Abstract
Mutations that alter the phenotypic behavior of the Escherichia coli mechanosensitive channel of small conductance (MscS) have been identified; however, most of these residues play critical roles in the transition between the closed and open states of the channel and are not directly involved in lipid interactions that transduce the tension response. In this study, we use molecular dynamic simulations to predict critical lipid interacting residues in the closed state of MscS. The physiological role of these residues was then investigated by performing osmotic downshock assays on MscS mutants where the lipid interacting residues were mutated to alanine. These experiments identified seven residues in the first and second transmembrane helices as lipid-sensing residues. The majority of these residues are hydrophobic amino acids located near the extracellular interface of the membrane. All of these residues interact strongly with the lipid bilayer in the closed state of MscS, but do not face the bilayer directly in structures associated with the open and desensitized states of the channel. Thus, the position of these residues relative to the lipid membrane appears related to the ability of the channel to sense tension in its different physiological states.
Introduction
Bacterial mechanosensitive channels function as pressure relief valves, alleviating the high intracellular pressure caused by hypoosmotic shock and preventing bacterial lysis. In Escherichia coli (E. coli), four mechanosensitive channels have been identified, cloned, and characterized: the mechanosensitive channel of large conductance (MscL), the mechanosensitive channel of small conductance (MscS), the mechanosensitive channel of potassium-dependant small conductance (MscK), and YbdG (1–4). MscS and MscL are the best studied of the bacterial mechanosensitive channels (5,6). These two channels are found in the plasma membrane, activated by mechanical tension on the lipid membrane, and rescue bacteria from cell lysis under hypoosmotic conditions. Hypoosmotic shock, or downshock, causes bacteria to swell due to an influx of water across the cellular membrane in order to equilibrate the osmotic strength of the internal and external environments. This swelling results in membrane tension and can induce cell lysis. Mechanosensitive channels in bacteria gate in response to this tension and release small osmolytes and even proteins to allow for rapid equilibration of the internal and external osmotic strengths.
The response of MscS to membrane tension has been characterized using osmotic downshock assays (2,7–9) and patch-clamp electrophysiology (10–12). In addition, molecular dynamic simulations of MscS under tension have been carried out in an attempt to understand MscS's tension response (13,14). Previous work has identified nine residues that are important for MscS gating: I37, A51, D62, F68, L86, L111, L115, R128, and R131 (12,15–17). Three of these residues, D62, R128, and R131, are postulated to work in concert to stabilize the open state conformation of the channel under tension through electrostatic interactions (16). Removal of one of these two positively charged arginines decreases the stability of the open state conformation. Based on the MscS crystal structures, this cluster of residues is predicted to be located in the loop between the first and second transmembrane domains (TM) and the cytoplasmic vestibule at the protein-lipid-water interface (16,18).
The remaining residues are within the TM domains and have been postulated to play important roles during the transition between closed and open states (12,15,16). Two pairs of residues, I37/L86 and A51/F68, are postulated to be involved with the displacement of TM1 and TM2 from the central axis during gating (12) and are predicted not to interact with the lipid bilayer based on the MscS crystal structures (12,18). Mutation of F68, L111, and L115 to serine has suggested that these residues are involved in the desensitization or inactivation of MscS (17). These three residues are thought to transduce mechanical stress to the gate through hydrophobic interactions. The removal of these hydrophobic interactions results in rapid desensitization (17). Furthermore, these hydrophobic interactions are postulated to be involved in the coupling of TM2 and TM3 because they come into contact with one another during the transduction of the force to the gate. Although these nine residues clearly play critical roles in MscS channel function, they are not involved in tension sensation or response from direct interaction with surrounding lipid.
Using the initial MscS crystal structure solved by Bass et al. (18) (1MXM/2OAU) as a starting point, Nomura et al. (12) employed asparagine scanning mutagenesis of the lipid-facing amino-acid side chains in an attempt to identify the residues critical for tension sensation. This study revealed a number of amino acids that phenotypically alter MscS channel function, including I37, A51, F68, and L86. The study of these mutations revealed that these residues take part in important intermolecular interactions that are involved in the open-to-closed transition state (12). These four residues interact as two pairs, I37/L86 and A51/F68, which work in concert to open the channel under mechanical stress. However, few if any of the residues identified in this study seem to be involved in tension sensation through the lipid bilayer. This may not be surprising because 1MXM (2OAU), the initial MscS crystal structure, is thought to represent a desensitized or inactivated state of MscS (13,19,20). In the desensitized state, the MscS channel is insensitive to tension. This insensitivity is potentially due to occlusion of the tension-sensing or tension-responsive residues in the desensitized state, leading to decreased interactions with the membrane. This is consistent with the assertion that changes to the lipid membrane can alter the desensitization behavior of MscS as observed upon lysophosphatidylcholine application, MscS reconstitution by the “sucrose method,” and in lipid exchange (13,21,22).
We have previously used a combination of extensive random mutagenesis and molecular dynamic simulations to show that many MscL residues important for sensing bilayer tension interact strongly with the lipid tail groups (23,24). A large library of MscL mutations, created by random mutagenesis, was used to identify critical residues involved in tension sensation (24). This analysis showed that mutations to the lipid lining residues of MscL had a high propensity of being phenotypically loss of function (LOF) (24). A LOF phenotype has been demonstrated to correspond to an increase in gating tension for bacterial mechanosensitive channels (25). In a subsequent study, we conducted molecular dynamic (MD) simulations on the closed state structure of MscL and analyzed the protein-lipid interaction energies. This analysis indicated that the majority of lipid-lining LOF mutations identified in our random mutagenesis study corresponded to residues with significant lipid interactions. Together, these observations implied that channel residues that interact with lipid are critical in tension sensation (23,24).
Unfortunately, a crystal structure of the closed state of MscS has yet to be determined to allow an analysis of the lipid-lining residues in the closed state similar to that carried out for the desensitized state (12). However, an experimentally derived electron paramagnetic resonance structure for the carbon-alpha (Cα) chain of MscS has been developed by Vásquez et al. (26). This structure cannot be directly analyzed for lipid bilayer interactions, because it is missing the amino-acid side chains. Nonetheless, this structure does provide an excellent starting point for the type of analysis that we have previously carried out on MscL (23).
Here, we explore the residues critical for tension sensation in MscS and evaluate how these residues move through the closed, open, and desensitized states. Starting from a computational analysis, we identified potentially critical residues in MscS tension sensation based on the closed state of MscS. The central role of these residues for tension sensation was then confirmed using in vivo downshock assays of alanine mutations (Fig. 1). Moreover, we note that the seven residues identified here are mostly distinct from those previously identified by Nomura et al. (12). By considering these residues and their relative location in the closed, open, and desensitized states of MscS, we gain significant insight into how MscS channels sense and respond to membrane tension.
Figure 1.
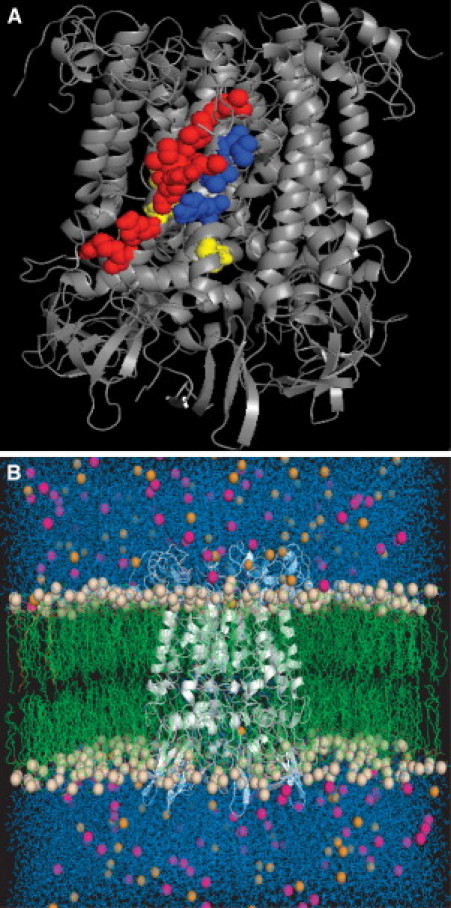
Closed state MscS model at the end of the MD simulation, as seen from the side (A) and with lipids, chloride ions, sodium ions, and water included in the simulation (B). The residues highlighted as a space-filling model in panel A correspond to (red) predicted lipid-interacting residues in TM1 (L35, I39, L42, I43, R46, N50, R54); (blue) predicted lipid interacting residues in TM2 (R74, I78, L82); and (yellow) noninteracting residues used as controls in this study (I44, I48, F68).
Materials and Methods
Molecular dynamics simulations
MODELLER 9.1 (27,28) was used to construct an all-atom structure of MscS based on the three-dimensional Cα model of the channel in its closed state developed by Vásquez et al. (26). Because the model does not represent the entire MscS protein, sequence alignments for modeling were produced using the ALIGN feature of MODELLER and the Vásquez structure was used as the modeling template. Three-hundred models were produced, and the model with the lowest DOPE energy was used as a starting structure for MD simulations.
MD simulations of MscS were performed using the GROMACS MD simulations suite and the GROMOS force field (29). The channel model was embedded in a preequilibrated palmitoyloleoyl-phosphatidylethanolamine (POPE) membrane using the method of Kandt et al. (30). The all-atom MscS structure obtained from MODELLER was superimposed onto a preequilibrated POPE bilayer with the desired orientation relative to the membrane, such that the structure overlapped with lipids. The bilayer was then expanded by translating lipid molecules and scaling the lateral dimensions of the box size by a factor of four, and the protein was centered in the new bilayer plane. A series of compression steps were performed using a scaling factor of 0.95 to slowly reequilibrate the lipid density and properly embed the channel in the bilayer. During this process, the protein was constrained and the system energy was minimized after each scaling step. The area per lipid was calculated at each step with scaling and minimization steps performed until the area per lipid reached 0.4990 nm2/lipid, the reference value of a POPE membrane (31). After reequilibration of the membrane, the system was hydrated, sufficient chloride ions were added to neutralize the net charge on the protein, sodium and chloride ions were added to yield an ionic strength of 100 mM, and the system was minimized using 50 steps of steepest-descents minimization. Any lipids that had significant overlap with the channel after the steepest-descents minimization were removed. The final system was composed of the protein, 451 POPE lipids, 40,205 water molecules, 127 sodium ions, and 148 chloride ions (Fig. 1).
Two MD simulations were carried out starting from the minimized system, using different initial random velocities. In both cases, the systems were initially heated to 310 K over 20 ps with the (α)-carbons of the protein restrained with a force constant of 1000 kJ/mol × nm2. These restraints were maintained for an additional 480 ps, at which time the restraints were removed and the trajectories were extended for a total of 50 ns. Simulations were carried out using a 2-fs timestep with water geometries constrained using SETTLE (32) and all other bonds constrained using LINCS (33). Temperature was held constant using a time constant (τt) of 0.1 ps for three groups: protein, lipid, and water/ions. Pressure was maintained at 1.0 atm using a time constant (τp) of 1.0 ps with anisotropic coupling. Short-range electrostatics and Lennard-Jones interactions were cut off at 1.0 nm and long-range electrostatics were calculated with particle-mesh Ewald (34), using a grid spacing of 0.10 nm and cubic interpolation. The force-field parameters from Berger et al. (35) for POPE lipids were used with GROMOS parameters for the oleoyl double bond, and GROMACS parameters were used for the protein, ions, and water. Fluctuations and lipid interaction energies were obtained as an average of the last 10 ns of the simulations, and protein-lipid interaction energies were calculated as described previously for MscL by Elmore and Dougherty (23). Analyses were performed using tools in the GROMACS suite, and figures were made using PyMol (36).
Strains and plasmids
The E. coli strain, MJF465 (MscS, MscL, MscK null), was used for osmotic downshock assays and expression experiments (2,37). Mutational cloning was conducted using the Top10F′ E. coli strain (Invitrogen, Carlsbad, CA). All mutants were created in the pB10b vector with a C-terminal six-His tag (4,37,38) under the control of the LacUV5 promoter.
Site-directed mutagenesis
Residues predicted to be involved in protein-lipid interactions as well as several controls were mutated to alanine using either Megaprimer mutagenesis (39) or QuikChange mutagenesis (Agilent, Santa Clara, CA). All mutations in MscS were made in the pB10B vector. For Megaprimer mutagenesis, forward and reverse primers located in the vector were used as the exterior primers and primers for mutagenesis were designed using Stratagene's QuikChange Primer design tool. Primer sequences are given in Table S1 in the Supporting Material. Mutations were verified by enzymatic digestion and sequences confirmed using automated sequencing (Big Dye v3.1; Applied Biosystems, Carlsbad, CA).
Osmotic downshock experiments
Downshock experiments were conducted as previously described in Booth et al. (40) and Caldwell et al. (41), with the following modifications. A single colony was used to inoculate an overnight culture in Luria Broth (LB Broth; BD Biosciences, San Jose, CA) supplemented with ampicillin (100 μg/mL), with the overnight culture subsequently used to inoculate (1:20) a culture in LB Broth with 250 mM NaCl and ampicillin. The resulting culture was grown to an OD600 of ∼0.5 and induced with 0.1 mM isopropyl-β-D-thiogalactopyranoside for 30 min. After 30 min of induction, the culture was diluted (1:40) into 1:1 LB Broth and deionized water or isotonically into LB Broth with 250 mM NaCl, and allowed to recover for 30 min in a shaking incubator. After hypoosmotic downshock or isotonic dilution, bacterial cultures were serially diluted and plated on LB plates supplemented with ampicillin (100 μg/mL). Plates containing between 25 and 250 colonies were used to determine the colony-forming units (CFU) per milliliter of media. Percent recovery was defined as the CFU of the downshocked culture divided by the CFU of the isotonic dilution. Six trials for each mutation were conducted.
Bacterial expression analysis
To verify bacterial expression of MscS mutants, cultures were grown as described above and pelleted for 15 min at 16,000×g after 30 min of induction. The supernatant was removed and the pellets resuspended in an equal volume of a buffer containing 50 mM Tris, 75 mM NaCl, 0.1% Fos-Choline-14 (Anatrace, Maumee, OH), and protease complete inhibitor (Roche, Basel, Switzerland). Samples were detergent-solubilized using a probe sonicator for three cycles of 15 s on and 45 s off. After lysis and solubilization, insoluble material was removed by pelleting for 30 min at 16,000×g. The supernatant was combined with sodium dodecyl sulfate polyacrylamide-gel electrophoresis (SDS-PAGE) loading dye, and the samples were boiled for 5 min. Samples were then run on a 12.5% poly-acrylamide gel using a TRIS-glycine running buffer. Equivalent volumes of the supernatant were loaded to allow for comparisons of protein expression levels. Proteins were transferred to a 0.2-μm nitrocellulose membrane using a semidry transfer system and Cashini's Buffer (0.1 M Tris, 0.02% SDS, 0.2 M glycine, 5% methanol). The protein expression of the C-terminally 6-His-tagged MscS mutants was probed using a primary mouse-anti-His monoclonal IgG (Covance, Emeryville, CA) and a secondary HRP-conjugated goat anti-mouse antibody (Jackson ImunoResearch Laboratories, West Grove, PA).
Results and Discussion
Computational analysis
MD simulations were conducted starting with the closed state model of MscS developed by Vásquez et al. (26) from distance constraints obtained by electron paramagnetic resonance. This particular model was selected because it was directly based on experimentally obtained constraints (26,42). However, this closed state model contains only Cα atoms for MscS, which does not allow it to be used directly for the identification of side chain-lipid bilayer interactions. To circumvent this problem, we built side chains into the model using the MODELLER suite (27,28) and allowed these side chains to equilibrate into a native closed state conformation using MD simulations with an explicit POPE lipid bilayer, water, and ions.
Two 50-ns simulations of MscS embedded in an explicit POPE membrane were performed (Fig. 1). POPE lipid was chosen for the membrane because of the prevalence of phosphatidylethanolamine lipids in bacterial membranes (43) and its use in previous simulations considering the lipid interactions of MscL (23). Both simulations gave similar results and showed structural equilibration within the first half of the trajectories (Fig. 2). As well, the residues in the lipid-exposed TM1 and TM2 domains were relatively stable during the final 10 ns of the trajectories, as shown by their low root mean-square (RMS) Cα fluctuations (Fig. 3).
Figure 2.
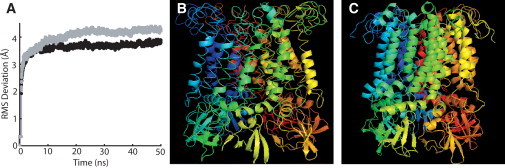
Structural deviations during the MD simulation. (A) RMS deviation of the MscS structure over the course of the MD simulation; Simulation 1 is shown in solid and Simulation 2 is shown in shaded representation. (B) Initial closed state all-atom MscS model generated with MODELLER. (C) MscS all-atom closed state model from Simulation 1 after 50 ns.
Figure 3.
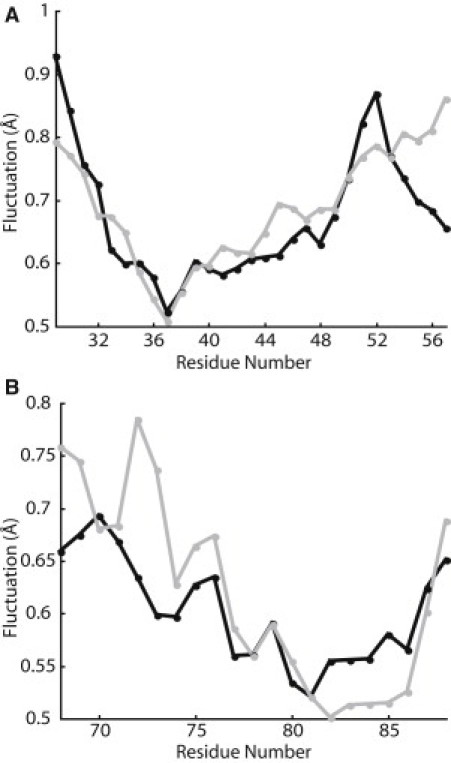
Average Cα RMS fluctuation per residue over the final 10 ns of the MD simulations for TM1 (A) and TM2 (B); Simulation 1 is shown in solid and Simulation 2 is shown in shaded representation.
The total protein-lipid interaction energy for each residue was calculated over the last 10 ns of each simulation (Fig. 4). A similar analysis previously conducted for MscL identified eight residues as having significant protein-lipid interactions (23). A comparison with random mutagenesis data showed that mutations to these residues were more likely to cause phenotypic changes in MscL than mutations to other residues, including residues with significant intersubunit interactions (23,24). Thus, in this study the protein-lipid interaction energy for TM1 and TM2 residues of MscS were analyzed to predict residues involved in tension sensation.
Figure 4.
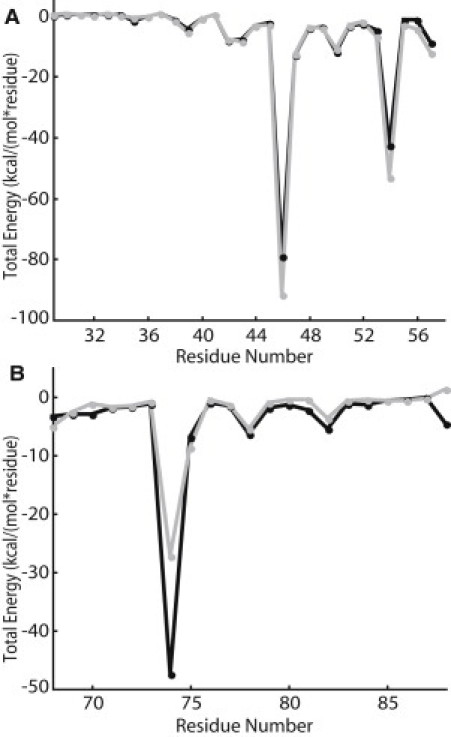
Protein-lipid interactions energies averaged over the final 10 ns of the MD simulations for TM1 (A) and TM2 (B); Simulation 1 is shown in solid and Simulation 2 is shown in shaded representation.
Based on our previous work with MscL, residues that are involved in tension sensation produce local minima in the protein-lipid interaction analysis (23). Local minima in this analysis correspond to strong-protein lipid interactions. These local minima are important in predicting tension sensation because channel opening involves a global structural rearrangement in which entire helices twist. Thus, these global structural rearrangements would require an entire lipid-exposed face of the protein to sense the tension on the lipid membrane. Based on our total protein-lipid interaction energy analysis (Fig. 4), we predicted residues in TM1 and TM2 to be involved in tension sensation. In TM1, residues that gave local minima in the analysis are L35, I39, L42, I43, R46, N50, and R54 (red residues in Fig. 1 a) and in TM2, residues that gave local minima are R74, I78, and L82 (blue residues in Fig. 1 a). As predicted, these residues lie on the outer face of the MscS channel in the closed state of the structure.
Functional analysis
To determine whether the predicted protein-lipid interacting residues were important for tension sensation, we individually mutated each of the residues identified by our protein-lipid interaction energy analysis (Fig. 4) to alanine. For hydrophobic residues, which could potentially interact with the lipid tails groups via the hydrophobic effect, mutation to alanine should greatly reduce the extent of interaction due to the small size of the alanine side chain. For charged residues, mutation to alanine both decreases the size of the residue and eliminates the possibility of electrostatic interaction (either hydrogen-bonding or ion pairs). Thus, alanine mutations were prepared by site-directed mutagenesis for each of the residues identified using our protein-lipid interactions energy analysis.
The function of the mutant channels was then tested using an osmotic downshock assay (Fig. 5). For each mutation six trials were conducted, and percent recovery values were compared using the t-test. A partial LOF phenotype was observed for L35A, I39A, L42A, I43A, N50A, I78A, and L82A, and R46A, R54A, and R74A were phenotypically wild-type. Mutants were defined as having a partial LOF phenotype if they were statistically different from wild-type MscS at >95%, as determined using the Student's t-test. In Fig. 5 a, phenotypically partial LOF mutants are light-shaded and phenotypically wild-type mutants are dark-shaded. Additionally, all mutations were confirmed not to be gain-of-function by measuring their stationary phase optical densities (see Table S2). The partial LOF phenotypic differences were not due to decreased channel expression levels, because all mutants and controls were expressed at levels comparable to wild-type MscS (Fig. 5 b). Thus, an observed partial LOF phenotype corresponds to the inability of a site-directed MscS mutant to gate in response to tension with a comparable gating threshold to wild-type MscS.
Figure 5.
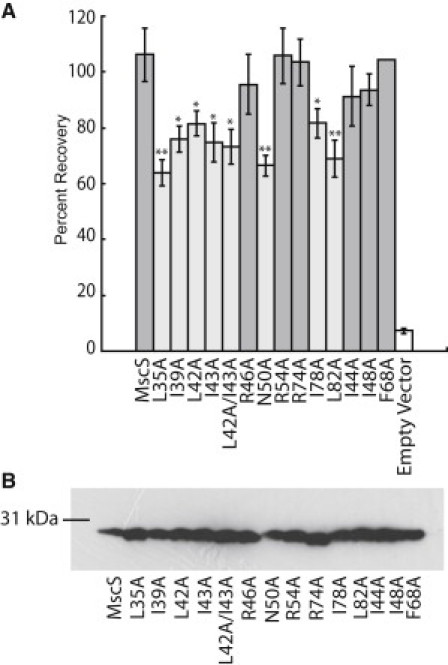
Functional analysis of MscS alanine mutations. (A) Osmotic downshock assay results of wild-type MscS and alanine mutants. Error bars represent the standard error of the mean for six independent trials. Statistically partial loss of function mutations (light shaded bar) were determined by comparison with wild-type MscS using a Student's t-test (∗∗ = p < 0.01 and ∗ = p < 0.05). (B) Western blot analysis of protein expression levels for wild-type MscS and alanine mutants under expression conditions identical to those used for the osmotic downshock assay.
Similar trends are observed in the MscS and MscL residues that are necessary for tension sensing. In this study of MscS, six of the seven alanine mutations that produced a partial LOF phenotype were very hydrophobic residues, and the three charged residues with significant protein-lipid interactions in MD simulations were insensitive to mutation (R46, R54, and R74). This trend is similar to that observed in MscL (44). In the analogous series of studies on MscL comparing MD simulations with random mutagenesis data, we also identified several LOF mutations upon substitutions of hydrophobic amino acids (23,24). Moreover, analyses of the effects of lipid composition on MscL have implied that specific electrostatic interactions, such as hydrogen-bonding, with lipid headgroups is generally less important for MscL mechanosensation than hydrophobic interactions with lipid tail groups (45). As well, almost all mutations that led to a significant reduction in channel function for MscS were clustered near the periplasmic interface of the channel as observed for MscL (23). This suggests that the tension sensor for both channels is located in hydrophobic lipid-interacting residues at the periplasmic interface. These regions of TM1 and TM2 also had lower RMS fluctuations than the cytoplasmic sides of the helices (Fig. 3), implying that these important lipid interactions lead to increased structural stability in this region of the channel.
To determine whether the effects observed for mutation of hydrophobic residues were specific to lipid-interacting residues, we also studied three additional MscS alanine mutations (I44A, I48A, and F68A) (Fig. 5). For these control mutations, we selected residues I44 and I48 in TM1 and F68 in TM2 (yellow residues in Fig. 1 a). All of these residues are very hydrophobic, akin to many of the residues that were identified as important lipid-interacting residues, but these residues are not predicted by our computational analysis of lipid interactions to be important in tension sensation (Fig. 4). None of these mutations led to a significant effect on channel function, as they were phenotypically wild-type (Fig. 5 a). These observations provide further evidence that lipid-interacting residues are particularly central to tension sensation in MscS and that, in order to interact with the lipids, the amino-acid side chains must be on the exterior side of the transmembrane helix. The nontension-sensing hydrophobic residues in the transmembrane helices are rotated slightly outward in the open and desensitized crystal structures, and therefore likely play a role in stabilizing these states through hydrophobic interactions with the lipid tail groups. This is reflected in the physiological differences observed for I48N and F68N by Nomura et al. (12).
One pair of lipid-interacting residues, L42 and I43, is particularly interesting because they are adjacent in the structure. Alanine mutations at both sites are statistically different from MscS (Fig. 5 a). A double-mutant of L42A/I43A was created to determine whether the removal of two adjacent lipid-interacting residues would create a unique phenotypic mutant with different properties from the single mutation. Interestingly, the double-mutation showed behavior similar to that of the two single mutations, implying that a disruption of one of these interactions is sufficient to disrupt channel function.
Role of lipid-interacting residues in tension sensation
The data presented above demonstrates that a series of hydrophobic residues lying along the outer face of MscS interact with the lipid tail groups of the bilayer in the closed state and are essential for effective tension sensation. Interestingly, these residues do not maintain their full lipid interactions throughout the channel's gating transition (Fig. 6). In the closed state model of the channel and in our simulations, these residues face directly toward the surrounding lipid membrane. However, in the open state of the channel, as observed in the crystal structure of A106V MscS from Wang et al. (46), these residues have turned such that they no longer face the membrane directly. The rotation of these residues during the transition from the closed state to the open state is likely driven by these residues finding new intraprotein hydrophobic contacts, as tension on the bilayer pulls the membrane away from these residues, breaking up their hydrophobic interactions with the lipid tail groups. Intriguingly, these residues also do not directly face the lipid in the MscS structure from Bass et al. (18) that has been interpreted as the desensitized state of the channel (18,47). The fact that these residues have lost their direct lipid contact in this structure likely leads to its insensitivity to gating by tension. Moreover, these observations also show that future studies attempting to identify the tension sensor of a mechanosensitive channel should perform the analysis of protein-lipid interactions on the closed state of the channel.
Figure 6.
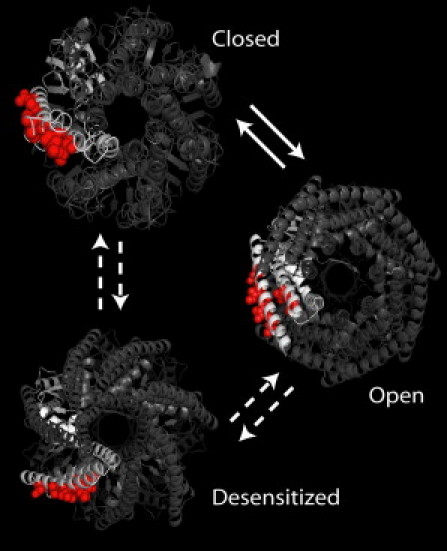
Positions of the tension-sensing residues are shown in the closed (50-ns simulation), open (2VV5 (46)), and desensitized (2OAU (18,47)) states of MscS.
Conclusions
Here, we have identified the critical residues that compose the region of MscS involved with directly sensing membrane tension through lipid interactions. This tension sensor is composed of lipid-facing hydrophobic resides within the first and second transmembrane domains of MscS. In the closed state of the channel, these residues interact with the lipid tail groups. Upon transition from the closed state to the open state of the channel, these residues turn inward to form intraprotein hydrophobic interactions. Finally, when MscS undergoes desensitization, the pore of the channel becomes occluded while these residues remain in intraprotein hydrophobic interactions.
Acknowledgments
The authors acknowledge Paul Blount (University of Texas Southwestern) for providing the MscS-6XHis in pB10b clone and Ian Booth (University of Aberdeen) for providing the MJF465 bacterial strain.
This work was supported by Washington University (to J.A.M.) and by a Wellesley Educational Research and Development Award (to D.E.E.).
Supporting Material
References
- 1.Li C., Edwards M.D., Booth I.R. Identification of mutations that alter the gating of the Escherichia coli mechanosensitive channel protein, MscK. Mol. Microbiol. 2007;64:560–574. doi: 10.1111/j.1365-2958.2007.05672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levina N., Tötemeyer S., Booth I.R. Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: identification of genes required for MscS activity. EMBO J. 1999;18:1730–1737. doi: 10.1093/emboj/18.7.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schumann U., Edwards M.D., Booth I.R. YbdG in Escherichia coli is a threshold-setting mechanosensitive channel with MscM activity. Proc. Natl. Acad. Sci. USA. 2010;107:12664–12669. doi: 10.1073/pnas.1001405107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sukharev S.I., Blount P., Kung C. A large-conductance mechanosensitive channel in E. coli encoded by MscL alone. Nature. 1994;368:265–268. doi: 10.1038/368265a0. [DOI] [PubMed] [Google Scholar]
- 5.Hurst A.C., Petrov E., Martinac B. MscS, the bacterial mechanosensitive channel of small conductance. Int. J. Biochem. Cell Biol. 2008;40:581–585. doi: 10.1016/j.biocel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Kloda A., Petrov E., Martinac B. Mechanosensitive channel of large conductance. Int. J. Biochem. Cell Biol. 2008;40:164–169. doi: 10.1016/j.biocel.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Vásquez V., Cortes D.M., Perozo E. An optimized purification and reconstitution method for the MscS channel: strategies for spectroscopical analysis. Biochemistry. 2007;46:6766–6773. doi: 10.1021/bi700322k. [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen T., Edwards M.D., Booth I.R. Tryptophan in the pore of the mechanosensitive channel MscS: assessment of pore conformations by fluorescence spectroscopy. J. Biol. Chem. 2010;285:5377–5384. doi: 10.1074/jbc.M109.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koprowski P., Grajkowski W., Kubalski A. Genetic screen for potassium leaky small mechanosensitive channels (MscS) in Escherichia coli: recognition of cytoplasmic β domain as a new gating element. J. Biol. Chem. 2011;286:877–888. doi: 10.1074/jbc.M110.176131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sukharev S.I., Martinac B., Kung C. Two types of mechanosensitive channels in the Escherichia coli cell envelope: solubilization and functional reconstitution. Biophys. J. 1993;65:177–183. doi: 10.1016/S0006-3495(93)81044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akitake B., Anishkin A., Sukharev S. The “dashpot” mechanism of stretch-dependent gating in MscS. J. Gen. Physiol. 2005;125:143–154. doi: 10.1085/jgp.200409198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nomura T., Sokabe M., Yoshimura K. Lipid-protein interaction of the MscS mechanosensitive channel examined by scanning mutagenesis. Biophys. J. 2006;91:2874–2881. doi: 10.1529/biophysj.106.084541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vásquez V., Sotomayor M., Perozo E. A structural mechanism for MscS gating in lipid bilayers. Science. 2008;321:1210–1214. doi: 10.1126/science.1159674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anishkin A., Kamaraju K., Sukharev S. Mechanosensitive channel MscS in the open state: modeling of the transition, explicit simulations, and experimental measurements of conductance. J. Gen. Physiol. 2008;132:67–83. doi: 10.1085/jgp.200810000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Machiyama H., Tatsumi H., Sokabe M. Structural changes in the cytoplasmic domain of the mechanosensitive channel MscS during opening. Biophys. J. 2009;97:1048–1057. doi: 10.1016/j.bpj.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nomura T., Sokabe M., Yoshimura K. Interaction between the cytoplasmic and transmembrane domains of the mechanosensitive channel MscS. Biophys. J. 2008;94:1638–1645. doi: 10.1529/biophysj.107.114785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belyy V., Anishkin A., Sukharev S. The tension-transmitting ‘clutch’ in the mechanosensitive channel MscS. Nat. Struct. Mol. Biol. 2010;17:451–458. doi: 10.1038/nsmb.1775. [DOI] [PubMed] [Google Scholar]
- 18.Bass R.B., Strop P., Rees D.C. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science. 2002;298:1582–1587. doi: 10.1126/science.1077945. [DOI] [PubMed] [Google Scholar]
- 19.Martinac B. Structural plasticity in MS channels. Nat. Struct. Mol. Biol. 2005;12:104–105. doi: 10.1038/nsmb0205-104. [DOI] [PubMed] [Google Scholar]
- 20.Anishkin A., Sukharev S. Water dynamics and dewetting transitions in the small mechanosensitive channel MscS. Biophys. J. 2004;86:2883–2895. doi: 10.1016/S0006-3495(04)74340-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Battle A.R., Petrov E., Martinac B. Rapid and improved reconstitution of bacterial mechanosensitive ion channel proteins MscS and MscL into liposomes using a modified sucrose method. FEBS Lett. 2009;583:407–412. doi: 10.1016/j.febslet.2008.12.033. [DOI] [PubMed] [Google Scholar]
- 22.Belyy V., Kamaraju K., Sukharev S. Adaptive behavior of bacterial mechanosensitive channels is coupled to membrane mechanics. J. Gen. Physiol. 2010;135:641–652. doi: 10.1085/jgp.200910371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elmore D.E., Dougherty D.A. Investigating lipid composition effects on the mechanosensitive channel of large conductance (MscL) using molecular dynamics simulations. Biophys. J. 2003;85:1512–1524. doi: 10.1016/S0006-3495(03)74584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maurer J.A., Dougherty D.A. Generation and evaluation of a large mutational library from the Escherichia coli mechanosensitive channel of large conductance, MscL: implications for channel gating and evolutionary design. J. Biol. Chem. 2003;278:21076–21082. doi: 10.1074/jbc.M302892200. [DOI] [PubMed] [Google Scholar]
- 25.Maurer J.A., Dougherty D.A. A high-throughput screen for MscL channel activity and mutational phenotyping. Biochim. Biophys. Acta. 2001;1514:165–169. doi: 10.1016/s0005-2736(01)00390-x. [DOI] [PubMed] [Google Scholar]
- 26.Vásquez V., Sotomayor M., Perozo E. Three-dimensional architecture of membrane-embedded MscS in the closed conformation. J. Mol. Biol. 2008;378:55–70. doi: 10.1016/j.jmb.2007.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madhusudhan M.S., Marti-Renom M.A., Sali A. Comparative protein structure modeling. In: Walker J.M., editor. The Proteomics Protocols Handbook. Humana Press; Totowa, NJ: 2005. [Google Scholar]
- 28.Sali A., Blundell T.L. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 29.Lindahl E., Hess B., van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 2001;7:306–317. [Google Scholar]
- 30.Kandt C., Ash W.L., Tieleman D.P. Setting up and running molecular dynamics simulations of membrane proteins. Methods. 2007;41:475–488. doi: 10.1016/j.ymeth.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Rand R.P., Parsegian V.A. Hydration forces between phospholipid bilayers. Biochim. Biophys. Acta. 1989;988:351–376. [Google Scholar]
- 32.Miyamoto S., Kollman P.A. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 33.Hess B., Bekker H., Fraaiji J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 34.Darden T., York D., Pedersen L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 35.Berger O., Edholm O., Jähnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeLano W.L. DeLano Scientific; San Carlos, CA: 2002. The PyMOL Molecular Graphics System. [Google Scholar]
- 37.Wild J., Altman E., Gross C.A. DnaK and DnaJ heat shock proteins participate in protein export in Escherichia coli. Genes Dev. 1992;6:1165–1172. doi: 10.1101/gad.6.7.1165. [DOI] [PubMed] [Google Scholar]
- 38.Ou X., Blount P., Kung C. One face of a transmembrane helix is crucial in mechanosensitive channel gating. Proc. Natl. Acad. Sci. USA. 1998;95:11471–11475. doi: 10.1073/pnas.95.19.11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshimura K., Batiza A., Kung C. Hydrophilicity of a single residue within MscL correlates with increased channel mechanosensitivity. Biophys. J. 1999;77:1960–1972. doi: 10.1016/S0006-3495(99)77037-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Booth I.R., Edwards M.D., Miller S. Physiological analysis of bacterial mechanosensitive channels. Meth. Enzymol. 2007;428:47–61. doi: 10.1016/S0076-6879(07)28003-6. [DOI] [PubMed] [Google Scholar]
- 41.Caldwell D.B., Malcolm H.R., Maurer J.A. Identification and experimental verification of a novel family of bacterial cyclic nucleotide-gated (bCNG) ion channels. Biochim. Biophys. Acta. 2010;1798:1750–1756. doi: 10.1016/j.bbamem.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Anishkin A., Akitake B., Sukharev S. Characterization of the resting MscS: modeling and analysis of the closed bacterial mechanosensitive channel of small conductance. Biophys. J. 2008;94:1252–1266. doi: 10.1529/biophysj.107.110171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elmore D.E., Dougherty D.A. Molecular dynamics simulations of wild-type and mutant forms of the Mycobacterium tuberculosis MscL channel. Biophys. J. 2001;81:1345–1359. doi: 10.1016/S0006-3495(01)75791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshimura K., Sokabe M. Mechanosensitivity of ion channels based on protein-lipid interactions. J. R. Soc. Interface. 2010;7(Suppl 3):S307–S320. doi: 10.1098/rsif.2010.0095.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moe P., Blount P. Assessment of potential stimuli for mechano-dependent gating of MscL: effects of pressure, tension, and lipid headgroups. Biochemistry. 2005;44:12239–12244. doi: 10.1021/bi0509649. [DOI] [PubMed] [Google Scholar]
- 46.Wang W., Black S.S., Booth I.R. The structure of an open form of an E. coli mechanosensitive channel at 3.45 Å resolution. Science. 2008;321:1179–1183. doi: 10.1126/science.1159262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steinbacher S., Bass R., Rees D.C. Structures of the prokaryotic mechanosensitive channels MscL and MscS. Curr. Top. Membr. Mechanosens. Ion Chan. A. 2007;58:1–24. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


