Abstract
Microvesicles (exosomes) shed from both normal and cancerous cells may serve as means of intercellular communication. These microvesicles carry proteins, lipids and nucleic acids derived from the host cell. Their isolation and analysis from blood samples have the potential to provide information about state and progression of malignancy and should prove of great clinical importance as biomarkers for a variety of disease states. However, current protocols for isolation of microvesicles from blood require high-speed centrifugation and filtration, which are cumbersome and time consuming. In order to take full advantage of the potential of microvesicles as biomarkers for clinical applications, faster and simpler methods of isolation will be needed. In this paper, we present an easy and rapid microfluidic immunoaffinity method to isolate microvesicles from small volumes of both serum from blood samples and conditioned medium from cells in culture. RNA of high quality can be extracted from these microvesicles providing a source of information about the genetic status of tumors to serve as biomarkers for diagnosis and prognosis of cancer.
Introduction
Many cell types actively shed small microvesicles, also known as exosomes, into the extracellular space including blood and other body fluids such as urine, milk, saliva, amniotic fluid, and malignant effusions.1 These membrane vesicles range in size from 30 to 100 nm in diameter. Exosomes can arise by invagination of the limiting membrane of late endosomes, which leads to the formation of multivesicular bodies (MVBs) whose contents are released into the extracellular milieu upon fusion of the MVB with the plasma membrane; they can also bud directly from the plasma membrane.2,3 Hence, exosomes contain cellular components and expose the extracellular domain of receptors at their surface. Their molecular composition is influenced by the type and activation state of the cell of origin. In addition to a set of membrane and cytosolic molecules common among donor cell types, exosomes can harbor unique and selected subsets of proteins, mRNA and microRNAs (miRNAs) associated with specific cell type-associated functions and genomic state. Exosomes were first identified through their involvement in the elimination of excess proteins,4 but several recent findings indicate that exosomes constitute a potential mode of targeted intercellular transfer of molecules.5–7 Following interaction with the recipient cell by fusion, adhesion or direct binding, exosomes can confer different functions on the recipient cell. The physiologic functions of exosomes vary with the cell of origin and include modulation of immune status8,9 and stimulation of angiogenesis,10 with many functions as yet to be revealed. Exosomes are increasingly recognized to play a role in inflammation,11 cancer,12 thrombosis,13 cell polarization,6 development14 and neurological disorders.15
Several advantages of the exosomal-acellular mode of communication should aid in the development of diagnostic and therapeutic strategies. Microvesicles contain sorted sets of molecules involved in many different cellular processes, have the capacity to transmit signaling and genetic information, and can be obtained non-invasively from body fluids. For example, some tumor cells are very active at shedding exosomes into the circulating vasculature.16 These tumor exosomes and their constituent RNAs present unique genetic information about the tumor concerning its presence, cellular type, state of malignancy and susceptibility to therapeutic treatment. Isolation of RNA from serum exosomes can yield up to 60 times greater concentrations of high integrity RNA as compared to that extracted directly from blood, serum or plasma (unpublished). Therefore exosomal RNA analysis provides a marked increase in the diagnostic sensitivity of transcriptome analysis and a powerful tool to identify potential disease biomarkers.
Although several methods have been developed to purify exosomes, none of these can clearly discriminate between exosomes and other shed membranes, lipid structures, or retrovirus particles, also found in bodily fluids, which are similar in terms of size and density.17 A common procedure to purify exosomes from cell culture supernatants or body fluids involves a series of centrifugations and filtration to remove dead cells, large debris and other cellular contaminants resulting from cell lysis, followed by a final high-speed ultracentrifugation to pellet small membrane vesicles.18 A typical flow chart is shown in Fig. 1c. This technique, however, is lengthy (4–5 h), requires an ultracentrifuge and yields a relatively low recovery of exosomes, ranging from 5–25% of the starting exosome MHC class II concentration,19 making it difficult for application in clinical practice. Contaminating material, such as protein aggregates, apoptotic vesicles or nucleosomal fragments that are released by apoptotic cells, can be separated from exosomes by flotation on continuous sucrose gradient.18 Exosomes ‘float’ to a density close to 1.13 g ml−1, but this may vary among different cells of origin depending on the content of the microvesicles. Recently, an alternative method of exosome isolation based on ultrafiltration and centrifugation in sucrose-deuterium oxide (D2O) cushions has been developed.19 This method is capable of preparing clinical-grade purified exosomes; however, it still takes 4–6 h to perform, and has modest exosome recovery rate of 36–65% based on the starting exosome MHC class II concentration.19 Microvesicles of tumor origin can also be isolated utilizing adherence to magnetic beads coated with antibodies against tumor-associated markers, such as epithelial cell adhesion molecule (EpCAM).20 However, the purification takes greater than 3 h and an ultracentrifugation step is required to recover exosomes diluted in the elution from magnetic beads. Hence, a rapid and reproducible purification method for exosome samples is essential to exploit them as a new diagnostic and therapeutic tool, as well as to carry out basic research on exosome functions.
Fig. 1.
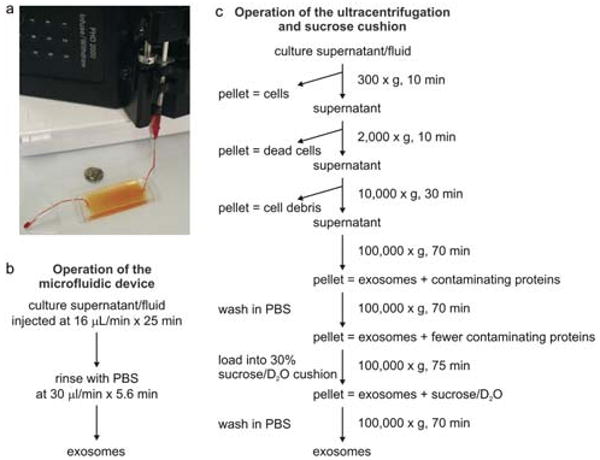
Experimental setup of microfluidic devices and ultracentrifugation procedures. (a) Image of a device with a syringe pump. (b) The operation procedure of the microfluidic device for isolation of microvesicles. (c) Flow chart for a typical microvesicle purification procedure based on differential ultracentrifugation and sucrose cushion (adapted from ref. 18).
Microfluidics and miniaturized lab-on-a-chip-type devices are attractive for medical diagnostics and blood analysis. The small dimensions of microfluidic devices, small sample sizes, and minimal amounts of reagents needed allow for faster reaction times, increased sensitivity, and reduced procedural costs. In this paper, we present the development of a microfluidic device that rapidly and specifically isolates exosomes from human sera or cell culture supernatants without requiring ultracentrifugation or resolution on sucrose gradients. Operation of the device is based on the selective binding of exosomes to antibody-coated surfaces. We demonstrate the purification and extraction of exosomal RNA within an hour from 100–400 μL serum samples, supporting the potential of this method as a rapid, point-of-care tool for the diagnosis of cancer.
Experimental
Materials
3-Mercaptopropyl trimethoxysilane was purchased from Gelest (Morrisville, PA). Ethanol (200 proof) was obtained from Fisher Scientific (Fair Lawn, NJ). For chamber fabrication, SU-8 photoresist and developer were obtained from MicroChem (Newton, MA); polydimethylsiloxane (PDMS) and curing agent were obtained from Dow Corning (Midland, MI). Phosphate buffered saline (PBS) was obtained from Mediatech (Herndon, VA). Lyophilized bovine serum albumin (BSA) was obtained from Aldrich Chemical Co. (Milwaukee, WI). The coupling agent GMBS (N-γ-maleimidobutyryloxy succinimide ester) and NeutrAvidin were obtained from Pierce Biotechnology (Rockford, IL). Biotinylated mouse anti-human anti-CD63 (clone AHN16.1/46-4-5) and biotinylated normal mouse control IgG were obtained from Ancell (Bayport, MN).
Collection of serum and tumor samples
Blood samples from healthy and glioblastoma-confirmed subjects were obtained through the Cancer Center Amsterdam, VU University Medical Center, Amsterdam, Netherlands under IRB approved protocols. Samples of 10 mL of peripheral blood were collected by venipuncture in serum separation tubes (BD Biosciences, Franklin Lakes, NJ) at the time of surgery and processed according to the manufacturer's protocol (<2 h from time of collection until freezing of serum), followed by passage through a 0.8 μm filter. These samples were kept at −80 °C until use. For primary cell culture, brain tumor specimens from patients diagnosed by a neuropathologist as glioblastoma multiforme (GBM) were taken directly from surgery and placed in cold sterile Neurobasal media (Invitrogen, Carlsbad, CA). The specimens were dissociated into single cells within 1 h from the time of surgery using a neural Tissue Dissociation Kit (Miltenyi Biotech, Bergisch Gladbach, Germany) and plated in DMEM supplemented with 5% microvesicle-depleted FBS (dFBS, prepared by ultracentrifugation at 110 000 × g for 16 h to remove bovine microvesicles), and penicillin–streptomycin (10 IU mL−1 and 10 mg mL−1, respectively, Sigma-Aldrich, St Louis, MO).10
Isolation of microvesicles by differential centrifugation from conditioned medium
Primary glioblastoma cells at passages 1–15 were cultured in DMEM supplemented with 5% dFBS. The conditioned medium was removed after 48 h of cell growth. The microvesicles in the conditioned medium were purified by a series of centrifugations at 4 °C First, the medium was centrifuged at 300 × g for 10 min to eliminate any cell contamination, followed by a second centrifugation of 16 500 × g for 20 min, and passage through a 0.22 μm filter. Microvesicles were then pelleted by ultracentrifugation at 110 000 × g for 70 min. The microvesicle pellets were washed in 13 mL of PBS, re-pelleted, and re-suspended in PBS.
Microfluidic channel design and fabrication
The microfluidic device used in this study was a straight flow channel of 19 mm width, 20 μm depth and 4.5 cm length with herringbone groves on its ceiling that were 50 μm wide, 10 μm deep for processing samples of volumes of 400 μL. Another device of dimensions 5 cm (L) × 4 mm (W) × 30 μm (H) was used for processing samples of volumes smaller than 100 μL and SEM imaging. The herringbone or slanted grooves in microchannels have been previously utilized for mixing of fluids/particles,21 separation and manipulation of particles.22,23 We have found these microstructures could be exploited to increase the capture efficiency (unpublished data), presumably due to the increased surface area and enhanced contact of particles with surfaces of the microchannel. The devices were fabricated in PDMS and bonded permanently to plain PDMS slabs after being treated with oxygen plasma, as described.24
Surface modification
Freshly fabricated devices were modified using the method described.25,26 Briefly, the microchannels were pretreated with 4% (v/v) solution of 3-mercaptopropyl trimethoxysilane in ethanol for 30 min at room temperature, followed by an incubation of 0.01 μmol mL−1 GMBS in ethanol for 15 min at room temperature. Afterwards, the microchannel was incubated with 10 μg mL−1 NeutrAvidin solution in PBS for 1 h at 4 °C. Finally, 10 μg mL−1 biotinylated anti-CD63, control IgG, or anti-CD4 solution in PBS containing 1% (w/v) BSA and 0.09% (w/v) sodium azide was injected to react with NeutrAvidin at room temperature for 15 min. After each step, the surfaces were rinsed with 8 device volumes of either ethanol or PBS, depending on the solvent used in the previous step, to flush away unreacted molecules.
Microfluidic isolation of microvesicles
An image of the experimental setup is shown in Fig. 1a. The operating procedure for microfluidic isolation of microvesicles is summarized in Fig. 1b. 10–400 μL serum samples were flowed into microchannels at optimized flow rates (16 μL min−1 and 4 μL min−1 for devices of a smaller footprint) based on the dimensions of the microchannels and the estimated diffusivity of microvesicles.27 Samples used in this study include serum from healthy donors or GBM patients, and microvesicle preparations obtained from differential centrifugations of the glioblastoma cell culture supernatant. After rinsing with PBS containing 1% BSA and 1 mM EDTA (168 μL at a flow rate of 30 μL min−1 and 50 μL at 10 μL min−1 for devices of a smaller footprint), microvesicles adherent to the surface were fixed for scanning electromicrograph (SEM) or lysed for RNA extraction.
Scanning electromicrographs
Microvesicles captured on PDMS microchannels were fixed in 0.5 × Karnovsky's fixative and then washed for 2 × 5 min with PBS. The samples were dehydrated in 35% ethanol for 10 min, 50% ethanol for 2 × 10 min, 70% ethanol for 2 × 10 min, 95% ethanol for 2 × 10 min, 100% ethanol for 4 × 10 min. The samples were then coated with palladium/gold in a GATAN Model 681 High Resolution Ion Beam Coater, and examined using Jeol 5600LV scanning electron microscope (Jeol Ltd., Tokyo, Japan) at the Whitehead Institute (Cambridge, MA).
Image analysis
The size distributions of captured microvesicles in SEM images were analyzed using an image processing program (ImageJ, National Institutes of Health). Briefly, images were converted to binary images by manually setting light intensity thresholds after subtracting the background. Projected area diameters were calculated from the projected area assuming circular geometry. Feret's diameters were determined as the greatest distance possible between any two points along the image boundary of individual microvesicles.
RNA isolation
Total RNA was purified using the mirVana RNA isolation Kit (Ambion, Austin, TX) according to the manufacturer's protocol. In brief, 300 μL of lysis/binding buffer followed by an equal amount of air was passed through the microchannel at a flow rate of 25 μL min−1 to lyse microvesicles captured on the chip. The lysate was collected in a collection tube via 2 inches of Teflon tubing (Small Parts, Inc., Miramar, FL). 30 μL homogenate provided in the isolation kit was added to the lysate. Total RNA was extracted by phenol–chloroform separation and precipitated with 100% ethanol and collected in the elution solution. RNA was further purified and concentrated using Qiagen MinElute RNA cleanup kit (Qiagne Inc., Valencia, CA) according to the manufacture's protocol. The quality of RNA was examined on a Eukaryote Total RNA Pico chip with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
RT-PCR and nested PCR
Total RNA purified was then converted into cDNA using Sensiscript RT kit (Qiagen Inc, Valencia, CA) with a mix of oligo dT and random nonamers according to the manufacture's protocol. The mRNA for GAPDH (glyceraldehyde 3-phosphate dehydrogenase) was selected as a control because it is usually stable and constitutively expressed at high levels in most tissues, cells and microvesicles.10 IDH-1 (isocitrate dehydrogenase 1) mRNA was analyzed as being of particular interest and as 12% GBM patients have a specific point mutation in this gene, which favors a better prognosis.28,29 The sequences of primers used in the PCR were: GAPDH primers—forward 5′-CAG CCT CAA GAT CAT CAG CA-3′, reverse 5′-TGT GGT CAT GAG TCC TTC CA-3′; IDH-1 primers—forward 5′-CGG TCT TCA GAG AAG CCA TT-3′, reverse 5′-TAT TGA TCC CCA TAA GCA TGA T-3′. PCR protocol: 95 °C 3 min; 95 °C 20 s, 59 °C 20 s, 72 °C 30 s × 35 cycles; 72 °C 7 min.
Results
Development of microfluidic devices for immunoaffinity-based isolation of microvesicles from cell-free supernatants or sera
One of the most abundant protein families that are found in exosomes comprises the tetraspanins and several members of this family, including CD9, CD63, CD81 and CD82, are highly enriched in exosomes from virtually any cell type.2 Anti-human CD63 IgG was chosen for isolating microvesicles from all cell origins because of its high expression levels. However, CD63 antigens are also expressed on the surface membrane of platelets, granulocytes and monocytes at low levels.30,31 We first examined the serum preparation by flowing serum samples into anti-CD63 IgG-coated microchannels and found there were few, if any, platelets and nucleated cells detected (see ESI†: SI_No_Cellular_Contamination_in_Serum_samples). Next, we confirmed microfluidic capture of microvesicles by injecting 10 μL PKH67-labeled microvesicle preparations obtained from differential centrifugations of the glioblastoma cell culture supernatant into microchannels coated with anti-CD63 antibodies or anti-CD4 antibodies as negative controls (see ESI†: SI_Microfluidic_Capture_of_PKH67-labled_Microvesicles). As shown in Fig. S2 (ESI†), fluorescence intensity inside the microchannel was higher in anti-CD63 IgG-coated microchannels, as compared to anti-CD4-coated channels and increased in the case of anti-CD63 with increasing concentration of microvesicles.
SEM images showed that surfaces of anti-CD63 antibodies coated microchannels were covered by microvesicles separated from 10 μL of microvesicle-containing medium obtained from primary glioblastoma cell cultures (Fig. 2a) or serum from a GBM patient (Fig. 2b–d). The size distributions of microvesicles captured from these two samples (shown in Fig. 2e and f, respectively) were well within the range reported in the literature. Only 3% of the projected area diameters of microvesicles from serum were greater than 100 nm (Fig. 2c and f). However, more than 13% of microvesicles from medium had a projected area diameter greater than 100 nm (Fig. 2a and e).
Fig. 2.
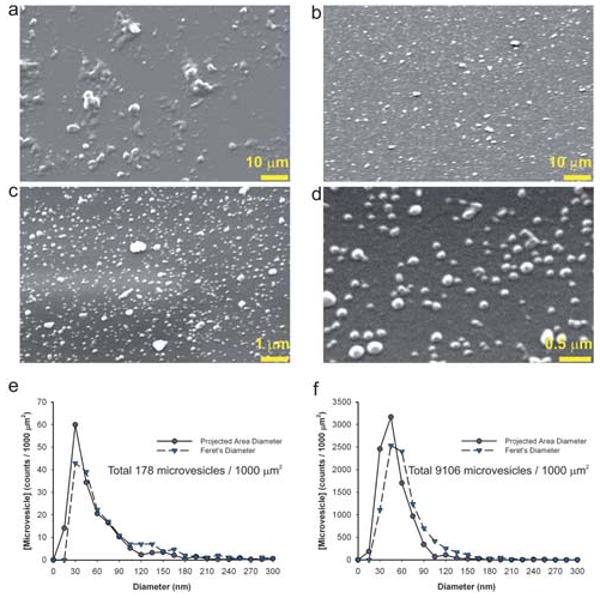
Scanning EM images of microvesicles captured in microchannels. (a) Scanning EM image showing the microvesicles prepared from classical sequential centrifugations of cell culture supernatant bound to the microchannel surface. (b) Scanning EM image showing the microvesicles bound to the microchannel surface after 10 μL of GBM patient serum was passed through the microchannel. Higher magnification images of (b) are shown in (c) and (d). The distributions of both the projected area diameters and the Feret's diameters of microvesicles in images (a) and (c) are shown in (e) and (f), respectively. Projected area diameters were calculated from the projected area assuming circular geometry while Feret's diameters were determined as the greatest distance possible between any two points along the boundary of a region of interest.
Extraction of RNA from captured microvesicles
RNAs from microvesicles from serum captured on microchannels as well as from the corresponding serum samples were extracted and analyzed with a Bioanalyzer, showing that the microvesicles contained a broad range of RNA sizes consistent with a variety of mRNAs and miRNAs (Fig. 3), but lacked the ribosomal peaks characteristic of cellular RNA. There appeared to be fewer microvesicles and less RNA in the normal serum compared to the GBM patient serum as indicated by RNA concentrations measured with a Bioanalyzer (Fig. 3 and Table 1). In addition, more RNAs were extracted from lysates from anti-CD63-coated chips than from both GBM patient/normal control sera directly.
Fig. 3.
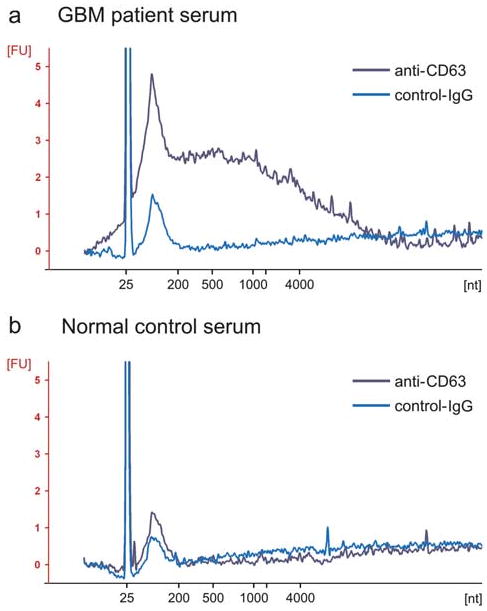
Microfluidically captured microvesicles contain RNA. Bioanalyzer data showing the intensities and size distributions of fluorescently labeled total RNA extracted from microvesicles captured on anti-CD63 or IgG antibody-coated microchannels injected with 100 μL of GMB patient serum (a) and 400 μL normal control serum (b), respectively. The lowest migrating peak at 25 nt represents an internal standard. The x-axis indicates the length of the RNA in nucleotides (nt) while the y-axis indicates fluorescence intensity in arbitrary units. Note that the two ribosomal RNA peaks (18S and 28S) from cellular RNA profile were absent in both (a) and (b).
Table 1.
Total RNA extracted from 400 μL of serum samples. Summary of total RNA extracted from captured microvesicles from 400 μL GBM and normal control serum samples on microchannels coated with either anti-human CD63 or negative control IgG. RNA concentrations were analyzed with a Bioanalyzer
| Serum | Antibody-coated on-chip | Average RNA extracted/ng per 400 μL serum |
|---|---|---|
| Glioblastoma multiforme patient (n = 4) | Anti-human CD63 | 30.88 |
| Negative control IgG | 4.04 | |
| Normal control (n = 4) | Anti-human CD63 | 4.14 |
| Negative control IgG | 1.96 |
RT-PCR analysis showed that the mRNAs for GAPDH (106 bp product) and IDH-1 (100 bp product) were present in the microvesicles captured from both GBM patient and normal control sera on anti-CD63-coated microchannels (Fig. 4, lane 1 and 3), but little were present in lysates from control IgG-coated microchannels (Fig. 4, lane 2 and 4).
Fig. 4.
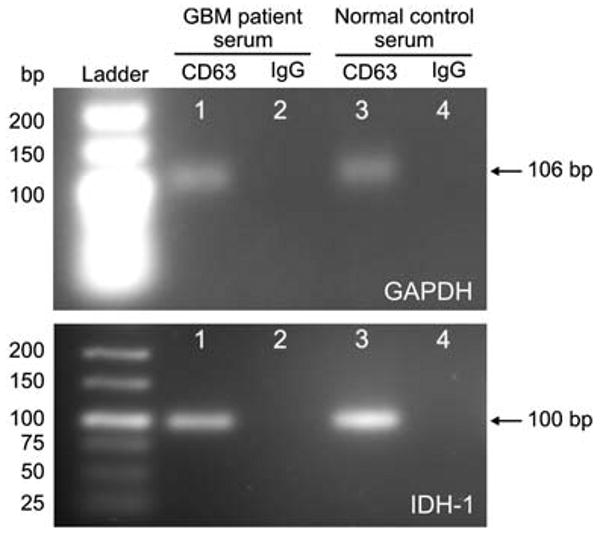
RT-PCR performed on RNA extracted from microfluidically isolated microvesicles. Agarose gels showing RT-PCR products performed on RNA extracted from microvesicles captured from both GBM patient and normal control sera on anti-CD63 or control IgG-coated microchannels. The RT-PCR product of GAPDH mRNA appears as a band at 106 bp, while the nested RT-PCR product of wild-type IDH-1 mRNA appears as a band at 100 bp.
Discussion
Microvesicles shed from cells provide a means whereby cells can communicate with cells nearby or at a distance and appear to play important roles in immunology and tumor development. In this study we developed a microfluidic apparatus that is capable of extracting antigen-specific microvesicles from biologically complex samples, such as serum and conditioned medium from cultured cells. Compared to the current standard protocols for isolating microvesicles, this microfluidic approach is faster, cheaper, requires smaller volumes of microvesicle-containing fluid and fewer reagents, can potentially isolate microvesicles of specific cell origin and is compatible with clinical laboratory procedures. In addition, most microvesicles isolated microfluidically maintained their native morphology (Fig. 2b–d). In contrast, a larger fraction of microvesicles isolated by differential centrifugation of the conditioned medium appeared aggregated (Fig. 2a). It is also likely that some microvesicles fuse to each other during the ultracentrifugation procedure, thus accounting for a larger diameter. On the other hand, for the microfluidic isolation, only one short, low-speed centrifugation was applied to the serum sample.
Other groups have analyzed RNA isolated directly from serum; however, we believe the microvesicle isolation method has a strong advantage over this direct approach for several reasons: (1) we can specifically pull out microvesicles shed by tumors using tumor-specific cell surface markers, for example, mutant/variant cell surface protein like EGFRvIII32 and other tumor-enriched proteins;33 we could, of course, also isolate organ-specific microvesicles when organ-specific surface markers are present; (2) the half-life of free RNA in serum is seconds to minutes,34 whereas RNA is protected from RNases in microvesicles;10 thus RNA extracted directly from serum includes a larger fraction of degraded RNA and more RNA from dead normal cells, as compared to living tumor cells; and (3) extraction of microvesicles from serum is scalable. It is more difficult to extract high quality RNA from large volumes of complex liquids like plasma and serum, since there are many components, e.g. lipids, proteins, etc., that are co-isolated with the RNA from serum and can inhibit PCR reactions.35–38 With the microvesicle microfluidic procedure, we can easily extract high quality RNA from small volumes of serum to increase the sensitivity of tumor detection and mutational/quantitative evaluation of tumor-derived RNAs. It has been found that a subset of GBM patients have a point mutation in the IDH-1 mRNA.28 As shown in Fig. 4, RT-PCR products of IDH-1 mRNA were present in the microvesicles captured from both GBM patient and healthy volunteer sera on anti-CD63-coated microchannels, demonstrating the quality and quantity of RNA were sufficient for mutation-specific PCR or sequence analysis to potentially identify the point mutation of the IDH-1 transcript. The presence of RNA in the effluent was most likely related to the fact that not all the target microvesicles in the sample were captured, and a fraction of them were lost into the effluent. The percentage recovery of microvesicles ranged from 42–94% (n = 4) based on the total RNA amounts extracted on chip and from the effluent. We are currently investigating approaches to improve the capture yield of microvesicle-chip by increasing microvesicle–surface interactions within the chip, such as modifying the dimensions of the microchannel, incorporating structures inside the microchannel,39–41 augmenting transverse flow,21 and increasing the coverage of active antibodies on the surface.42
In addition, the microfluidic isolation of microvesicles is unique in that it sorts microvesicles directly from serum in a single step. This contrasts with magnetic-bead-based systems20 that require multiple open-system preparation steps (incubation, washing, centrifugation and ultracentrifugation), which can result in fusion and loss of a significant proportion of microvesicles, and are difficult to validate for clinical manufacturing. Owing to its simplicity, the microvesicle-chip is readily adaptable for potential use in point-of-care and clinical settings. Based on similar immunoaffinity techniques, we have previously developed different microfluidic devices for isolation of CD4 + T cells,24 lymphocytes,43 circulating tumor cells (CTCs),39,44 and neutrophils45 directly from whole blood. The information gained from these captured living cells can also be critical to the diagnosis and treatment of disease. We were, for example, able to detect genetic markers for lung cancer from isolated CTCs and utilize this information to identify the best treatment for particular patients and monitor response to therapy.39 In comparison, microvesicles can be readily purified without any cellular contamination and are continuously shed by tumor cells into the circulation while the accumulation of microvesicles of non-tumor cell origins is rarely observed,46 which facilitates the genetic and proteomic analysis of tumor-derived microvesicles. Although microvesicles do not mirror the exact transcriptome of the tumor cells of origin, rather a subset composition of these cells, tumor-derived microvesicles can have enhanced expression of tumor antigens and unique genetic information,10,47 which present a distinctive opportunity for early diagnosis and monitoring of cancer.
Conclusion
Exosomes shed from both normal and cancerous cells provide a means of intercellular communication allowing for exchange of charged molecules, including RNA and non-secreted proteins between cells. In this paper, we present an easy and rapid microfluidic immunoaffinity method to isolate exosomes from both serum and cell culture medium. High quality RNA can be extracted in a single wash-through procedure with RT-PCR of mRNA and miRNA providing biomarkers for mutational status and expression profiles for diagnosis and prognosis of tumors.
Supplementary Material
Acknowledgments
We thank Octavio Hurtado for valuable assistance with microfabrication. This work was supported in part by the NIBIB P41 EB002503 (MT), NCI CA86355 (XOB), NCI CA69246 (XOB), Wenner-Gren Foundation (JS) and Stiftelsen Olle Engkvist Byggmästare (JS).
Footnotes
Electronic supplementary information (ESI) available: The supplementary materials show no cellular contamination in the serum sample and capture of fluorescently labeled microvesicles on the chip.
References
- 1.Andre F, Schartz NEC, Movassagh M, Flament C, Pautier P, Morice P, Pomel C, Lhomme C, Escudier B, Le Chevalier T, Tursz T, Amigorena S, Raposo G, Angevin E, Zitvogel L. Lancet. 2002;360:295–305. doi: 10.1016/S0140-6736(02)09552-1. [DOI] [PubMed] [Google Scholar]
- 2.Thery C, Zitvogel L, Amigorena S. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 3.Booth AM, Fang Y, Fallon JK, Yang JM, Hildreth JEK, Gould SJ. J Cell Biol. 2006;172:923–935. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan BT, Johnstone RM. Cell. 1983;33:967–977. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 5.Fevrier B, Raposo G. Curr Opin Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Lakkaraju A, Rodriguez-Boulan E. Trends Cell Biol. 2008;18:199–209. doi: 10.1016/j.tcb.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Nat Cell Biol. 2007;9:654–U672. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 8.Andre F, Chaput N, Schartz NEC, Flament C, Aubert N, Bernard J, Lemonnier F, Raposo G, Escudier B, Hsu DH, Tursz T, Amigorena S, Angevin E, Zitvogel L. J Immunol. 2004;172:2126–2136. doi: 10.4049/jimmunol.172.4.2126. [DOI] [PubMed] [Google Scholar]
- 9.Taylor DD, Akyol S, Gercel-Taylor C. J Immunol. 2006;176:1534–1542. doi: 10.4049/jimmunol.176.3.1534. [DOI] [PubMed] [Google Scholar]
- 10.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Carter BS, Krichevsky AM, Breakefield XO. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SH, Lechman ER, Bianco N, Menon R, Keravala A, Nash J, Mi ZB, Watkins SC, Gambotto A, Robbins PD. J Immunol. 2005;174:6440–6448. doi: 10.4049/jimmunol.174.10.6440. [DOI] [PubMed] [Google Scholar]
- 12.Hegmans J, Bard MPL, Hemmes A, Luider TM, Kleijmeer MJ, Prins JB, Zitvogel L, Burgers SA, Hoogsteden HC, Lambrecht BN. Am J Pathol. 2004;164:1807–1815. doi: 10.1016/S0002-9440(10)63739-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zwaal RFA, Comfurius P, Bevers EM. Biochim Biophys Acta. 1992;1180:1–8. doi: 10.1016/0925-4439(92)90019-j. [DOI] [PubMed] [Google Scholar]
- 14.Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. Traffic. Vol. 3. Oxford; U K: 2002. pp. 321–330. [DOI] [PubMed] [Google Scholar]
- 15.Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, Laude H, Raposo G. Proc Natl Acad Sci U S A. 2004;101:9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor DD, Gercel-Taylor C. Br J Cancer. 2005;92:305–311. doi: 10.1038/sj.bjc.6602316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantin R, Diou J, Belanger D, Tremblay AM, Gilbert C. J Immunol Methods. 2008;338:21–30. doi: 10.1016/j.jim.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Thery C, Clayton A, Amigorena S, Raposo G. In: Current Protocols in Cell Biology. Morgan K, editor. John Wiley; New York: 2006. p. UNIT 3.22. [DOI] [PubMed] [Google Scholar]
- 19.Lamparski HG, Metha-Damani A, Yao JY, Patel S, Hsu DH, Ruegg C, Le Pecq JB. J Immunol Methods. 2002;270:211–226. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- 20.Taylor DD, Gercel-Taylor C. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 21.Stroock AD, Dertinger SKW, Ajdari A, Mezic I, Stone HA, Whitesides GM. Science. 2002;295:647–651. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- 22.Hsu CH, Di Carlo D, Chen CC, Irimia D, Toner M. Lab Chip. 2008;8:2128–2134. doi: 10.1039/b813434k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi S, Park JK. Lab Chip. 2007;7:890–897. doi: 10.1039/b701227f. [DOI] [PubMed] [Google Scholar]
- 24.Cheng XH, Irimia D, Dixon M, Sekine K, Demirci U, Zamir L, Tompkins RG, Rodriguez W, Toner M. Lab Chip. 2007;7:170–178. doi: 10.1039/b612966h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murthy SK, Sin A, Tompkins RG, Toner M. Langmuir. 2004;20:11649–11655. doi: 10.1021/la048047b. [DOI] [PubMed] [Google Scholar]
- 26.Usami S, Chen HH, Zhao YH, Chien S, Skalak R. Ann Biomed Eng. 1993;21:77–83. doi: 10.1007/BF02368167. [DOI] [PubMed] [Google Scholar]
- 27.Squires TM, Messinger RJ, Manalis SR. Nat Biotechnol. 2008;26:417–426. doi: 10.1038/nbt1388. [DOI] [PubMed] [Google Scholar]
- 28.Parsons D, Jones S, Zhang X, Lin J, Leary R, Angenendt P, Mankoo P, Carter H, Siu I, Gallia G, Olivi A, McLendon R, Rasheed B, Keir S, Nikolskaya T, Nikolsky Y, Busam D, Tekleab H, Diaz L, Hartigan J, Smith D, Strausberg R, Marie S, Shinjo S, Yan H, Riggins G, Bigner D, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu V, Kinzler K. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan H, Parsons DW, Jin GL, McLendon R, Rasheed BA, Yuan WS, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tschoepe D, Schultheiss HP, Kolarov P, Schwippert B, Dannehl K, Nieuwenhuis HK, Kehrel B, Strauer B, Gries FA. Circulation. 1993;88:37–42. doi: 10.1161/01.cir.88.1.37. [DOI] [PubMed] [Google Scholar]
- 31.Tohami T, Drucker L, Radnay J, Shapira H, Lishner M. Tissue Antigens. 2004;64:235–242. doi: 10.1111/j.1399-0039.2004.00271.x. [DOI] [PubMed] [Google Scholar]
- 32.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 33.Graner M, Alzate O, Dechkovskaia A, Keene J, Sampson J, Mitchell D, Bigner D. FASEB J. 2009;23:1541–1557. doi: 10.1096/fj.08-122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hasselmann D, Rappl G, Tilgen W, Reinhold U. Clin Chem. Vol. 47. Washington, D C: 2001. pp. 1488–1489. [PubMed] [Google Scholar]
- 35.Abu Al-Soud W, Jonsson LJ, Radstrom P. J Clin Microbiol. 2000;38:345–350. doi: 10.1128/jcm.38.1.345-350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akane A, Matsubara K, Nakamura H, Takahashi S, Kimura K. J Forensic Sci. 1994;39:362–372. [PubMed] [Google Scholar]
- 37.Powell HA, Gooding CM, Garrett SD, Lund BM, McKee RA. Lett Appl Microbiol. 1994;18:59–61. [Google Scholar]
- 38.Wilson IG. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia EJ, Hart AJ, Wardle BL, Slocum AH. Adv Mater. 2007;19:2151–2156. [Google Scholar]
- 41.Wardle B, Saito D, Garcia E, Hart A, de Villoria R, Verploegen E. Adv Mater. 2008;20:2707–2714. doi: 10.1002/adma.200800295. [DOI] [PubMed] [Google Scholar]
- 42.Peluso P, Wilson DS, Do D, Tran H, Venkatasubbaiah M, Quincy D, Heidecker B, Poindexter K, Tolani N, Phelan M, Witte K, Jung LS, Wagner P, Nock S. Anal Biochem. 2003;312:113–124. doi: 10.1016/s0003-2697(02)00442-6. [DOI] [PubMed] [Google Scholar]
- 43.Russom A, Sethu P, Irimia D, Mindrinos MN, Calvan SE, Iris GA, Finnerty C, Tannahill C, Abouhamze A, Wilhelmy J, Lopez MC, Baker HV, Herndon DN, Lowry SF, Maier RV, Davis RW, Moldawer LL, Tompkins RG, Toner M. Clin Chem. Vol. 54. Washington, D C: 2008. pp. 891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, Digumarthy S, Muzikansky A, Irimia D, Settleman J, Tompkins RG, Lynch TJ, Toner M, Haber DA. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agrawal N, Toner M, Irimia D. Lab Chip. 2008;8:2054–2061. doi: 10.1039/b813588f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor DD, Lyons KS, Gercel-Taylor C. Gynecol Oncol. 2002;84:443–448. doi: 10.1006/gyno.2001.6551. [DOI] [PubMed] [Google Scholar]
- 47.Taylor DD, Taylor CG, Jiang C, Black PH. Int J Cancer. 1988;41:629–635. doi: 10.1002/ijc.2910410425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


