fujita et al. 2007; Zhang and Rock 2008). As pointed out before, an increase in fatty acid production has been achieved already in E. coli by deleting fadD in combination with the overexpression of other key genes of the fatty acid pathway (Lu et al. 2008). In addition, long-chain acyl-CoA seems also to control the expression of genes encoding enzymes of fatty acid catabolism by interacting with the FadR transcriptional regulator. Previous results indicated that FadR is able to bind to specific DNA sequences inhibiting transcription of genes involved in fatty acid catabolism. If however, long-chain acyl-CoA is attached to FadR it is released from the DNA, allowing gene expression (Schujman and de Mendoza 2005; Zhang and Rock 2008). Another strategy to optimise the lipid content in the cell is to reduce lipid degradation through β-oxidation. These approaches have already been studied in plants and yeasts. However, β-oxidation is one of the major energy sources for the cells and mutation of genes involved in this pathway could lead to a decrease of the organism performance. In addition, many enzymes involved in the lipid metabolism possess common activities constraining the possibilities of eliminating single steps. In yeast, deletion of the fatty acid catabolism-encoding genes shows a fatty acids rise in the cells. The fatty acid secretion seems to be also stimulated in these mutants (Radakovits et al. 2010). Thus, similar strategies could be developed in cyanobacteria.
Any extra amount of energy or C produced during the cell growth is accumulated in storage products in the organisms. Although, as mentioned earlier, glycogen is the main carbohydrate reserve in cyanobacteria, these organisms also produced polyhydroxyalkanoates to store their excess of energy and C. There are three main enzymes participating in the cyanobacterial glycogen synthesis, an ADP-glucose pyrophosphorylase (agp (glgC), EC 2.7.7.27), a synthetase (glgA, EC 2.4.1.21) and a branching enzyme (glgB, EC 2.4.1.18). In Synechocystis sp. PCC 6803, it seems that mutation of the agp gene is linked to a higher accumulation of PHB compared to the wild type during photoautotrophic growth (Wu et al. 2002). Mixotrophic growth in presence of either glc or acetate resulted in an enhancement of the glycogen (in the wild type and increased growth in both wild type and mutant) and PHB (in mutant and wild type) contents, respectively. Since acetate provides the acetyl subunits required to form acetyl-CoA, the levels of PHB are higher when compared to photoautotrophic conditions. Nevertheless, acetate unlike glc did not seem to stimulate the cellular growth in a significant way. It has been suggested, that the produced acetyl-CoA is not incorporated into pathways that contributes to the basic functions of the cells. The participation of acetyl-CoA in the rate-limiting step of the lipid biosynthesis catalysed by ACCase might provide a partial explanation to these findings. Another study on agp-deficient mutants suggested that low light intensities (45 μmol photon m−2 s−1) increase moderately the cell growth, whereas higher intensities (82 μmol photon m−2 s−1) reduce the growth (Miao et al. 2003b). These results indicate that agp mutants are able to harness the chemical energy from photosynthesis more efficiently at low light conditions; at higher intensities impaired glycogen synthesis would cause photosynthesis inhibition (feedback inhibition). The same authors later found that during salt stress, these modified organisms produce sucrose rather than GG as osmoprotectant (Miao et al. 2003a). This example further illustrates the complexity of metabolic networks and their modulation.
In S. elongatus PCC 7942, deletions in the glgC and glgA genes strongly repressed glycogen accumulation (Suzuki et al. 2010). Under constant conditions of light, the growth, photosynthesis and respiration rates were particularly low in these mutants. Although during salt and oxidative stress they were able to produce sucrose (PCC 7942 does not accumulate GG), these modified organisms presented an additional growth reduction compared to wild type. These results indicate that glycogen is indirectly linked to the strains ability to successfully deal with stress conditions, probably due to the correlation between glycogen and ATP availability. Previous studies in eukaryotic algae have shown that the inactivation of genes involved in the biosynthesis of starch, another storage compound, causes an enhanced lipid production (Ramazanov and Ramazanov 2006). Unfortunately, the lipid and protein contents were not examined in the glycogen-deficient mutants of Synechococcus.
Amino acids synthesis via the TCA cycle
The TCA cycle is in most organisms an essential aerobic pathway for the final oxidation of carbohydrates and fatty acids. Its major role is to supply reducing power (NADH and FADH2) to produce ATP and intermediates required by other biosynthetic reactions. In contrast, cyanobacteria possess a non-complete cycle mainly orientated to the 2-oxoglutarate (2-OG) synthesis, which is involved in amino acid biosynthesis and N2 fixation. The 2-OG is derived from citrate in a reaction catalyzed by isocitrate dehydrogenase (icd, EC 1.1.1.42). Thus, in cyanobacteria, the reducing power is mainly generated during photosynthesis (Muro-Pastor and Florencio 1992).
The first enzymatic step of the cycle is mediated by a citrate synthase (gltA, EC 2.3.3.5) which condenses acetyl-CoA and OAA to produce citrate. The acetyl-CoA participates in the TCA cycle is derived from glycolysis and β-oxidation, whereas the OAA has originated from PEP in an irreversible step mediated by PEPC. OAA serves as a precursor of the biosynthesis of several amino acids including aspartate. It should be pointed out that PEPC lacks oxygenase activity and is therefore more effective in the C assimilation than Rubisco where carboxylase and oxygenase activities compete with each other (Gillion 1998). This fact could explain why C flux, in cyanobacteria, is primarily allocated to the protein biosynthesis. Usually, TCA intermediates cause the suppression of pyruvate kinase activity, the main modulating enzyme in carbohydrate catabolism (Lin et al. 1989).
As stated above, cyanobacteria possess an incomplete TCA cycle where two key enzymes are missing, the 2-oxoglutarate dehydrogenase (ogdh, EC 1.2.4.2), involved in the transformation of 2-OG into succinyl-CoA, and the succinyl-CoA synthetase (sucL, EC 6.2.1.4, 6.2.1.5), for the formation of succinate (Pearce et al. 1969; Stanier and Cohenbazire 1977). Previous modelling studies in Synechocystis PCC6803 evaluated the addition of these missing enzymes to the metabolomic network (Shastri and Morgan 2005). Their results suggested that the lack of a completed TCA cycle does not diminish the growth significantly in this microorganism.
It seems that in cyanobacteria, succinate can originate from 2-OG using a different route (Cooley et al. 2000). In these microorganisms, 2-OG is directly involved in the N2 fixation via the NH3-assimilating mechanisms, GDH (gdhA, EC 1.4.1.14) (Chávez and Candau 1991) and GS-GOGAT route (Meeks et al. 1978). This process is directly related to amino acid biosynthesis including glutamate and glutamine or indirectly leading (via glutamate) to alanine and aspartate derived from pyruvate and OAA, respectively. The GS-GOGAT system seems to be able to produce 2-OG as well. However, the production of 2-OG by this mechanism is rather insignificant if compared with IDH activity (Muro-Pastor and Florencio 1992).
It has been suggested that 2-OG plays a fundamental role in the amino acid biosynthesis. This theory has been supported in Synechocystis by experiments where the external addition of ammonium to N2-free medium enhanced the 2-OG levels as well as the glutamate and glutamine production (Merida et al. 1991). Furthermore, in cyanobacteria, 2-OG seems to be implicated in the heterocyst differentiation. Previous studies have indicated that the activity of key enzymes in the heterocyst development, including NtcA and PII, is subjected to 2-OG concentrations (Zhang et al. 2005). In the same way, this compound is also considered to modulate the N and C metabolism in non-nitrogen-assimilating cyanobacteria (Eisenhut et al. 2008; Muro-Pastor et al. 2005). However, proving the role of 2-OG as a signalling compound is still challenging since it is quickly diverted into different metabolic processes.
Conclusions
The renewed interest in alternative energies derived from biomass has been recently triggered by the prediction of a reduction in the crude oil production 10 years earlier than speculated by experts (Nashawi et al. 2010). In this context, cyanobacteria have received significant consideration stimulated by the fact that these microorganisms seem able to cope with some of the major difficulties encountered with preceding biofuel generations. Furthermore, cyanobacteria offer a promising biomass feedstock for various organic (ethanol, CH4 and biodiesel) and inorganic (H2 and electricity) biofuels. As the examples in “Cyanobacteria as a source of renewable energy” section illustrate, cyanobacteria are a potential source of a wide range of valuable biofuels using different substrates for their production. Many of the cited strategies are still under development and their energy yield may not be economically feasible yet at industrial production levels. Therefore, the metabolic network needs to be optimized to generate an efficient and economic biofuel production system extrapolatable to a commercial scale.
A detailed study of the biosynthetic routes in cyanobacteria would assist us to evaluate the impact of genetic manipulations and its limitations in the entire metabolic network. Thus, this approach would further facilitate the design by genetic engineering of an optimized metabolic network for biofuel production in cyanobacteria.
Previous work has shown that a minimum of the C derived from photosynthesis is directed to pathways involved in the biofuel production (Lindberg et al. 2010). Thus, in order to optimize the energy production by these microorganisms, new strategies of pathway engineering need to be proposed to redistribute the C flux among the biosynthetic pathways of other fuel feedstocks.
Future directions in genetic engineering have been suggested along with this paper not only for an efficient energy production from sugars and lipids but also to expand the spectrum of the products targeted as bioenergy feedstocks (isoprene, propanol and butanol). Both considerations are crucial factors to properly implement cyanobacteria in a future large-scale system of biofuel production.
As indicated previously, metabolic engineering of metabolic pathways could cause unplanned and unforeseen deleterious effects on cellular function. However, engineered organisms could still be a valuable tool for bioenergy production in case the manipulated genes are ligated to specific promoters that can be turned on after the organism reaches some pre-established desirable conditions. Previous studies revealed that temperature-modulated promoters are suitable for controlling ethanol production in Synechococcus (Wood et al. 2004).
In conclusion, the success of future generation of biofuels will rely on the advances in metabolic engineering to optimize the existing energy-related biosynthetic pathways and to reduce the stress on the genetically modified organisms. An efficient and cost-effective fuel production from biomass should decrease our current dependence on conventional energies which are both scarce and polluting.
Acknowledgments
This work was supported by the Dutch Ministry of Economic Affairs (EOSLT 07039).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Abed RMM, Dobretsov S, Sudesh K. Applications of cyanobacteria in biotechnology. J Appl Microbiol. 2009;106:1–12. doi: 10.1111/j.1365-2672.2008.03918.x. [DOI] [PubMed] [Google Scholar]
- Afgan NH, Carvalho MG. Multi-criteria assessment of new and renewable energy power plants. Energy. 2002;27:739–755. [Google Scholar]
- Ananyev G, Carrieri D, Dismukes GC. Optimization of metabolic capacity and flux through environmental cues to maximize hydrogen production by the cyanobacterium “Arthrospira (Spirulina) maxima”. Appl Environ Microbiol. 2008;74:6102–6113. doi: 10.1128/AEM.01078-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermayr SA, Hellingwerf KJ, Lindblad P, de Mattos MJT. Energy biotechnology with cyanobacteria. Curr Opin Biotechnol. 2009;20:257–263. doi: 10.1016/j.copbio.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Asthana RK, Nigam S, Maurya A, Kayastha AM, Singh SP. Trehalose-producing enzymes MTSase and MTHase in Anabaena 7120 under NaCl stress. Curr Microbiol. 2008;56:429–435. doi: 10.1007/s00284-008-9121-0. [DOI] [PubMed] [Google Scholar]
- Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- Atsumi S, Higashide W, Liao JC. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat Biotechnol. 2009;12:1177–80. doi: 10.1038/nbt.1586. [DOI] [PubMed] [Google Scholar]
- Avonce N, Mendoza-Vargas A, Morett E, Iturriaga G. Insights on the evolution of trehalose biosynthesis. BMC Evol Biol. 2006;6:109–124. doi: 10.1186/1471-2148-6-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, Price GD, Long BM, Woodger FJ. The environmental plasticity and ecological genomics of the cyanobacterial CO2 concentrating mechanism. J Exp Bot. 2006;57:249–265. doi: 10.1093/jxb/eri286. [DOI] [PubMed] [Google Scholar]
- Ball SG, Morell MK. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol. 2003;54:207–233. doi: 10.1146/annurev.arplant.54.031902.134927. [DOI] [PubMed] [Google Scholar]
- Ballicora MA, Iglesias AA, Preiss J. ADP-glucose pyrophosphorylase, a regulatory enzyme for bacterial glycogen synthesis. Microbiol Mol Biol Rev. 2003;67:213–225. doi: 10.1128/MMBR.67.2.213-225.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera BK, Balasundaram R, Gadgil K, Sharma DK. Photobiological production of hydrogen from Spirulina for fueling fuel cells. Enerc Sourc. 2007;29:761–767. [Google Scholar]
- Billi D, Wright DJ, Helm RF, Prickett T, Potts M, Crowe JH. Engineering desiccation tolerance in Escherichia coli. Appl Environ Microbiol. 2000;66:1680–1684. doi: 10.1128/aem.66.4.1680-1684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañizares-Villanueva RO, Dominguez AR, Cruz MS, Riosleal E. Chemical composition of cyanobacteria grown in diluted, aereated swine waste-water. Bioresour Technol. 1995;51:111–116. [Google Scholar]
- Carrieri D, Momot D, Brasg IA, Ananyev G, Lenz OB, Bryant DA, Dismukes GC. Boosting autofermentation rates and product yields with sodium stress cycling: application to renewable fuel production by cyanobacteria. Appl Environ Microbiol. 2010;76:6455–6462. doi: 10.1128/AEM.00975-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez S, Candau P. An NAD-specific glutamate dehydrogenase from cyanobacteria. Identification and properties. FEBS Lett. 1991;285:35–38. doi: 10.1016/0014-5793(91)80719-j. [DOI] [PubMed] [Google Scholar]
- Chellapandim P, Prabaharan D, Uma L. Evaluation of methanogenic activity of biogas plant slurry for monitoring codigestion of ossein factory wastes and cyanobacterial biomass. Appl Biochem Biotechnol. 2010;162(2):524–535. doi: 10.1007/s12010-009-8834-2. [DOI] [PubMed] [Google Scholar]
- Chen LM, Omiya T, Hata S, Izui K. Molecular characterization of a phosphoenolpyruvate carboxylase from a thermophilic cyanobacterium, Synechococcus vulcanus with unusual allosteric properties. Plant Cell Physiol. 2002;43:159–169. doi: 10.1093/pcp/pcf019. [DOI] [PubMed] [Google Scholar]
- Chisti Y. Biodiesel from microalgae. Biotechnol Adv. 2007;25:294–306. doi: 10.1016/j.biotechadv.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Chisti Y. Biodiesel from microalgae beats bioethanol. Trends Biotechnol. 2008;26:126–131. doi: 10.1016/j.tibtech.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Ciferri O. Spirulina, the edible microorganism. Microbiol Rev. 1983;47(4):551–578. doi: 10.1128/mr.47.4.551-578.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converti A, Oliveira RPS, Torres BR, Lodi A, Zilli M. Biogas production and valorization by means of a two-step biological process. Bioresour Technol. 2009;100:5771–5776. doi: 10.1016/j.biortech.2009.05.072. [DOI] [PubMed] [Google Scholar]
- Cooley JW, Howitt CA, Vermaas WFJ. Succinate: quinol oxidoreductases in the cyanobacterium Synechocystis sp strain PCC 6803: presence and function in metabolism and electron transport. J Bacteriol. 2000;182:714–722. doi: 10.1128/jb.182.3.714-722.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE, Waldrop GL. Multi-subunit acetyl-CoA carboxylases. Prog Lipid Res. 2002;41:407–435. doi: 10.1016/s0163-7827(02)00007-3. [DOI] [PubMed] [Google Scholar]
- Curatti L, Porchia AC, Herrera-Estrella L, Salerno GL. A prokaryotic sucrose synthase gene (susA) isolated from a filamentous nitrogen-fixing cyanobacterium encodes a protein similar to those of plants. Planta. 2000;211:729–735. doi: 10.1007/s004250000343. [DOI] [PubMed] [Google Scholar]
- Curatti L, Giarrocco LE, Cumino AC, Salerno GL. Sucrose synthase is involved in the conversion of sucrose to polysaccharides in filamentous nitrogen-fixing cyanobacteria. Planta. 2008;228:617–625. doi: 10.1007/s00425-008-0764-7. [DOI] [PubMed] [Google Scholar]
- Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S. Phospholipid : diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci USA. 2000;97:6487–6492. doi: 10.1073/pnas.120067297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D, Veziroglu TN. Hydrogen production by biological processes: a survey of literature. Int J Hydrogen Energ. 2001;26:13–28. [Google Scholar]
- Davis MS, Solbiati J, Cronan JE. Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J Biol Chem. 2000;275:28593–28598. doi: 10.1074/jbc.M004756200. [DOI] [PubMed] [Google Scholar]
- Dawar S, Behara BK, Mohanty P. Development of a low-cost oxy-hydrogen bio-fuel cell for generation of electricity using Nostoc as a source of hydrogen. Int J Energ Res. 1998;22:1019–1028. [Google Scholar]
- Deng MD, Coleman JR. Ethanol synthesis by genetic engineering in cyanobacteria. Appl Environ Microbiol. 1999;65:523–528. doi: 10.1128/aem.65.2.523-528.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats P, Folco E, Salerno GL. Sucrose may play an additional role to that of an osmolyte in Synechocystis sp PCC 6803 salt-shocked cells. Plant Physiol Biochem. 2005;43:133–138. doi: 10.1016/j.plaphy.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Dewinder B, Stal LJ, Mur LR. Crinalium epipsammum sp. nov.: a filamentous cyanobacterium with trichomes composed of elliptical cells and containing poly-β-(1,4) glucar (cellulose) J Gen Microbiol. 1990;136:1645–1653. [Google Scholar]
- Dexter J, Fu PC. Metabolic engineering of cyanobacteria for ethanol production. Energ Environ Sci. 2009;2:857–864. [Google Scholar]
- Dutta D, De D, Chaudhuri S, Bhattacharya SK. Hydrogen production by Cyanobacteria. Microb Cell Fact. 2005;4:1–11. doi: 10.1186/1475-2859-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, Huege J, Schwarz D, Bauwe H, Kopka J, Hagemann M. Metabolome phenotyping of inorganic carbon limitation in cells of the wild type and photorespiratory mutants of the Cyanobacterium Synechocystis sp strain PCC 6803. Plant Physiol. 2008;148:2109–2120. doi: 10.1104/pp.108.129403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco EC, Débora BN, Eduardo JL, Franco TT. Microalgae as feedstock for biodiesel production: carbon dioxide sequestration, lipid production and biofuel quality. J Chem Technol Biotechnol. 2010;85:395–403. [Google Scholar]
- Fujita Y, Matsuoka H, Hirooka K. Regulation of fatty acid metabolism in bacteria. Mol Microbiol. 2007;66:829–839. doi: 10.1111/j.1365-2958.2007.05947.x. [DOI] [PubMed] [Google Scholar]
- Gantzer CJ, Maier WJ. Biological degradation of cyanide by nitrogen-fixing cyanobacteria. Cincinnati: National Service Center for Environmental Publications; 1990. [Google Scholar]
- Garcia M, Myouga F, Takechi K, Sato H, Nabeshima K, Nagata N, Takio S, Shinozaki K, Takano H. An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Plant J. 2008;53:924–934. doi: 10.1111/j.1365-313X.2007.03379.x. [DOI] [PubMed] [Google Scholar]
- Gibson JL, Tabita FR. The molecular regulation of the reductive pentose phosphate pathway in Proteobacteria and Cyanobacteria. Arch Microbiol. 1996;166:141–150. doi: 10.1007/s002030050369. [DOI] [PubMed] [Google Scholar]
- Gillion JS. Carbon isotope discrimination in terresital plants. Guildford: Griffiths; 1998. [Google Scholar]
- Goldemberg J. Ethanol for a sustainable energy future. Science. 2007;315:808–810. doi: 10.1126/science.1137013. [DOI] [PubMed] [Google Scholar]
- Gomez-Casati DF, Cortassa S, Aon MA, Iglesias AA. Ultrasensitive behavior in the synthesis of storage polysaccharides in cyanobacteria. Planta. 2003;216:969–975. doi: 10.1007/s00425-002-0949-4. [DOI] [PubMed] [Google Scholar]
- Gressel J. Transgenics are imperative for biofuel crops. Plant Science. 2008;174:246–263. [Google Scholar]
- Griffiths MJ, Harrison STL. Lipid productivity as a key characteristic for choosing algal species for biodiesel production. J Appl Physiol. 2009;21:493–507. [Google Scholar]
- Grobbelaar JU. Physiological and technological considerations for optimising mass algal cultures. J Appl Physiol. 2000;12:201–206. [Google Scholar]
- Hagemann M, Schoor A, Jeanjean R, Zuther E, Joset F. The stpA gene from Synechocystis sp. strain PCC 6803 encodes the glucosylglycerol-phosphate phosphatase involved in cyanobacterial osmotic response to salt shock. J Bacteriol. 1997;179:1727–1733. doi: 10.1128/jb.179.5.1727-1733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DO, Moss PA. Biomass for energy in developing countries. Geojournal. 1983;7(1):5–14. [Google Scholar]
- Hall DO, Markov SA, Watanabe Y, Rao KK. The potential applications of cyanobacterial photosynthesis for clean technologies. Photosynth Res. 1995;46:159–167. doi: 10.1007/BF00020426. [DOI] [PubMed] [Google Scholar]
- Hanumantha Rao YV, RSV Sitarama Raju AV, Nageswara Reddy P (2009) Experimental investigations on jatropha biodiesel and additive in diesel engine. Indian Journal of Science and Technology 2(4):25–31
- Heath RJ, Rock CO. Regulation of malonyl-CoA metabolism by acyl-acyl carrier protein and beta-ketoacyl-acyl carrier protein synthases in Escherichia coli. J Biol Chem. 1995;270:15531–15538. doi: 10.1074/jbc.270.26.15531. [DOI] [PubMed] [Google Scholar]
- Heath RJ, Rock CO. Inhibition of beta-ketoacyl-acyl carrier protein synthase III (FabH) by acyl-acyl carrier protein in Escherichia coli. J Biol Chem. 1996;271:10996–11000. doi: 10.1074/jbc.271.18.10996. [DOI] [PubMed] [Google Scholar]
- Heath RJ, Rock CO. Regulation of fatty acid elongation and initiation by acyl acyl carrier protein in Escherichia coli. J Biol Chem. 1996;271:1833–1836. doi: 10.1074/jbc.271.4.1833. [DOI] [PubMed] [Google Scholar]
- Heath RJ, Jackowski S, Rock CO. Guanosine tetraphosphate inhibition of fatty acid and phospholipid synthesis in Escherichia coli is relieved by overexpression of glycerol-3-phosphate acyltransferase (plsB) J Biol Chem. 1994;269:26584–26590. [PubMed] [Google Scholar]
- Hellingwerf KJ, de Mattos MJT. Alternative routes to biofuels: light-driven biofuel formation from CO2 and water based on the ‘photanol’ approach. J Biotechnol. 2009;142:87–90. doi: 10.1016/j.jbiotec.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Heyer H, Krumbein WE. Excretion of fermentation products in dark and anaerobically incubated cyanobacteria. Arch Microbiol. 1991;155:284–287. [Google Scholar]
- Higo A, Katoh H, Ohmori K, Ikeuchi M, Ohmori M. The role of a gene cluster for trehalose metabolism in dehydration tolerance of the filamentous cyanobacterium Anabaena sp PCC 7120. Microbiology-Sgm. 2006;152:979–987. doi: 10.1099/mic.0.28583-0. [DOI] [PubMed] [Google Scholar]
- Hill J, Nelson E, Tilman D, Polasky S, Tiffany D. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc Natl Acad Sci USA. 2006;103:11206–11210. doi: 10.1073/pnas.0604600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiczyk E, Hansel A. Cyanobacterial cell walls: news from an unusual prokaryotic envelope. J Bacteriol. 2000;182:1191–1199. doi: 10.1128/jb.182.5.1191-1199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SJ, Lee CG. Evaluation of central metabolism based on a genomic database of Synechocystis PCC6803. Biotechnol Bioproc Eng. 2007;12:165–173. [Google Scholar]
- Howell K. Biofuels: Exxon sinks $600M into algae in major strategy shift. Washington, D.C.: E&E Publishing, LLC; 2009. [Google Scholar]
- Huang HH, Camsund D, Lindblad P, Heidorn T. Design and characterization of molecular tools for a synthetic biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 2010;38:2577–2593. doi: 10.1093/nar/gkq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izui K, Yoshinag T, Morikawa M, Katsuki H. Activation of phosphoenolpyruvate carboxylase of Escherichia coli by free fatty acids or their coenzyme A derivatives. Biochem Biophys Res Commun. 1970;40:949–956. doi: 10.1016/0006-291x(70)90995-2. [DOI] [PubMed] [Google Scholar]
- Kaczmarzyk D, Fulda M. Fatty Acid activation in cyanobacteria mediated by acyl-acyl carrier protein synthetase enables Fatty Acid recycling. Plant Physiol. 2010;152(3):1598–610. doi: 10.1104/pp.109.148007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalscheuer R, Stolting T, Steinbuchel A. Microdiesel: Escherichia coli engineered for fuel production. Microbiology-Sgm. 2006;152:2529–2536. doi: 10.1099/mic.0.29028-0. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- Kaplan A, Hagemann M, Bauwe H, Kahlon S, Ogawa T. Carbon acquisition by cyanobacteria: mechanisms, comparative genomics and evolution. In: Herrero A, Flores E, editors. The cyanobacteria: molecular biology, genomics and evolution. Sevilla: Caister Academic Press; 2008. [Google Scholar]
- Kaygusuz K. Bioenergy as a Clean and Sustainable Fuel. Energy sources, part A: recovery, utilization, and environmental effects. Energ Sourc. 2009;31:1069–1080. [Google Scholar]
- Kenyon CN. Fatty acid composition of unicellular strains of blue-green algae. J Bacteriol. 1972;109:827–834. doi: 10.1128/jb.109.2.827-834.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CN, Stanier RY, Rippka R. Fatty acid comopostion and physiological properties of some filamentous blue-green algae. Arch Mikrobiol. 1972;83:216–236. doi: 10.1007/BF00645123. [DOI] [PubMed] [Google Scholar]
- Knothe G. “Designer” biodiesel: optimizing fatty ester composition to improve fuel properties. Energ Fuels. 2008;22:1358–1364. [Google Scholar]
- Koshland DK (2009) Methods and compositions for production of methane gas. In: UC Berkeley, Field and Francis LLP B, editors. PatentsDoc. USA
- Lin M, Turpin DH, Plaxton WC. Pyruvate-kinase isozymes from the green-alga, Selenastrum minutum. 1. Purification and physical and immunological characterization. Arch Biochem Biophys. 1989;269:219–227. doi: 10.1016/0003-9861(89)90103-3. [DOI] [PubMed] [Google Scholar]
- Lindberg P, Park S, Melis A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab Eng. 2010;12:70–79. doi: 10.1016/j.ymben.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Liu XJ, Jiang Y, Chen F. Fatty acid profile of the edible filamentous cyanobacterium Nostoc flagelliforme at different temperatures and developmental stages in liquid suspension culture. Process Biochem. 2005;40:371–377. [Google Scholar]
- Liu X, Brune D, Vermaas D, Curtiss R. Production and secretion of fatty acids in genetically engineered cyanobacteria. Proc Natl Acad Sci USA. 2010;107:1–6. doi: 10.1073/pnas.1001946107. [DOI] [PubMed] [Google Scholar]
- Liu X, Brune D, Vermaas D, Curtiss R. Retraction: production and secretion of fatty acids in genetically engineered cyanobacteria. Proc Natl Acad Sci USA. 2010b;107:13189. doi: 10.1073/pnas.1001946107. [DOI] [PubMed] [Google Scholar]
- Liu T, Vora H, Khosla C. Quantitative analysis and engineering of fatty acid biosynthesis in E. coli. Metab Eng. 2010c;12:378–386. doi: 10.1016/j.ymben.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Lu X. A perspective: photosynthetic production of fatty acid-based biofuels in genetically engineered cyanobacteria. Biotechnol Adv. 2010;28:742–746. doi: 10.1016/j.biotechadv.2010.05.021. [DOI] [PubMed] [Google Scholar]
- Lu YJ, Zhang YM, Grimes KD, Qi JJ, Lee RE, Rock CO. Acyl-phosphates initiate membrane phospholipid synthesis in gram-positive pathogens. Mol Cell. 2006;23:765–772. doi: 10.1016/j.molcel.2006.06.030. [DOI] [PubMed] [Google Scholar]
- Lu XF, Vora H, Khosla C (2008). Overproduction of free fatty acids in E. coli: implications for biodiesel production. Metab Eng 10(6):333–339 [DOI] [PubMed]
- Luinenburg I, Coleman JR. Expression of Escherichia coli phosphoenolpyruvate carboxylase in a cyanobacterium. Functional complementation of Synechococcus PCC 7942. Plant Physiol. 1993;101:121–126. doi: 10.1104/pp.101.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Hu Z, Choi DG, Thomas VM, Realff MJ, Chance RR. Life cycle energy and greenhouse gas emissions for an ethanol production process based on blue-green algae. Environ Sci Technol. 2010;44:8670–8677. doi: 10.1021/es1007577. [DOI] [PubMed] [Google Scholar]
- Lykidis A, Ivanova N (2008) Genomic prospecting for microbial biodiesel production. Lawrence Berkeley National Laboratory. LBNL Paper LBNL-301E. Retrieved from: http://www.escholarship.org/uc/item/04h4d5dp
- Marin K, Zuther E, Kerstan T, Kunert A, Hagemann M. The ggpS gene from Synechocystis sp. strain PCC 6803 encoding glucosyl-glycerol-phosphate synthase is involved in osmolyte synthesis. J Bacteriol. 1998;180:4843–4849. doi: 10.1128/jb.180.18.4843-4849.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata TM, Martins AA, Caetano NS. Microalgae for biodiesel production and other applications: a review. Renew Sustain Energ Rev. 2010;14:217–232. [Google Scholar]
- Matsunaga T, Matsumoto M et al (2009) Characterization of marine microalga, Scenedesmus sp. strain JPCC GA0024 toward biofuel production. Biotechnol Lett 31(9):1367–1372 [DOI] [PubMed]
- McKinlay JB, Harwood CS. Curr Opin Biotechnol. 2010;21:244–251. doi: 10.1016/j.copbio.2010.02.012. [DOI] [PubMed] [Google Scholar]
- McNeely K, Xu Y, Bennette N, Bryant DA, Dismukes GC. Redirecting reductant flux into hydrogen production via metabolic engineering of fermentative carbon metabolism in a cyanobacterium. Appl Environ Microbiol. 2010;76:5032–5038. doi: 10.1128/AEM.00862-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks JC, Wolk CP, Lockau W, Schilling N, Shaffer PW, Chien WS. Pathways of assimilation of [13N]N2 and 13NH4+ by Cyanobacteria with and without heterocysts. J Bacteriol. 1978;134:125–130. doi: 10.1128/jb.134.1.125-130.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merida A, Candau P, Florencio FJ. In vitro reactivation of in vivo amium-inactivated glutamine-synthase from Synechocystis sp PCC 6803. Biochem Biophys Res Commun. 1991;181:780–786. doi: 10.1016/0006-291x(91)91258-e. [DOI] [PubMed] [Google Scholar]
- Miao XL, Wu QY. Biodiesel production from heterotrophic microalgal oil. Bioresour Technol. 2006;97:841–846. doi: 10.1016/j.biortech.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Miao XL, Wu QY, Wu GF, Zhao NM. Sucrose accumulation in salt-stressed cells of agp gene deletion-mutant in cyanobacterium Synechocystis sp PCC 6803. FEMS Microbiol Lett. 2003;218:71–77. doi: 10.1111/j.1574-6968.2003.tb11500.x. [DOI] [PubMed] [Google Scholar]
- Miao XL, Wu QY, Wui GF, Zhaol NM. Changes in photosynthesis and pigmentation in an agp deletion mutant of the cyanobacterium Synechocystis sp. Biotechnol Lett. 2003;25:391–396. doi: 10.1023/a:1022446330284. [DOI] [PubMed] [Google Scholar]
- Moriuchi T, Morishima K, Furukawa Y. Improved power capability with pyrolyzed carbon electrodes in micro direct photosynthetic/metabolic bio-fuel cell. Int J Precis Eng Manuf. 2008;9:23–27. [Google Scholar]
- Muro-Pastor MI, Florencio FJ. Putificatiom amd properties of NADP-Isocitrate dehydrogenase from the unicellular cyanobacterium Synechocystis sp PCC 6803. Eur J Biochem. 1992;203:99–105. doi: 10.1111/j.1432-1033.1992.tb19833.x. [DOI] [PubMed] [Google Scholar]
- Muro-Pastor MI, Reyes JC, Florencio FJ. Ammonium assimilation in cyanobacteria. Photosynth Res. 2005;83:135–150. doi: 10.1007/s11120-004-2082-7. [DOI] [PubMed] [Google Scholar]
- Nagarkar S, Williams GA, Subramanian G, Saha SK. Cyanobacteria-dominated biofilms: a high quality food resource for intertidal grazers. Hydrobiologia. 2004;512:89–95. [Google Scholar]
- Nashawi IS, Malallah A, Al-Bisharah M. Forecasting world crude oil production using multicyclic Hubbert model. Energ Fuels. 2010;24:1788–1800. [Google Scholar]
- Nelson N (2010) New energy source from the common pea: scientists create a solar energy device from a plant protein structure. ScienceDaily: American Friends of Tel Aviv University
- Nicolaisen K, Mariscal V, Bredemeier R, Pernil R, Moslavac S, Lopez-Igual R, Maldener I, Herrero A, Schleiff F, Flores E. The outer membrane of a heterocyst-forming cyanobacterium is a permeability barrier for uptake of metabolites that are exchanged between cells. Mol Microbiol. 2009;74:58–70. doi: 10.1111/j.1365-2958.2009.06850.x. [DOI] [PubMed] [Google Scholar]
- Nobles DR, Brown RM. Transgenic expression of Gluconacetobacter xylinus strain ATCC 53582 cellulose synthase genes in the cyanobacterium Synechococcus leopoliensis strain UTCC 100. Cellulose. 2008;15:691–701. [Google Scholar]
- Nobles DR, Romanovicz DK, Brown RM. Cellulose in Cyanobacteria. Origin of vascular plant cellulose synthase? Plant Physiol. 2001;127:529–542. [PMC free article] [PubMed] [Google Scholar]
- Ono E, Cuello JL. Carbon dioxide mitigation using thermophilic Cyanobacteria. Biosystems Eng. 2007;96:129–134. [Google Scholar]
- Osanai T, Kanesaki Y, Nakano T, Takahashi H, Asayama M, Shirai M, Kanehisa M, Suzuki I, Murata N, Tanaka K. Positive regulation of sugar catabolic pathways in the cyanobacterium Synechocystis sp PCC 6803 by the group 2 sigma factor sigE. J Biol Chem. 2005;280:30653–30659. doi: 10.1074/jbc.M505043200. [DOI] [PubMed] [Google Scholar]
- Osanai T, Azuma M, Tanaka K. Sugar catabolism regulated by light- and nitrogen-status in the cyanobacterium Synechocystis sp PCC 6803. Photochem Photobiol Sci. 2007;6:508–514. doi: 10.1039/b616219n. [DOI] [PubMed] [Google Scholar]
- Owttrim GW, Colman B. Purification and characterization of phosphoenolpyruvate carboxylase from a cyanobacterium. J Bacteriol. 1986;168:207–212. doi: 10.1128/jb.168.1.207-212.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page-Sharp M, Behm CA, Smith GD. Cyanophycin and glycogen synthesis in a cyanobacterial Scytonema species in response to salt stress. FEMS Microbiol Lett. 1998;160:11–15. [Google Scholar]
- Paoletti L, Lu YJ, Schujman GE, de Mendoza D, Rock CO. Coupling of fatty acid and phospholipid synthesis in Bacillus subtilis. J Bacteriol. 2007;189:5816–5824. doi: 10.1128/JB.00602-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce J, Leach CK, Carr NG. The incomplete tricarboxylic acid cycle in the blue-green alga Anabaena variabilis. J Gen Microbiol. 1969;55:371–378. doi: 10.1099/00221287-55-3-371. [DOI] [PubMed] [Google Scholar]
- Pereira S, Zille A, Micheletti E, Moradas-Ferreira P, De Philippis R, Tamagnini P. Complexity of cyanobacterial exopolysaccharides: composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol Rev. 2009;33:917–941. doi: 10.1111/j.1574-6976.2009.00183.x. [DOI] [PubMed] [Google Scholar]
- Pinzi S, Garcia IL, Lopez-Gimenez FJ, Luque de Castro MD, Dorado G, Dorado MP. The ideal vegetable oil-based biodiesel composition: a review of social, economical and technical implications. Energ Fuels. 2009;23:2325–2341. [Google Scholar]
- Radakovits R, Jinkerson RE, Darzins A, Posewitz MC. Genetic engineering of algae for enhanced biofuel production. Eukaryot Cell. 2010;9:486–501. doi: 10.1128/EC.00364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radakovits R, Eduafo PM, Posewitz MC. Genetic engineering of fatty acid chain length in Phaeodactylum tricornutum. Metab Eng. 2011;13:89–95. doi: 10.1016/j.ymben.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Ramazanov A, Ramazanov Z. Isolation and characterization of a starchless mutant of Chlorella pyrenoidosa STL-PI with a high growth rate, and high protein and polyunsaturated fatty acid content. Physiol Res. 2006;54:255–259. [Google Scholar]
- Ranalli P. Improvement of crop plants for industrial end uses. Dordrecht: Springer; 2007. pp. 83–127. [Google Scholar]
- REN21 (2007) Global status report. Ren21 1–52
- REN21 (2009) Globlal status report. Ren21 1–31
- Rittmann BE. Opportunities for renewable bioenergy using microorganisms. Biotechnol Bioeng. 2008;100(2):203–212. doi: 10.1002/bit.21875. [DOI] [PubMed] [Google Scholar]
- Gallagher E. The Gallagher review of the indirect effects of biofuel production. UK: St Leonards-on-Sea; 2008. [Google Scholar]
- Rodolfi L, Zittelli GC, Bassi N, Padovani G, Biondi N, Bonini G, Tredici MR. Microalgae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol Bioeng. 2009;102:100–112. doi: 10.1002/bit.22033. [DOI] [PubMed] [Google Scholar]
- Roessler, PYC, Liu B, Dodge C (2009) Secretion of fatty acids by photosynthetic organisms. WO Patent WO 2009/076,559
- Salerno GL, Curatti L. Origin of sucrose metabolism in higher plants: when, how and why? Trends Plant Sci. 2003;8:63–69. doi: 10.1016/S1360-1385(02)00029-8. [DOI] [PubMed] [Google Scholar]
- Salerno GL, Porchia AC, Vargas WA, Abdian PL. Fructose-containing oligosaccharides: novel compatible solutes in Anabaena cells exposed to salt stress. Plant Science. 2004;167:1003–1008. [Google Scholar]
- Samson R, Leduy A. Biogas production from anaerobic-digestion of Spirulina algal biomass. Biotechnol Bioeng. 1982;24:1919–1924. doi: 10.1002/bit.260240822. [DOI] [PubMed] [Google Scholar]
- Samson R, Leduy A. Detailed study of anaerobic digestion of Spirulina maxima algal biomass. Biotechnol Bioeng. 1986;28:1014–1023. doi: 10.1002/bit.260280712. [DOI] [PubMed] [Google Scholar]
- Savage DF, Afonso B, Chen AH, Silver PA. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science. 2010;327:1258–1261. doi: 10.1126/science.1186090. [DOI] [PubMed] [Google Scholar]
- Schirmer A, Rude MA, Xuezhi L, Popova E, del Cardayre SB. Microbial biosynthesis of alkanes. Science. 2010;329:559–562. doi: 10.1126/science.1187936. [DOI] [PubMed] [Google Scholar]
- Schujman GE, de Mendoza D. Transcriptional control of membrane lipid synthesis in bacteria. Curr Opin Microbiol. 2005;8:149–153. doi: 10.1016/j.mib.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Schütz K, Happe T, Troshina O, Lindblad P. Cyanobacterial H2 production—a comparative analysis. Planta. 2004;218:350–359. doi: 10.1007/s00425-003-1113-5. [DOI] [PubMed] [Google Scholar]
- Scragg AH, Illman AM, et al. Growth of microalgae with increased calorific values in a tubular bioreactor. Biomass and Bioenergy. 2002;23(1):67–73. [Google Scholar]
- SERI (1984) Fuel option from microalgae with representative chemical composition
- Shastri AA, Morgan JA. Flux balance analysis of photoautotrophic metabolism. Biotechnol Prog. 2005;21:1617–1626. doi: 10.1021/bp050246d. [DOI] [PubMed] [Google Scholar]
- Sheehan J, Dunahay T, Benemann J, Roessler P (1998) A look back at the U.S. Department of Energy’s Aquatic Species Program-Biodiesel from Algae Report
- Sialve B, Bernet N, Bernard O. Anaerobic digestion of microalgae as a necessary step to make microalgal biodiesel sustainable. Biotechnol Adv. 2009;27:409–416. doi: 10.1016/j.biotechadv.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Singh AK, Sherman LA. Pleiotropic effect of a histidine kinase on carbohydrate metabolism in Synechocystis sp strain PCC 6803 and its requirement for heterotrophic growth. J Bacteriol. 2005;187:2368–2376. doi: 10.1128/JB.187.7.2368-2376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder DS, Brahamsha B, Azadi P, Palenik B. Structure of compositionally simple lipopolysaccharide from marine Synechococcus. J Bacteriol. 2009;191:5499–54509. doi: 10.1128/JB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D, Hou L, Shi D. Exploitation and utilization of rich lipids-microalgae, as new lipids feedstock for biodiesel production—a review. Sheng Wu Gong Cheng Xue Bao. 2008;24:341–348. doi: 10.1016/s1872-2075(08)60016-3. [DOI] [PubMed] [Google Scholar]
- Stal LJ, Moezelaar R. Fermentation in cyanobacteria. FEMS Microbiol Rev. 1997;21:179–211. [Google Scholar]
- Stanier RY, Cohenbazire G. Phototrophic prokaryotes—Cyanobacteria. Annu Rev Microbiol. 1977;31:225–274. doi: 10.1146/annurev.mi.31.100177.001301. [DOI] [PubMed] [Google Scholar]
- Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, del Cardayre SB, Keasling JD. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463:559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- Stockel J, Welsh EA, Liberton M, Kunnvakkam R, Aurora R, Pakrasi HB. Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes. Proc Natl Acad Sci USA. 2008;105:6156–6161. doi: 10.1073/pnas.0711068105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki E, Ohkawa H, Moriya K, Matsubara T, Nagaike Y, Iwasaki I, Tsuziki M, Fujiwara S, Nakamura Y. Carbohydrate metabolism in the mutants of the cyanobacterium Synechococcus elongatus PCC 7942 defective in glycogen synthesis. Appl Environ Microbiol. 2010;76:3153–3159. doi: 10.1128/AEM.00397-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabei Y, Okada K, Makita N, Tsuzuki M. Light-induced gene expression of fructose 1,6-bisphosphate aldolase during heterotrophic growth in a cyanobacterium, Synechocystis sp PCC 6803. FEBS J. 2009;276:187–198. doi: 10.1111/j.1742-4658.2008.06772.x. [DOI] [PubMed] [Google Scholar]
- Timmerman L (2009) Targeted growth plots future as agricultural biotech, Cleantech Pionee. Xconomy. http://www.xconomy.com/seattle/2009/07/24/targeted-growth-plots-future-as-agricultural-biotech-cleantech-pioneer/
- Tsujimura S, Wadano A, Kano K, Ikeda T. Photosynthetic bioelectrochemical cell utilizing cyanobacteria and water-generating oxidase. Enzyme Microb Technol. 2001;29:225–231. [Google Scholar]
- Vanderoost J, Bulthuis BA, Feitz S, Krab K, Kraayenhof R. Fermentation metabolism of the unicellular cyanobacterium Cyanothece PCC 7822. Arch Microbiol. 1989;152:415–419. [Google Scholar]
- Varel VH, Chen TH, Hashimoto AG. Thermophilic and mesophilic methane production from anaerobic degradation of the cyanobacterium Spirulina maxima. Resour Conservat Recycl. 1988;1:19–26. [Google Scholar]
- Vargas MA, Rodriguez H, Moreno J, Olivares H, Del Campo JA, Rivas J, Guerrero MG. Biochemical composition and fatty acid content of filamentous nitrogen-fixing cyanobacteria. J Phycol. 1998;34:812–817. [Google Scholar]
- Vargas W, Cumino A, Salerno GL. Cyanobacterial alkaline/neutral invertases. Origin of sucrose hydrolysis in the plant cytosol? Planta. 2003;216:951–960. doi: 10.1007/s00425-002-0943-x. [DOI] [PubMed] [Google Scholar]
- Vermaas WFJ (2001) Photosynthesis and respiration in Cyanobacteria. Encyclopedia of life sciences. Wiley, New York 1–7
- Walsh K, Jones GJ, Hugh DR. Effect of irradiance on fatty acid, carotenoid, total protein composition and growth of Microcystis aeruginosa. Phytochemistry. 1997;44:817–824. [Google Scholar]
- Weier D, Muller C, Gaspers C, Frentzen M. Characterisation of acyltransferases from Synechocystis sp PCC6803. Biochem Biophys Res Commun. 2005;334:1127–1134. doi: 10.1016/j.bbrc.2005.06.197. [DOI] [PubMed] [Google Scholar]
- Weyman PD (2010) Expression of oxygen-tolerant hydrogenases in Synechococcus elongatus. 10th Cyanobacterial Molecular Biology Workshop. June 11–15, Lake Arrowhead, USA
- Wood RP, Coleman JR, de Deng M. Genetically modified cyanobacteria for the production of ethanol, the constructs and method thereof. USA: Enol Energy Inc; 2004. [Google Scholar]
- Wu GF, Shen ZY, Wu QY. Modification of carbon partitioning to enhance PHB production in Synechocystis sp PCC6803. Enzyme Microb Technol. 2002;30:710–715. [Google Scholar]
- Wu B, Zhang B, Feng X, Rubens JR, Huang R, Hicks LM, Pakrasi HB, Tang YJ. Alternative isoleucine synthesis pathway in cyanobacterial species. Microbiology. 2010;156:596–602. doi: 10.1099/mic.0.031799-0. [DOI] [PubMed] [Google Scholar]
- Yagishita T, Sawayama S, Tsukahara K, Ogi T. Photosynthetic big-fuel cells using cyanobacteria. New York: Elsevier; 1996. pp. 958–961. [Google Scholar]
- Yang C, Hua Q, Shimizu K. Metabolic flux analysis in Synechocystis using isotope distribution from 13C-labeled glucose. Metab Eng. 2002;4:202–216. doi: 10.1006/mben.2002.0226. [DOI] [PubMed] [Google Scholar]
- Yen CLE, Stone SJ, Koliwad S, Harris C, Farese RV. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008;49:2283–22301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Keppel C, Spalding M, Jane JL. Effects of growth condition on the structure of glycogen produced in cyanobacterium Synechocystis sp PCC6803. Int J Biol Macromol. 2007;40:498–504. doi: 10.1016/j.ijbiomac.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Zhang YM, Rock CO. Thematic review series: glycerolipids. Acyltransferases in bacterial glycerophospholipid synthesis. J Lipid Res. 2008;49:1867–1874. doi: 10.1194/jlr.R800005-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CC, Laurent S, Bédu S. Molecular signal of nitrogen starvation, insight from the cyanobacterium Anabaena PCC 7120. Netherlands: Springer; 2005. pp. 77–78. [Google Scholar]
- Zhang K, Sawaya MR, Eisenberg DS, Liao JC. Expanding metabolism for biosynthesis of nonnatural alcohols. Proc Natl Acad Sci USA. 2008;105:20653–20658. doi: 10.1073/pnas.0807157106. [DOI] [PMC free article] [PubMed] [Google Scholar]