Abstract
Recent advances in the biology of stem cells has resulted in significant interest in the development of normal epithelial cell lines from the intestinal mucosa, both to exploit the therapeutic potential of stem cells in tissue regeneration and to develop treatment models of degenerative disorders of the digestive tract. However, the difficulty of propagating cell lines of normal intestinal epithelium has impeded research into the molecular mechanisms underlying differentiation of stem/progenitor cells into the various intestinal lineages. Several short-term organ/organoid and epithelial cell culture models have been described. There is a dearth of long-term epithelial and/or stem cell cultures of intestine. With an expanding role of stem cells in the treatment of degenerative disorders, there is a critical need for additional efforts to develop in vitro models of stem/progenitor epithelial cells of intestine. The objective of this review is to recapitulate the current status of technologies and knowledge for in vitro propagation of intestinal epithelial cells, markers of the intestinal stem cells, and gene and protein expression profiles of the intestinal cellular differentiation.
Introduction
The mucosa of the intestine absorbs essential nutrients from the lumen for the body and produces mucous and cytokines with protective and signaling characteristics. The two intestinal segments—small and large (colon) intestine—exhibit differences, with regard to disease processes as well as susceptibility to environmental factors including carcinogens. Small intestinal cells appear highly resistant to injury and if damaged undergo rapid apoptosis and are sloughed off into the lumen. However, replicating cells in the colon may undergo mutations after damage by toxic agents resulting in carcinogenesis. The high incidence of colon cancer has engendered considerable effort to develop in vitro models of colon epithelial cells [1–7] that have contributed to the current knowledge of mechanisms involved in colon cancer as well as normal colonic growth and differentiation. In contrast, there have been far fewer studies of small intestine. There is a lack of suitable in vitro models for long-term cultures, although several studies have described primary and short-term cultures [8–10].
As a result of recent advances in identification and culture of putative stem cells, interest in in vitro models of intestinal epithelial cells has been rejuvenated. In particular, there is considerable interest in the regenerative potential of intestinal stem/progenitor cells that populate the tissue. These efforts can contribute to the treatment of degenerative diseases such as colitis, inflammatory bowel disease, intestinal infections, and radiation injury [11–13]. Furthermore, identification of factors and pathways involved in controlled differentiation of stem/progenitor cells into tissue-specific lineages is expected to contribute immensely to effective clinical protocols.
Intestinal epithelium cellular replication and differentiation show spatial boundaries characterized by distinct phenotypes [12,14]. The small intestinal epithelium consists of villi extending into the lumen. The epithelium is composed mainly of terminally differentiated mucous and absorptive cells, with crypts located at the base of villi where replicating cells including putative stem cells are residents [15,16]. Stem cells constitute the replication units of villi. The daughter cells migrate mainly upward into villi and differentiate into mucous goblet, absorptive, and endocrine cells. The paneth cells—that also reportedly originate from the stem cells migrate below the replicative units—secrete antimicrobial peptides, that is lysozymes and certain growth factors [17]. The colonic mucosa is essentially similar to small intestine except that it does not have villi and paneth cells.
Studies to identify and characterize intestinal stem cells are hampered by the lack of definitive markers for these populations, although investigators have recently identified an array of candidate genes. For example, genes such as EphB2 and EphB3 [18], CD44 [19], Fgfr3 [20], and Sox9 [21] are expressed in crypt cells, but not in villi. In contrast, expression of Hes1 increases progressively from villi downward to the stem cell zone [22]. Analysis of the Wnt and Notch signaling pathways associated with development and cell replication have identified Musashi1 as a possible marker as this is expressed mainly in crypt base cells [22–24]. Recent work by Barker et al. [25] has identified a leucine-rich orphan G protein-coupled receptor, Lgr5/GPR49, a putative stem cell marker, which is expressed mainly in mouse small intestine stem cells. However, additional characterization is pertinent because these genes are broadly expressed in other actively replicating cells. Therefore, there is a critical need to establish in vitro models of stem/progenitor cells of intestinal epithelial mucosa.
Several earlier studies demonstrated long-term propagation of primary epithelial cell cultures from rodent and human colon [1,3,4,6,7]. Some showed long-term replication potential and functional differentiation, suggesting that the cultures may have contained stem cell populations, although such characterization was not performed. More recently, intestinal epithelial cells expressing certain stem cell markers have been described, but the cells failed to develop into cell lines and died after 3–4 passages [26–28]. The difficulty in producing stable cell lines likely relates to imperfect knowledge of tissue dissociation methods, culture conditions, and inclusion of appropriate growth factor supplements. Epithelial cell lines derived from intestinal mucosa should be invaluable for elucidating appropriate regulation of growth and differentiation. In addition, such culture models are highly desirable for biomedical and clinical applications such as toxicity testing, drug development, and transplantation. This review recapitulates current status of in vitro propagation and maintenance of cells derived from intestinal epithelium including methods for isolation, culture, and characterization.
In Vitro Maintenance of Intestinal Mucosa
Organ culture
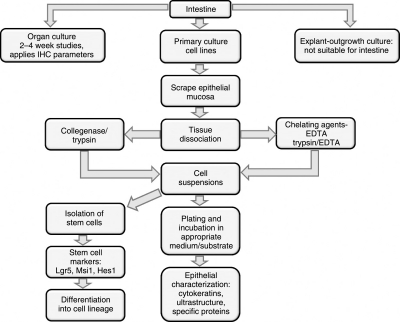
During the past 40 years, in vitro methods applied to investigate structure and function of intestinal mucosa include explant/organ culture and isolation and culture of epithelial cells using chelating agents and proteolytic enzymes (Fig. 1). Occasionally, these methods are combined with flow cytometry or differential centrifugation to isolate different epithelial cell types. Organ culture was the original approach for studying physiological activities of intestinal lineage outside of an organism. Small (1–4 mm2) intestinal explants from fetal tissues of various species can be maintained with normal tissue organization for up to 3 weeks, allowing investigation of growth, differentiation, and ion transport across the epithelium [29–33]. In contrast, adult intestinal mucosa in organ culture shows degeneration after about 48 h [34]. Subsequently, it was observed that adult epithelial mucosa regenerated and remained viable for several weeks [35,36]. Minced fragments of neonatal and adult intestine embedded in collagen gel have been cultured in three-dimensional organization at air–liquid interphase [37]. The viability and replication of the organoids were enhanced by treatment with Wnt agonist adenovirus RSpo1-Fc fusion protein and the cultures could be maintained for up to 28 days after which the viability was reduced. Without the treatment with adenovirus RSpo1-Fc fusion protein, the viability was restricted to 7–10 days. Organ culture techniques, however, have limited appeal in biological applications because these do not allow studies on specific epithelial lineage differentiation and carcinogenesis independent of other cells. Using an explant outgrowth procedure, epithelial cell populations from several human tissues (eg, prostate, bronchus, and salivary gland) have been generated [38–40]. Small tissue explants are held in a dish by a plasma clot and cultured in a suitable medium. After 6–7 days, outgrowths consisting of replicating epithelial cells are formed around the explants that enlarge to 10 mm in diameter. The explants are then removed and the outgrowths passaged to generate epithelial cell lines. However, the explant outgrowth procedure has not worked well for intestinal mucosa. Possible reasons for the difficulty include a more stringent requirement of the cells for extracellular matrix components and certain growth factors not yet defined.
FIG. 1.
In vitro methods to investigate intestinal epithelial cells.
Tissue dissociation
The most frequently used approaches to propagate primary cultures and cell lines apply tissue dissociation techniques using chelating agents and/or proteolytic enzymes. Ethylenediaminetetraacetic acid (EDTA) is a widely used chelating agent for the dissociation and culture of intestinal epithelial cells (Fig. 1). The concentration of EDTA, incubation temperature, and time required for optimal dissociation vary for different tissues [41]. However, when this technique was applied to intestinal epithelia the dissociated cells failed to attach to plastic culture substrate and rapidly degenerated [6,42]. This failure is ostensibly due to the treatment of tissues with EDTA in divalent ion-free medium, which disrupts the functions of extracellular matrix molecules, that is, integrins and cadherins [43,44], thereby preventing attachment of cells and inducing apoptosis. Rapidly plating the chelated cells on suitable substrates including collagen matrix or fibroblast feeder layers can prevent apoptosis and allow cells to replicate [45,46]. Isolated mouse crypts containing Lgr5-positive stem cells when propagated in lami-nin-rich matrigel replicated actively to produce additional crypts exhibiting cell lineage differentiation analogous to a crypt/villus structure [37]. Such organoids could be mechanically dissociated and subcultured to yield new crypt/villus structures. Furthermore, these organoids could also be generated from a single Lgr5-positive stem cell indicating that the crypt/villus structure is self-organizing and can be produced from a single stem cell and in the absence of any nonepithelial components [37].
To overcome these limitations of EDTA, proteolytic enzymes such as trypsin and collagenase have been applied for dissociation of intestinal epithelial cells. Enzymatic digestion of tissues allows retention of significant amounts of the extracellular matrix components improving attachment and survival of epithelial cells. One of us, DC, has used trypsin and collagenase for isolation of epithelial cells from the colon of suckling rat [2] and human fetus [4]. The isolated cells attached readily to plastic substrate and yielded epithelial cell lines, which retained many phenotypes of the colon cells. Trypsin was also employed to isolate various types of colonocytes (ie, proliferative, mucous, and absorptive) according to their location along the villus–crypt axis, by repeated time dissociation of colonic mucosa of adult rat [47]. Quaroni et al. also used collagenase to isolate and culture epithelial cells from small intestine of rat [8] and pig [9]. Gibson et al. [48] employed a combination of collagenase and dispase to dissociate colonic mucosa, although the isolated crypts could not be propagated to yield cell lines. One of the major difficulties encountered in isolating replicative cells from intestine is the presence of mucus and DNA released from damaged nuclei that cause clumping of cells and prevent their attachment. DNAse 1 is often used to minimize cell clumping during dissociation [49]. Recently, collage-nase and dispase were used to isolate a stem cell population from mouse jejunum [28]. Despise the diversity of protocols involving enzymatic isolation of epithelial cells for in vitro propagation, use of trypsin and collagenase predominates. Trypsin appears to be more efficient in dissociating the epithelial mucosa, but may cause irreversible damage. Collagenase is less disruptive, providing clumps of cells and maintaining some intact intracellular matrix.
Primary and long-term intestinal epithelial cell culture
A variety of culture media, growth supplements, and substrates have been employed to promote replication of primary and long-term cultures of intestinal epithelial cells. Although manipulation of culture conditions may have some physiological basis, those relating to growth supplements and substrates are selected empirically to achieve optimal growth and tissue-specific phenotypes. The most widely used basal media for intestinal epithelium include MEM [1,2,3,9], DMEM [7,8], and Ham's F-12 [50]. Siddiqui and Chopra [4] used 1:1 combination of DMEM and Ham's F-12 for culturing human fetal colon epithelial cells. Selection of one basal medium over another may be made to achieve stable osmolarity and reduce culture shock. To reduce significant alterations in pH during observations, medium is frequently supplemented with HEPES buffer [2,4]. An issue that is acute to intestinal cell culture, in particular to colon cells, is the avoidance of resident microbial contamination. The most commonly used concentrations of antibiotics and antifungal agents are penicillin (100 IU/mL), streptomycin (100 μg/mL), fungizone (2.5 μg/mL), and/or gentamicin (10 μg/mL). Although the specific effects of antimicrobial agents on growth and physiology of intestinal cells have not been reported, inclusion of these agents in culture medium has successfully prevented contamination and yielded several primary and long-term cultures [1,2,4,8].
Earlier studies routinely supplemented basal medium with fetal bovine serum (FBS) to promote cell replication. Inclusion of FBS was believed to protect primary cells from osmotic shock, stabilize pH, and provide certain growth factors, for example, EGF, insulin-like growth factor, and transforming growth factor. Subsequently, however, it was reported that FBS at high concentrations (10%–20%) inhibited epithelial cell replication due to TGF-β in the serum [51]. Chopra and Yeh [3] examined the effects of different concentrations of FBS on suckling rat colon epithelial cells and demonstrated that FBS at lower concentrations (2.5%) promoted replication, while higher concentrations (10%–15%) inhibited cell replication. Similar results were reported by Fukamachi [50] for rat fetal intestinal primary epithelial cultures. Consequently, when included in medium, FBS is usually heat-inactivated (56°C for 30 min) to destroy the growth inhibitory factors [52,53]. Inclusion of FBS in basal medium has other limitations associated with the presence of unknown components and variations among different batches of serum. Consequently, in recent years there has been a considerable effort to design chemically defined serum-free medium for epithelial cell culture.
Hormones and growth factors are frequently added to culture media to stimulate cell replication. Such factors are necessary in serum-free media although a variety of growth factors are also included in serum-supplemented media. Most frequently, supplemental factors for intestinal epithelial cells include insulin [4,5,9,53], hydrocortisone [4,7,54,55], EGF [4,5,53,56], transferrin [4,7,53], and cholera toxin [4,50]. Supplementing low serum media with insulin stimulated replication of suckling rat colon [3] and human fetal colon epithelial cells [5]. The concentrations of the growth factors used by different investigators vary considerably. Therefore, the optimal concentrations used for each growth factor must be determined for each combination of factors for each system. Media conditioned by mesothelial or epithelial cells have been used to promote attachment and replication of intestinal cells. Such conditioned media reportedly enhanced attachment and replication of early passage suckling rat colon epithelial cells [2]. Odedra et al. [57] used medium conditioned with mesothelial cells to promote growth of rat jejunum cells.
Fibroblasts frequently overgrow the desired epithelial cells, especially in serum-containing media. Different strategies have been used to attempt to eliminate the fibroblasts from epithelial cultures. Because fibroblasts appear to be more sensitive than epithelial cells to dissociation by trypsin/EDTA [58], differential trypsinization was applied to primary cultures of suckling rat colon epithelial cells [2]. This method, however, was ineffective because both the epithelial and fibroblast cells showed similar sensitivities to trypsin/EDTA. An alternative is substituted culture medium with d-valine instead of l-valine, as fibroblasts lack d-amino acid oxidase, preventing the utilization of d-valine by fibroblasts and inhibiting their growth [59]. This metabolic condition was unsuccessful because substituting d-valine for l-valine also inhibited the growth of epithelial cells [2]. The epithelial cells were selectively subcultured by the penicylinder method, which yielded almost pure populations of epithelial cells [2]. Fibroblast contamination of human colon epithelial cell cultures was reduced by differential rapid attachment of fibroblasts and maintaining the epithelial cells in serum-free medium [60].
Characterization of intestinal epithelial cell cultures
The major objective of intestinal epithelial cell culture has been the propagation of primary and long-term cultures that maintain active cellular replication and tissue-specific characteristics. Most published studies have not addressed the stem cell characteristics of primary or long-term cultures. The epithelial nature and tissue-specific origin of cultures from different species are established using structural, immunocytochemical (IHC), and functional properties (Fig. 1). Under light microscopy, the intestinal epithelial cells have polygonal cobble stone morphology and are intimately connected with each other [2,4,6–9,61–63]. At the ultrastructural level, the cells exhibit surface microvilli and are interconnected by desmosomes and apical tight junctions. The cytoplasm contains typical tonofilament bundles, rough endoplasmic reticulum, and Golgi complexes [2,4,6–9,61–63]. Immunocytochemical and biochemical analyses have demonstrated that the cells contain cytokeratins, including CK18, CK19, and E-cadherin [2,4,6–9,61–63]. Several studies have also reported functional properties of intestinal cells, including the expression of mucopolysaccharides and enzyme activities, for example, alkaline phosphatase, aminopeptidase, maltase, and α-glycosidase [2,8,50,61]. Gene expression analysis, the utility of which is covered in greater detail below, has indicated the expression of specific genes associated with intestinal structural and functional elements [61]. Most epithelial cultures lacked the expression of vimentin, a characteristic of mesothelial cells. However Rusu et al. [61] observed the expression of vimentin in bovine jejunum and colon epithelial cultures, the expression of which increased progressively with passaging of the cultures. It is also noteworthy that the functional properties of epithelial cells are commonly lost in high passage cultures, even though the cells continue to replicate [2,50,61]. This suggests that the functional properties observed in primary and low passage cultures may be due to the presence of differentiated cells in heterogeneous cell populations that are lost during passaging. A realistic concern is that a cell population may be derived from target tissue that is highly replicative, but in the artifactual in vitro situation are functionally estranged from the tissue of origin.
Gene expression profiling of intestinal cell differentiation using microarrays
Microarray analysis has become a ubiquitous component of biomedical research, allowing concurrent measurement of gene expression events on a genome-wide basis. This technology has been used with in vitro systems to identify transcriptional profiles associated with proliferation, differentiation, and migration of intestinal epithelial cells. Microarray-based investigations of intestinal cell differentiation in vitro have largely used the Caco-2 human colon carcinoma cell line. These model of colonic epithelial cell differentiation are associated with migration along the crypt–villus axis [64]. At confluence Caco-2 cells undergo contact-dependent differentiation to small intestine-like enterocytes, providing a system to study differentiation toward absorptive cells [65].
Mariadason et al. [66] used cDNA microarrays to characterize gene expression events in a post-confluence time course analysis of Caco-2 cells. Over a 21-day period [66], a total of 2,286 genes were identified as differentially expressed out of 17,280 assayed. Of these, 697 were up-regulated and 1,589 were down-regulated. The analysis revealed expression patterns correlated with cell differentiation marked by down-regulation of genes involved with cell cycle progression, DNA synthesis, and protein translation/folding. A decrease in activity of these biological processes is consistent with the expected reduction in proliferation associated with differentiating cells. Among the down-regulated genes involved in cell cycle control were cyclins A, B, B1, D1, D2, E, and F, along with a number of genes coding for cdc cell division cycle proteins and the cyclin-dependent kinases cdk-1 and cdk-2. Pathways enriched with up-regulated genes included extracellular matrix formation, lipid transport/metabolism, and xenobiotic metabolism. Phase II drug-metabolizing enzymes were among the most highly induced genes, including glutathione S-transferases A1-1, A3, and A4, along with sulfo-transferases and UDP glycosyltransferases.
The authors also observed that down-regulation of components of the β-catenin-TCF pathway was correlated with cell differentiation. The microarray analysis revealed that c-myc and cyclin D1, both important downstream targets of this pathway, were decreased in expression during the progression toward differentiation. This observation is consistent with the proposed involvement of these genes in controlling proliferation and differentiation of intestinal cells, including work demonstrating that TCF4 signaling is essential to the regulation of crypt stem cells in the small intestine [67–69].
A similar study was published shortly after the Mariadason report that used the brush border expressing subclone of Caco-2 cells [70]. Affymetrix oligonucleotide arrays were used to characterize gene expression changes in proliferating, post-proliferative, undifferentiated, and differentiated cells. A total of 1,150 genes were identified as differentially expressed in four independent samples across a 15-day period out of a total of 12,363 genes available on the microarray. The authors noted that 322 of the differentially expressed genes had previously been identified by Mariadason et al. [66]. However, gene expression-fold changes revealed a poor correlation (r2 = 0.01) between the two studies. The work by Fleet et al. [70] utilized an oligo-nucleotide array, providing improved specificity over the earlier generation of cDNA arrays, along with analysis of variance (ANOVA) on data from biological replicates.
However, while noting the discrepancy in the expression ratios observed, the two experiments were actually similar in terms of the signaling pathways and biological processes indicated to be associated with intestinal cell differentiation. Both groups reported decreased expression of genes involved in proliferation, including cyclins B1, D1, and D2. Other down-regulated processes noted by both groups were DNA synthesis/repair and protein translation/folding. The two reports also noted similar pathways enriched in up-regulated genes, including lipid metabolism and xenobiotic metabolism. Importantly, both studies reported repression of genes in the Wnt/β-catenin/TCF pathway associated with differentiation. Among the downstream targets of this pathway, c-myc and cyclin D1 were identified as down-regulated in both the studies.
Subramanian et al. [71] noted that comparisons of differentially expressed genes between similar microarray studies often result in very weak concordance in the identified gene lists. This is largely due to the traditional emphasis on statistical significance of individual genes, rather than on sets of genes involved in coordinated processes. Subtle gene expression changes associated with phenotypic differences may be obscured. However, as noted by the Subramanian group [71], a small change in expression of numerous genes in a pathway may have a dramatic biological effect while a much larger change in expression of a single gene may be much less important. Hence, Gene Set Enrichment Analysis (GSEA) considers the cumulative contribution of sets of genes within a biological process or pathway. For example, data from two independent microarray studies were compared assessing clinical outcome in lung cancer patients [72,73]. Of the 100 genes best correlated with clinical outcome in each experiment, only 12 were common to both sets. However, GSEA analysis revealed excellent consistency when considering underlying biological processes associated with gene expression changes. Similarly, the Mariadason et al. [66] and Fleet et al. [70] studies exhibited good overlap when entire signaling and metabolic pathways were considered. The search for reliable markers of intestinal progenitor stem cells and their differentiation may be improved by consideration of panels of genes, perhaps having subtle changes in expression, rather than focusing on highly regulated individual gene markers. A greater consideration of gene sets will seek to reveal clustering along activity in a biological pathway, with less likelihood of being distracted by huge changes in single genes that may indicate an artifactual response or false positive.
A more recent gene microarray analysis of Caco-2 cell differentiation was performed with samples grown in monolayers and taken at 11 time points over a 26-day period [74]. Triplicate samples were obtained at each time point, and five replicate experiments were performed. Expression profiles from the Caco-2 time course were also compared to expression profiles in normal colon and tumor tissue. The Caco-2 samples preceding the day 4 time point clustered tightly with the colon cancer samples, while later samples clustered closely with the normal colon samples. At day 4 there was a marked decrease in expression in numerous genes associated with cell cycle regulation. The cell cycle genes decreased in expression as the post-mitotic cells polarized. Similar to the findings of the Mariadason [66] and Fleet [70] groups, a decrease in Wnt pathway activity was correlated with differentiating cells. Genes previously identified as induced by TCF4 (including c-myc) had a well-correlated decrease in expression in the post-mitotic time points. Those genes identified as TCF4-repressed were induced as a function of time. Two other Wnt target genes identified in this study were CD44 and Sox9, both having been implicated in stem cell regulation. Both genes were expressed at higher levels in the proliferating Caco-2 cells and their expression decreased with cell differentiation. In addition to the Wnt pathway, Saaf et al. [74] identified coordinated gene expression changes in the Notch, Hedehog, BMP, and FGF pathways.
Velcich et al. [75] used microarrays to demonstrate that although two subclones of HT29 colon tumor cells (C116E and C119A) differentiate into different phenotypes, they exhibit similar gene expression programming during differentiation. C116E cells spontaneously differentiated into mucus-producing goblet cells, while C119A cells attain a deep-crypt secretory phenotype. cDNA microarrays consisting of 8,063 clones were used to identify 257 differentially expressed genes in the C116E cells and 330 genes in the C119A cells measured at six time points over a 20-day period. Velcich et al. [75] observed changes in numerous cell cycle regulation genes as well as decreased expression of c-myc correlating with the transition from proliferating to differentiated cells. The authors noted that while only 115 genes were common to the differentially expressed sets of the two cell lines, closer examination revealed that the overall expression profiles in the two clones were strikingly similar. The differences were substantially quantitative in terms of absolute levels of expression; only 31 genes showed opposite regulation. They concluded that subtle changes in expression can have a large effect on phenotypic outcome.
Proteomic profiling of intestinal stem cells
The proteome of mouse embryonic stem cells grown as a pure culture has been determined to a depth of over 5,000 proteins [76] and includes accepted stem cell markers that are expected to be low abundance proteins. The difficulty in expanding intestinal epithelial cells in culture has precluded an extensive cataloging of such cells. In order to identify differences in the proteomes of replicating and terminally differentiated cells from mouse small intestine, cells representing the villus and others representing crypts were isolated and subjected to two-dimensional fluorescence difference gel electrophoresis (2D DIGE) [77]. For this work, the cells were isolated from the crypts and the villi were labeled with either a Cy-3 or a Cy-5 dye and then mixed together before separation by 2D electrophoresis. 2D DIGE allows proteins that are of greater or lesser abundance in the two samples to be identified by the fluorescent signal from the spots on the 2D gel. Spots of interest were subjected to MALDI-MS/MS-based protein identification. Using the 2D DIGE technique, a total of 46 proteins were identified as being differentially expressed in the crypts and the villi. Of those 46 proteins only adenosine deaminase, which was 6.4-fold more abundant in the villi than in the crypts, had a fold change greater than 2.5. The inability to identify any proteins that are expressed exclusively in the crypt fraction may indicate that stem cells represent too small a part of that cell population to be effectively evaluated, or that the stem cell-specific proteins are not abundant enough to be identified after 2D electrophoresis. The inability of 2D electrophoresis to probe deeply into the proteome is indicated by the assignment of 93% of all the differentially expressed proteins being known high-abundance proteins. Application of additional cell selection techniques, such as laser capture micro-dissection or cell sorting using intestinal stem cell markers such as Lgr5, in combination with the most advanced mass spectrometry techniques, should allow a greater depth of the intestinal stem cell proteome to be cataloged and new intestinal stem cell markers to be identified.
Intestinal stem cells
The intestinal mucosa is a unique model for studies on stem cell biology because proliferative and differentiated cells occupy relatively distinct regions. The functional cells (ie, absorptive and mucous goblet) populate the villus and the replicating cells are localized mainly at the crypt base, a region that apparently also contains the stem cells. The putative intestinal stem cell should exhibit extensive proliferative and self-renewal capacity and the ability to produce the various differentiated cell lineages required to populate the intestinal tissue [12–15,24,25,78]. Stem cell proliferation classically occurs by asymmetric mitosis. Thus, one of the daughter cells retains stem cell properties while the second becomes a progenitor that continues to divide, migrating upward into the villus to generate differentiated cell lineages [79,80]. Until recently, based on DNA retention studies, stem cells were believed to occupy the +4 columnar cell position at the crypt base above the paneth cells [81]. Another view implies that stem cells constitute the small undifferentiated cells interdispersed among paneth cells at the crypt base [16,79]. Barker et al. [25] discovered Lgr5 as an exclusive stem cell marker in mouse small intestine and colon and demonstrated that Lgr5-positive replicating columnar stem cells occupy the crypt base and were different from the +4 columnar cells. Using an inducible Cre knock-in allele and the Rosa 26-lacZ reporter strain, they further demonstrated that the Lgr5-positive crypt base cells generated the differentiated intestinal cell lineage [25]. It should, however, be noted that Lgr5-positive stem cells are not specific to intestine and are present in other tissues. Further, animals lacking the Lgr5 gene develop normal intestinal mucosa but the deletion is lethal due to other abnormalities [82]. Although it is possible that other receptors may compensate for Lgr5 in the knockout animals, the role of Lgr5 gene in the maintenance and function of stem cells requires further clarification. However, Lgr5 is receiving broad acceptance as a marker to distinguish stem cells within the intestine from more mature cell types. More recently, Ascl2 was reported to be expressed along with Lgr5 in the putative stem cell in the intestine [83]. Analysis of the various data showed that Ascl2 was expressed uniquely in the stem cells of small intestine and colon where it appears to regulate stem cell fate.
The RNA-binding protein, Musashi1, is another widely applied stem cell marker apparently required for characteristic asymmetric cell division. Musashi1 was originally discovered in sensory organ precursor cells in Drosophila [84] but the gene is well conserved and Musashi1 homologs have been identified in mammals [85]. The gene is highly expressed in crypt base cells and also in a few cells above the paneth cells [24,86,87]. In human colon, Musashi1-positive cells were predominantly located between positions 3 and 4 although some labeled cells were seen up to position 10 in the crypts. Because Musashi1-positive cells are mostly localized in the proliferative stem cell zone, the protein is considered a pertinent intestinal stem cell marker [87]. However, others have reported the expression of Musashi1 to be less circumscribed, and this protein is perhaps better described as a marker of intestinal progenitor cells [88].
Cell surface proteins are considered important markers for cell identification. In intestine, epithelial cells attach to basement membrane through interaction between integrins on the epithelial cell surface and collagen, fibronectin, and laminin in the basement membrane. High expression of β1- and α2-integrins has been reported in the proliferative basal and middle crypt region of colon [89] and small intestine [90]. High expression of α2/β1-integrins has also been reported in stem cells of prostate [91,92] and epidermis [93]. Because integrins are expressed on the cell surface, these proteins may serve as pertinent markers for identifying and sorting of intestinal stem cells. However, whether the stem cells will endure the prolonged enzymatic and FACS processing and provide viable cells for in vitro propagation is an empirical question.
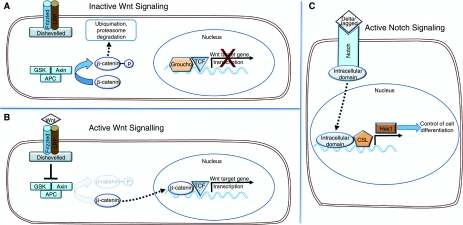
Studies of molecular pathways involved in the regulation of cell replication and differentiation in intestine have contributed to identification of stem cells. Two of the pathways most extensively explored in this respect are canonical Wnt β-catenin and Notch signaling, both of which are involved in development and morphogenesis as well as in tissue homeostasis (Fig. 2). An essential component of the Wnt pathway is the cytoplasmic signal transducer β-catenin [94]. The absence of Wnt signals causes phosphorylation of free cytosolic β-catenin, channeling it for degradation via the β-catenin destruction complex. The degradation complex is composed of tumor suppressor adenomatous polyposis coli (APC) and axin and also casein kinase 1 and glycogen synthase kinase 3β. After binding to the degradation complex, β-catenin is phosphorylated at Ser45 at the N-terminus resulting in its ubiquitination and proteosomal degradation. Consequently within the nucleus, lymphoid enhancer factor (LEF)/TCF remains bound to corepressor and serves to repress Wnt target genes [95,96].
FIG. 2.
Summary of Wnt and Notch signaling pathways involved in the regulation of replication and differentiation of intestinal stem cells. (A) Lack of Wnt signals causes phosphorylation of β-catenin and channels it for proteosomal degradation via the β-catenin degradation complex. (B) Following Wnt activation, the cytoplasmic protein Disheveled is activated blocking the activation of β-catenin degradation complex. Unphosphorylated β-catenin then enters the nucleus where it binds to the transcription factor TCF facilitating the transcription of Wnt target genes. (C) Activation of Notch by interactions with delta/jagged of neighboring cells results in the cleavage of intracellular domain of Notch that translocates to the nucleus forming a transcriptional complex with CSL and causing the transcription of downstream gene Hes 1, which in turn activates the transcription of genes involved in cellular differentiation.
Wnt ligand activation by binding to frizzled and low-density lipoprotein-related protein transmembrane receptors activates the cytoplasmic protein Disheveled, blocking the activation of β-catenin degradation complex. β-catenin then enters the nucleus where it binds to LEF/TCF transcription factor displacing the corepressor Groucho and facilitating the transcription of Wnt target genes such as c-myc and cyclin D1 [94,69]. Experimental manipulations have demonstrated that the Wnt β-catenin pathway predominantly regulates crypt cell proliferation and maintains stemness and the undifferentiated state of intestinal stem cells. For instance, β-catenin deletion rapidly blocks crypt cell replication and induces functional differentiation. Continuous crypt to villus migration of the cells subsequently results in crypt disappearance as most differentiated cells enter the villus. On the other hand, overexpression of β-catenin increased proliferation, causing crypt expansion and decreasing functional differentiation [97,98], possibly caused by expansion of the stem cell population. It is interesting to note that colorectal cancer is accompanied by mutations in APC, a member of Wnt signaling pathway, resulting in the formation of constitutive nuclear β-catenin/TCF complexes [99,100]. In intestine, Wnt signaling activity is restricted to crypt cells, the strongest expression being located in the putative stem cell zone [97]. Although the Wnt pathway is permissive for proliferation and stem cell self-renewal, it apparently functions in association with other cell cycle regulatory PTEN/PI3K signaling [101]. For instance, P-PTEN and P-AKT are predominantly expressed in the replicative stem cell zone and reportedly affect the transcription of β-catenin, as PTEN inhibition of AKT restricts the nuclear accumulation of β-catenin [102]. Similarly, activation of Wnt signaling has been reported in stem cells of other tissues, including epidermis [103], hematopoietic [104], and neural cells [105] in which the pathway regulates cell replication and self-renewal.
The Notch signaling pathway is required for maintenance of the crypt zone in a proliferative undifferentiated state. However, Notch proteins regulate mainly the development of intestinal differentiated cell lineage via paracellular interaction (Fig. 2). The transmembrane Notch receptor of transitional proliferative cells interacts with the ligand delta and jagged of adjacent cells resulting in cleavage of intracellular domain of Notch, which then translocates to the nucleus to bind to transcription factors and activates the Notch target gene hairy/enhancer of split (Hes1). Up-regulation of Hes1 results in inhibition of basic helix-loop-helix (bHLH) transcriptional activators such as Math1 and neurogenin3, which are required for endocrine cell differentiation [106,107]. Mice with Math1 deletion do not develop goblet, paneth, or enteroendocrine cells and produce only enterocytes in the small intestine [108]. In contrast, Hes1 null mice exhibit increased development of endocrine and goblet cells [109]. Further, Hes1 is normally restricted to the nonproliferating villus cells, indicating that the Notch pathway plays a major role in cell differentiation rather than proliferation.
One essential feature of stem cells is their replicative capacity. Over the years, several different intestinal cell lines have been reported from rodent and human tissues. Among them are cell lines from human fetal [4], adult [1], and neonatal colon [2]; rat small intestine [8]; and, dog small intestine [9]. Although most of these cell lines exhibited apparently unlimited proliferation, they were not characterized for stem cell markers. The cell lines were tested for functional phenotypes and demonstrated to be positive for brush border proteins [2,8] and mucopolysaccharides [2,4]. Even though certain cell lines showed potential functional markers, they should now be evaluated for the novel stem cell markers unavailable when the cell lines were established. Normal epithelial cells isolated from the human colon of a patient with colon carcinoma expressed stem cell markers such as β1 integrin when grown in soft agar [110]. These results should be viewed with caution because the cells were not tested for lack of tumor growth in nude mice or for diploid karyotype. Using fluorescence-activated cell sorting, putative stem cells were isolated as a “side population” from the mouse jejunum [28]. The stem cell-enriched population was positive for Musashi1 and β1 integrin but the cells could not be cultured, even though they apparently remained viable for 14 days.
Therefore, at present there is no in vitro model of intestinal stem cells in which to demonstrate differentiation of various cell lineages. In this respect, it is pertinent to evaluate the existing intestinal epithelial cell lines for the expression of stem cell markers and their capacity to differentiate. New initiatives should be undertaken to develop additional stem cell lines using the novel markers recently discovered.
Acknowledgment
This work was supported in part by NIEHS P30 EHS Center grant ES06639.
References
- 1.Moyer MP. Culture of human gastrointestinal epithelial cells. Proc Soc Exp Biol. 1983;174:12–15. doi: 10.3181/00379727-174-1-rc1. [DOI] [PubMed] [Google Scholar]
- 2.Yeh KY. Chopra DP. Epithelial cell culture from the colon of the suckling rat. In Vitro. 1980;16:976–986. doi: 10.1007/BF02619336. [DOI] [PubMed] [Google Scholar]
- 3.Chopra DP. Yeh KY. Long-term culture of epithelial cells from the normal rat colon. In Vitro. 1981;17:441–449. doi: 10.1007/BF02626745. [DOI] [PubMed] [Google Scholar]
- 4.Siddiqui KM. Chopra DP. Primary and long-term epithelial cell cultures from human fetal normal colonic mucosa. In Vitro. 1984;20:859–863. doi: 10.1007/BF02619632. [DOI] [PubMed] [Google Scholar]
- 5.Chopra DP. Siddiqui KM. Effects of insulin, transferrin, cholera toxin and epidermal growth factor on growth and differentiation of human normal colon epithelial cells. Gastroenterology. 1987;92:891–904. doi: 10.1016/0016-5085(87)90962-0. [DOI] [PubMed] [Google Scholar]
- 6.Deveney CW. Rand-Luby L. Rutten MJ. Luttropp CA. Fowler WM. Land J. Meichsner CL. Farahmand M. Sheppard BC. Crass RA. Deveney KE. Establishment of human colonic epithelial cells in long-term culture. J Surg Res. 1996;64:161–169. doi: 10.1006/jsre.1996.0323. [DOI] [PubMed] [Google Scholar]
- 7.Bartsch I. Zschaler I. Haseloff M. Steinberg P. Establishment of a long-term culture system for rat colon epithelial cells. In Vitro Cell Dev Biol Anim. 2004;40:278–284. doi: 10.1290/0404035.1. [DOI] [PubMed] [Google Scholar]
- 8.Quaroni A. Wands J. Trelstad RL. Isselbacher KJ. Epithelioid cell cultures from rat small intestine. Characterization by morphologic and immunologic criteria. J Cell Biol. 1979;80:248–265. doi: 10.1083/jcb.80.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weng XH. Beyenbach KW. Quaroni A. Cultured monolayers of the dog jejunum with the structural and functional properties resembling the normal epithelium. Am J Physiol Gastrointest Liver Physiol. 2005;288:G705–G717. doi: 10.1152/ajpgi.00518.2003. [DOI] [PubMed] [Google Scholar]
- 10.Ma TY. Hollander D. Bhalla D. Nguyen H. Krugliak P. IEC-18, a nontransformed small intestinal cell line for studying epithelial permeability. J Lab Clin Med. 1992;120:329–341. [PubMed] [Google Scholar]
- 11.Gulati AS. Ochsner SA. Henning SJ. Molecular properties of side population-sorted cells from mouse small intestine. Am J Physiol Gastrointest Liver Physiol. 2008;294:G286–G294. doi: 10.1152/ajpgi.00416.2007. [DOI] [PubMed] [Google Scholar]
- 12.Scoville DH. Sato T. He XC. Li L. Current view: intestinal stem cells and signaling. Gastroenterology. 2008;134:849–864. doi: 10.1053/j.gastro.2008.01.079. [DOI] [PubMed] [Google Scholar]
- 13.Montgomery RK. Breault DT. Small intestinal stem cell markers. J Anat. 2008;213:52–58. doi: 10.1111/j.1469-7580.2008.00925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rizvi AZ. Hunter JG. Wong MH. Gut-derived stem cells. Surgery. 2005;137:585–590. doi: 10.1016/j.surg.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Cheng H. Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. Am J Anat. 1974;141:537–561. doi: 10.1002/aja.1001410407. [DOI] [PubMed] [Google Scholar]
- 16.Bjerknes M. Cheng H. Clonal analysis of mouse intestinal epithelial progenitors. Gastroenterology. 1999;116:7–14. doi: 10.1016/s0016-5085(99)70222-2. [DOI] [PubMed] [Google Scholar]
- 17.Bry L. Falk P. Huttner K. Ouellette A. Midtvedt T. Gordon JI. Paneth cell differentiation in the developing intestine of normal and transgenic mice. Proc Natl Acad Sci USA. 1994;91:10335–10339. doi: 10.1073/pnas.91.22.10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batlle E. Henderson JT. Beghtel H. van den Born MM. Sancho E. Huls G. Meeldijk J. Robertson JJ. van de Wetering M. Pawson T. Clevers H. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- 19.Bettess MD. Dubois N. Murphy MJ. Dubey C. Roger C. Robine S. Trumpp A. c-Myc is required for the formation of intestinal crypts but dispensable for homeostasis of the adult intestinal epithelium. Mol Cell Biol. 2005;17:7868–7878. doi: 10.1128/MCB.25.17.7868-7878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vidrich A. Buzan JM. Ilo C. Bradley L. Skaar K. Cohn SM. Fibroblast growth factor receptor-3 is expressed in undifferentiated intestinal epithelial cells during murine crypt morphogen. Dev Dyn. 2004;230:114–123. doi: 10.1002/dvdy.20018. [DOI] [PubMed] [Google Scholar]
- 21.Blache P. van de Wetering M. Duluc I. Domon C. Berta P. Freund JN. Clevers H. Jay P. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J Cell Biol. 2004;166:37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kayahara T. Sawada M. Takaishi S. Fukui H. Seno H. Fukuzawa H. Suzuki K. Hiai H. Kageyama R. Okano H. Chiba T. Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett. 2003;535:131–135. doi: 10.1016/s0014-5793(02)03896-6. [DOI] [PubMed] [Google Scholar]
- 23.Gregorieff A. Pinto D. Begthel H. Destrée O. Kielman M. Clevers H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 24.Potten CS. Booth C. Tudor GL. Booth D. Brady G. Hurley P. Ashton G. Clarke R. Sakakibara S. Okano H. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 2003;71:28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- 25.Barker N. van Es JH. Kuipers J. Kujala P. van den Born M. Cozijnsen M. Haegebarth A. Korving J. Begthel H. Peters PJ. Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 26.Booth C. O'Shea JA. Potten CS. Maintenance of functional stem cells in isolated and cultured adult intestinal epithelium. Exp Cell Res. 1999;249:359–366. doi: 10.1006/excr.1999.4483. [DOI] [PubMed] [Google Scholar]
- 27.Booth C. Patel S. Bennion GR. Potten CS. The isolation and culture of adult mouse colonic epithelium. Epithelial Cell Biol. 1995;4:76–86. [PubMed] [Google Scholar]
- 28.Dekaney CM. Rodriguez JM. Graul MC. Henning SJ. Isolation and characterization of a putative intestinal stem cell fraction from mouse jejunum. Gastroenterology. 2005;129:1567–1580. doi: 10.1053/j.gastro.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 29.Dobbins WO., III Hijmans JC. McCarty KS. A light and electron microscopic study of duodenal epithelium of chick embryos cultured in the presence and absence of hydrocortisone. Gastroenterology. 1967;53:557–574. [PubMed] [Google Scholar]
- 30.Dolin R. Blacklow NR. Malmgren RA. Chanock RM. Establishment of human fetal intestinal organ cultures for growth of viruses. J Infect Dis. 1970;122:227–231. doi: 10.1093/infdis/122.3.227. [DOI] [PubMed] [Google Scholar]
- 31.Fukamachi H. Takayama S. Epithelial-mesenchymal interaction in differentiation of duodenal epithelium of fetal rats in organ culture. Experientia. 1980;36:335–336. doi: 10.1007/BF01952310. [DOI] [PubMed] [Google Scholar]
- 32.Quaroni A. Development of fetal rat intestine in organ and monolayer culture. J Cell Biol. 1985;100:1611–1622. doi: 10.1083/jcb.100.5.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pyke KW. Gogerly RL. Murine fetal colon in vitro: assays for growth factors. Differentiation. 1985;29:56–62. doi: 10.1111/j.1432-0436.1985.tb00292.x. [DOI] [PubMed] [Google Scholar]
- 34.Ferland S. Hugon JS. Organ culture of adult mouse intestine. I. Morphological results after 24 and 48 hours of culture. In Vitro. 1979;15:278–287. doi: 10.1007/BF02618952. [DOI] [PubMed] [Google Scholar]
- 35.Autrup H. Explant culture of human colon. Methods Cell Biol. 1980;21B:385–401. doi: 10.1016/s0091-679x(08)60694-9. [DOI] [PubMed] [Google Scholar]
- 36.Moorghen M. Chapman M. Appleton DR. An organ-culture method for human colorectal mucosa using serum-free medium. J Pathol. 1996;180:102–105. doi: 10.1002/(SICI)1096-9896(199609)180:1<102::AID-PATH613>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 37.Ootani A. Li X. Sangiorgi E. Ho QT. Ueno H. Toda S. Sugihara H. Fujimoto K. Weissman IL. Capecchi MR. Kuo CJ. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chopra DP. Xue-Hu IC. Reddy LV. Growth and gene expression in diploid epithelial cell lines derived from normal human parotid gland. Differentiation. 1995;58:241–251. doi: 10.1046/j.1432-0436.1995.5830241.x. [DOI] [PubMed] [Google Scholar]
- 39.Chopra DP. Grignon DJ. Joiakim A. Mathieu PA. Mohamed A. Sakr WA. Powell IJ. Sarkar FH. Differential growth factor responses of epithelial cell cultures derived from normal human prostate, benign prostatic hyperplasia, and primary prostate carcinoma. J Cell Physiol. 1996;169:269–280. doi: 10.1002/(SICI)1097-4652(199611)169:2<269::AID-JCP6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 40.Taylor GW. Chopra DP. Mathieu PA. Differences in secretory profiles of epithelial cell cultures derived from human tracheal and bronchial mucosa and submucosal glands. Epithelial Cell Biol. 1993;2:163–169. [PubMed] [Google Scholar]
- 41.Flint N. Cove FL. Evans GS. A low-temperature method for the isolation of small-intestinal epithelium along the crypt-villus axis. Biochem J. 1991;280(Pt 2):331–334. doi: 10.1042/bj2800331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evans GS. Flint N. Somers AS. Eyden B. Potten CS. The development of a method for the preparation of rat intestinal epithelial cell primary cultures. J Cell Sci. 1992;101:219–231. doi: 10.1242/jcs.101.1.219. [DOI] [PubMed] [Google Scholar]
- 43.Arnaout MA. Mahalingam B. Xiong JP. Integrin structure, allostery, and bidirectional signaling. Annu Rev Cell Dev Biol. 2005;21:381–410. doi: 10.1146/annurev.cellbio.21.090704.151217. [DOI] [PubMed] [Google Scholar]
- 44.Gooding JM. Yap KL. Ikura M. The cadherin–catenin complex as a focal point of cell adhesion and signalling: new insights from three-dimensional structures. Bioessays. 2004;26:497–511. doi: 10.1002/bies.20033. [DOI] [PubMed] [Google Scholar]
- 45.Kalabis J. Patterson MJ. Enders GH. Marian B. Iozzo RV. Rogler G. Gimotty PA. Herlyn M. Stimulation of human colonic epithelial cells by leukemia inhibitory factor is dependent on collagen-embedded fibroblasts in organotypic culture. FASEB J. 2003;179:1115–1117. doi: 10.1096/fj.02-0852fje. [DOI] [PubMed] [Google Scholar]
- 46.Wildrick DM. Lointier P. Nichols DH. Roll R. Quintanilla B. Boman BM. Isolation of normal human colonic mucosa: comparison of methods. In Vitro Cell Dev Biol Anim. 1997;331:18–27. doi: 10.1007/s11626-997-0017-2. [DOI] [PubMed] [Google Scholar]
- 47.Chopra DP. Yeh K. Brockman RW. Isolation and characterization of epithelial cell types from the normal rat colon. Cancer Res. 1981;41:168–175. [PubMed] [Google Scholar]
- 48.Gibson PR. van de Pol E. Maxwell LE. Gabriel A. Doe WF. Isolation of colonic crypts that maintain structural and metabolic viability in vitro. Gastroenterology. 1989;96:283–291. doi: 10.1016/0016-5085(89)91549-7. [DOI] [PubMed] [Google Scholar]
- 49.Brubaker PL. Vranic M. Fetal rat intestinal cells in monolayer culture: a new in vitro system to study the glucagon-like immunoreactive peptides. Endocrinology. 1987;120:1976–1985. doi: 10.1210/endo-120-5-1976. [DOI] [PubMed] [Google Scholar]
- 50.Fukamachi H. Proliferation and differentiation of fetal rat intestinal epithelial cells in primary serum-free culture. J Cell Sci. 1992;103(Pt 2):511–519. doi: 10.1242/jcs.103.2.511. [DOI] [PubMed] [Google Scholar]
- 51.Masui T. Wakefield LM. Lechner JF. LaVeck MA. Sporn MB. Harris CC. Type beta transforming growth factor is the primary differentiation-inducing serum factor for normal human bronchial epithelial cells. Proc Natl Acad Sci USA. 1986;83:2438–2442. doi: 10.1073/pnas.83.8.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freshney IR. Culture of Animal Cells. A Manual of Basic Techniques. 3rd. Wiley-Liss; New York: 1994. p. 486. [Google Scholar]
- 53.Booth C. Evans GS. Potten CS. Growth factor regulation of proliferation in primary cultures of small intestinal epithelium. In Vitro Cell Dev Biol Anim. 1995;31:234–243. doi: 10.1007/BF02639439. [DOI] [PubMed] [Google Scholar]
- 54.Kédinger M. Simon-Assmann P. Alexandre E. Haffen K. Importance of a fibroblastic support for in vitro differentiation of intestinal endodermal cells and for their response to glucocorticoids. Cell Differ. 1987;20:171–182. doi: 10.1016/0045-6039(87)90431-3. [DOI] [PubMed] [Google Scholar]
- 55.Hahn U. Tallmach A. Hahn EG. Riecken EO. Basement membrane components are potent promoters of rat intestinal epithelial cell differentiation in vitro. Gastroenterology. 1990;98:322–335. doi: 10.1016/0016-5085(90)90821-h. [DOI] [PubMed] [Google Scholar]
- 56.Flint N. Cove FL. Evans GS. Heparin stimulates the proliferation of intestinal epithelial cells in primary culture. J Cell Sci. 1994;107(Pt 2):401–411. doi: 10.1242/jcs.107.2.401. [DOI] [PubMed] [Google Scholar]
- 57.Odedra RM. Hart CA. Saunders JR. Getty B. van de Wall S. Sorensen SH. Embaye H. Batt RM. Growth and propagation of normal rat intestinal epithelial cells. In Vitro Cell Dev Biol Anim. 1996;32:107–115. doi: 10.1007/BF02723042. [DOI] [PubMed] [Google Scholar]
- 58.Owens RB. Smith HS. Hackett AJ. Epithelial cell cultures from normal glandular tissue of mice. J Natl Cancer Inst. 1974;53:261–269. doi: 10.1093/jnci/53.1.261. [DOI] [PubMed] [Google Scholar]
- 59.Gilbert SF. Migeon BR. d-Valine as a selective agent for normal human and rodent epithelial cells in culture. Cell. 1975;5:11–17. doi: 10.1016/0092-8674(75)90086-0. [DOI] [PubMed] [Google Scholar]
- 60.Baten A. Sakamoto K. Shamsuddin AM. Long-term culture of normal human colonic epithelial cells in vitro. FASEB J. 1992;6:2726–2734. doi: 10.1096/fasebj.6.9.1377141. [DOI] [PubMed] [Google Scholar]
- 61.Rusu D. Loret S. Peulen O. Mainil J. Dandrifosse G. Immunochemical, biomolecular and biochemical characterization of bovine epithelial intestinal primocultures. BMC Cell Biol. 2005;6:42–54. doi: 10.1186/1471-2121-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Föllmann W. Weber S. Birkner S. Primary cell cultures of bovine colon epithelium: isolation and cell culture of colonocytes. Toxicol In Vitro. 2000;14:435–445. doi: 10.1016/s0887-2333(00)00033-3. [DOI] [PubMed] [Google Scholar]
- 63.Seidelin JB. Horn T. Nielsen OH. Simple and efficient method for isolation and cultivation of endoscopically obtained human colonocytes. Am J Physiol Gastrointest Liver Physiol. 2003;285:G1122–G1128. doi: 10.1152/ajpgi.00533.2002. [DOI] [PubMed] [Google Scholar]
- 64.Mariadason JM. Rickard KL. Barkla DH. Augenlicht LH. Gibson PR. Divergent phenotypic patterns and commitment to apoptosis of Caco-2 cells during spontaneous and butyrate-induced differentiation. J Cell Physiol. 2000;1833:347–354. doi: 10.1002/(SICI)1097-4652(200006)183:3<347::AID-JCP7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 65.Ding QM. Ko TC. Evers BM. Caco-2 intestinal cell differentiation is associated with G1 arrest and suppression of CDK2 and CDK4. Am J Physiol. 1998;275:C1193–C1200. doi: 10.1152/ajpcell.1998.275.5.C1193. [DOI] [PubMed] [Google Scholar]
- 66.Mariadason JM. Arango D. Corner GA. Arañes MJ. Hotchkiss KA. Yang W. Augenlicht LH. A gene expression profile that defines colon cell maturation in vitro. Cancer Res. 2002;62:4791–4804. [PubMed] [Google Scholar]
- 67.Korinek V. Barker N. Moerer P. van Donselaar E. Huls G. Peters PJ. Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 68.Mariadason JM. Bordonaro M. Aslam F. Shi L. Kuraguchi M. Velcich A. Augenlicht LH. Divergent phenotypic patterns and commitment to apoptosis of Caco-2 cells during spontaneous and butyrate-induced differentiation. Cancer Res. 2001;61:3465–3471. [Google Scholar]
- 69.van de Wetering M. Sancho E. Verweij C. de Lau W. Oving I. Hurlstone A. van der Horn K. Batlle E. Coudreuse D. Haramis AP. Tjon-Pon-Fong M. Moerer P. van den Born M. Soete G. Pals S. Eilers M. Medema R. Clevers H. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 70.Fleet JC. Wang L. Vitek O. Craig BA. enberg HJ. Gene expression profiling of Caco-2 BBe cells suggests a role for specific signaling pathways during intestinal differentiation. Physiol Genomics. 2003;13:57–68. doi: 10.1152/physiolgenomics.00152.2002. [DOI] [PubMed] [Google Scholar]
- 71.Subramanian A. Tamayo P. Mootha VK. Mukherjee S. Ebert BL. Gillette MA. Paulovich A. Pomeroy SL. Golub TR. Lander ES. Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhattacharjee A. Richards WG. Staunton J. Li C. Monti S. Vasa P. Ladd C. Beheshti J. Bueno R. Gillette M. Loda M. Weber G. Mark EJ. Lander ES. Wong W. Johnson BE. Golub TR. Sugarbaker DJ. Meyerson M. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beer DG. Kardia SL. Huang CC. Giordano TJ. Levin AM. Misek DE. Lin L. Chen G. Gharib TG. Thomas DG. Lizyness ML. Kuick R. Hayasaka S. Taylor JM. Iannettoni MD. Orringer MB. Hanash S. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 74.Saaf AM. Halbleib JM. Chen X. Yuen ST. Leung SY. Nelson WJ. Brown PO. Parallels between global transcriptional programs of polarizing Caco-2 intestinal epithelial cells in vitro and gene expression programs in normal colon and colon cancer. Mol Biol Cell. 2007;18:4245–4260. doi: 10.1091/mbc.E07-04-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Velcich A. Corner G. Paul D. Zhuang M. Mariadason JM. Laboisse C. Augenlicht L. Quantitative rather than qualitative differences in gene expression predominate in intestinal cell maturation along distinct cell lineages. Exp Cell Res. 2005;304:28–39. doi: 10.1016/j.yexcr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 76.Graumann J. Hubner NC. Kim JB. Ko K. Moser M. Kumar C. Cox J. Schöler H. Mann M. Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol Cell Proteomics. 2008;7:672–683. doi: 10.1074/mcp.M700460-MCP200. [DOI] [PubMed] [Google Scholar]
- 77.Chang J. Chance MR. Nicholas C. Ahmed N. Guilmeau S. Flandez M. Wang D. Byun DS. Nasser S. Albanese JM. Corner GA. Heerdt BG. Wilson AJ. Augenlicht LH. Mariadason JM. Proteomic changes during intestinal cell maturation in vivo. J Proteomics. 2008;71:530–546. doi: 10.1016/j.jprot.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van der Flier LG. Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2008;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 79.Bjerknes M. Cheng H. Intestinal epithelial stem cells and progenitors. Methods Enzymol. 2006;419:337–383. doi: 10.1016/S0076-6879(06)19014-X. [DOI] [PubMed] [Google Scholar]
- 80.Marshman E. Booth C. Potten CS. The intestinal epithelial stem cell. Bioessays. 2002;24:91–98. doi: 10.1002/bies.10028. [DOI] [PubMed] [Google Scholar]
- 81.Potten CS. Kovacs L. Hamilton E. Continuous labelling studies on mouse skin and intestine. Cell Tissue Kinet. 1974;7:271–283. doi: 10.1111/j.1365-2184.1974.tb00907.x. [DOI] [PubMed] [Google Scholar]
- 82.Morita H. Mazerbourg S. Bouley DM. Luo CW. Kawamura K. Kuwabara Y. Baribault H. Tian H. Hsueh AJ. Neonatal lethality of LGR5 null mice is associated with ankyloglossia and gastrointestinal distension. Mol Cell Biol. 2004;24:9736–9743. doi: 10.1128/MCB.24.22.9736-9743.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van der Flier LG. van Gijn ME. Hatzis P. Kujala P. Haegebarth A. Stange DE. Begthel H. van den Born M. Guryev V. Oving I. van Es JH. Barker N. Peters PJ. van de Wetering M. Clevers H. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 84.Nakamura M. Okano H. Blendy JA. Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- 85.Sakakibara S. Imai T. Hamaguchi K. Okabe M. Aruga J. Nakajima K. Yasutomi D. Nagata T. Kurihara Y. Uesugi S. Miyata T. Ogawa M. Mikoshiba K. Okano H. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev Biol. 1996;176:230–242. doi: 10.1006/dbio.1996.0130. [DOI] [PubMed] [Google Scholar]
- 86.Asai R. Okano H. Yasugi S. Correlation between Musashi-1 and c-hairy-1 expression and cell proliferation activity in the developing intestine and stomach of both chicken and mouse. Dev Growth Differ. 2005;47:501–510. doi: 10.1111/j.1440-169X.2005.00825.x. [DOI] [PubMed] [Google Scholar]
- 87.Kayahara T. Sawada M. Takaishi S. Fukui H. Seno H. Fukuzawa H. Suzuki K. Hiai H. Kageyama R. Okano H. Chiba T. Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett. 2003;535:131–135. doi: 10.1016/s0014-5793(02)03896-6. [DOI] [PubMed] [Google Scholar]
- 88.Gregorieff A. Pinto D. Begthel H. Destrée O. Kielman M. Clevers H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 89.Fujimoto K. Beauchamp RD. Whitehead RH. Identification and isolation of candidate human colonic clonogenic cells based on cell surface integrin expression. Gastroenterology. 2002;123:1941–1948. doi: 10.1053/gast.2002.37065. [DOI] [PubMed] [Google Scholar]
- 90.Beaulieu JF. Differential expression of the VLA family of integrins along the crypt-villus axis in the human small intestine. J Cell Sci. 1992;102:427–436. doi: 10.1242/jcs.102.3.427. [DOI] [PubMed] [Google Scholar]
- 91.Lawson DA. Xin L. Lukacs RU. Cheng D. Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci USA. 2007;104:181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Collins AT. Habib FK. Maitland NJ. Neal DE. Identification and isolation of human prostate epithelial stem cells based on alpha(2)beta(1)-integrin expression. J Cell Sci. 2001;114:3865–3872. doi: 10.1242/jcs.114.21.3865. [DOI] [PubMed] [Google Scholar]
- 93.Jones PH. Watt FM. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell. 1993;21:713–724. doi: 10.1016/0092-8674(93)90251-k. [DOI] [PubMed] [Google Scholar]
- 94.Behrens J. Lustig B. The Wnt connection to tumorigenesis. Int J Dev Biol. 2004;48:477–487. doi: 10.1387/ijdb.041815jb. [DOI] [PubMed] [Google Scholar]
- 95.Cavallo RA. Cox RT. Moline MM. Roose J. Polevoy GA. Clevers H. Peifer M. Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 96.Roose J. Molenaar M. Peterson J. Hurenkamp J. Brantjes H. Moerer P. van de Wetering M. Destrée O. Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 97.Fevr T. Robine S. Louvard D. Huelsken J. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol Cell Biol. 2007;27:7551–7559. doi: 10.1128/MCB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sansom OJ. Reed KR. Hayes AJ. Ireland H. Brinkmann H. Newton IP. Batlle E. Simon-Assmann P. Clevers H. Nathke IS. Clarke AR. Winton DJ. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rubinfeld B. Albert I. Porfiri E. Fiol C. Munemitsu S. Polakis P. Binding of GSK3beta to the APC–beta-catenin complex and regulation of complex assembly. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 100.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 101.He XC. Zhang J. Tong WG. Tawfik O. Ross J. Scoville DH. Tian Q. Zeng X. He X. Wiedemann LM. Mishina Y. Li L. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt–beta-catenin signaling. Nat Genet. 2004;36:1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- 102.Persad S. Troussard AA. McPhee TR. Mulholland DJ. Dedhar S. Tumor suppressor PTEN inhibits nuclear accumulation of beta-catenin and T cell/lymphoid enhancer factor 1-mediated transcriptional activation. J Cell Biol. 2001;153:1161–1174. doi: 10.1083/jcb.153.6.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huelsken J. Vogel R. Erdmann B. Cotsarelis G. Birchmeier W. beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 104.Reya T. Duncan AW. Ailles L. Domen J. Scherer DC. Willert K. Hintz L. Nusse R. Weissman IL. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 105.Zechner D. Fujita Y. Hülsken J. Müller T. Walther I. Taketo MM. Crenshaw EB., III Birchmeier W. Birchmeier C. beta-Catenin signals regulate cell growth and the balance between progenitor cell expansion and differentiation in the nervous system. Dev Biol. 2003;258:406–418. doi: 10.1016/s0012-1606(03)00123-4. [DOI] [PubMed] [Google Scholar]
- 106.Jensen J. Pedersen EE. Galante P. Hald J. Heller RS. Ishibashi M. Kageyama R. Guillemot F. Serup P. Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 107.Schröder N. Gossler A. Expression of Notch pathway components in fetal and adult mouse small intestine. Gene Expr Patterns. 2002;2:247–250. doi: 10.1016/s1567-133x(02)00060-1. [DOI] [PubMed] [Google Scholar]
- 108.Yang Q. Bermingham NA. Finegold MJ. Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 109.van Den Brink GR. de Santa Barbara P. Roberts DJ. Development. Epithelial cell differentiation—a Mather of choice. Science. 2001;294:2115–2116. doi: 10.1126/science.1067751. [DOI] [PubMed] [Google Scholar]
- 110.Fujimoto K. Beauchamp RD. Whitehead RH. Identification and isolation of candidate human colonic clonogenic cells based on cell surface integrin expression. Gastroenterology. 2002;123:1941–1948. doi: 10.1053/gast.2002.37065. [DOI] [PubMed] [Google Scholar]