Abstract
Recent advances in the synthesis of polymeric colloids have opened the doors to new advanced materials. There is strong interest in using these new techniques to produce particles that mimic and/or interact with biological systems. An important characteristic of biological systems that has not yet been exploited in synthetic polymeric colloids is their wide range of deformability. A canonical example of this is the human red blood cell (RBC) which exhibits extreme reversible deformability under flow. Here we report the synthesis of soft polymeric colloids with sizes and shapes that mimic those of the RBC. Additionally, we demonstrate that the mechanical flexibility of the colloids can be reproducibly varied over a large range resulting in RBC-like deformability under physiological flow conditions. These materials have the potential to impact the interaction between biological and synthetic systems.
Keywords: crosslinking, deformable, hydrogel, microparticle, stop flow lithography
Introduction
Architecture is as important as the material of construction when considering the mechanical properties of structures. This is true on the macro scale as in the case of the Eiffel tower which derives its mechanical stability as much from its unique architecture as from the iron used in its construction. Likewise in biology, on the nano- to micro-scales, the hollow tube architecture of a microtubule leads to a large effective bending modulus for this important cellular scaffold material. On the colloidal length scale the RBC is an exquisite biological example of a structure which is highly deformable. The deformability of the RBC is a vital property allowing it to function.[1–3] While the unique mechanical properties of RBCs are related to the complex architecture of the cell membrane,[4] much of their general mechanical deformability may be captured by the intelligent design of a simple synthetic particle mimic.
Recently, there has been strong interest in fabricating and characterizing new polymeric particles with a variety of shapes, chemistries, and sizes [5–9] particularly on the cellular length scale which is important for interaction with biological systems. The interaction between synthetic and biological systems is one of today’s most vibrant areas of research and it is increasingly recognized that the physical properties of synthetic materials are as important as their chemical properties.[10–12]
As mentioned, mechanical properties on all scales are dependent upon the architecture of the object in question. To date, it has proven difficult to translate architectural concepts from the man-made macro scale, or the biologically self-assembled nano-scale, to the micro-scale for use in polymeric colloids. More generally, the idea of deformability in biological systems such as the RBC has not been translated to polymeric colloids. Recently, the concept of deformable particles was alluded to in the literature but no measure of particle flexibility was made.[13] Additionally, the reported studies in which the flexibility of polymeric colloids has been systematically modulated are mostly limited to rheological measurements of dense colloidal pastes.[14–15] Generally, these studies were performed with spherical particles and no measure was made of the mechanical properties of the colloids themselves. It is of strong interest to control the mechanical properties of polymeric particles independent of their chemical properties especially for interaction with biological systems.[10] There is increasing evidence that the physical properties of bio-compatible materials are important in regulating biological responses. Such an advance evokes visions of a new class of in vivo applications where cell-like synthetic colloids are used for cargo delivery, sensing, or patient diagnostics. As a first step toward the development of such materials, we synthesized polyethylene glycol (PEG) hydrogel particles with sizes and shapes reminiscent of RBCs. The motivation for choosing to mimic RBCs stems from the potential for in vivo uses of such particles where, like real RBCs, they would be expected to remain in circulation in the blood stream. Here we describe their synthesis and characterization and we demonstrate how control over both the nano- and micro-structure of the particles leads to an extremely large range of mechanical properties.
Experimental Part
Particle Synthesis
The monomer solution consisted of 65% v/v PEG mixture containing PEG diacrylate with a molecular weight of 700 (PEG(700)DA, Sigma–Aldrich) and PEG with a molecular weight of 200 (PEG 200, Sigma–Aldrich), 15% v/v photoinitiator 2-Hydroxy-2-methyl-1-phenyl-propan-1-one (Darocur 1173, Ciba), and 20% v/v TE buffer (100×TE, OmniPur diluted to 3× with deionized water). In the case of particles functionalized with rhodamine the solution contained 1% v/v of a 10 mg·mL−1 methacryloxyethylthiocarbamoyl rhodamine B (Polysciences) solution in PEG 200. The fraction of PEG(700)DA in the mixture was varied such that its total concentration in the monomer solution was between 10 and 40% v/v. The particles were synthesized in an all polydimethylsiloxane (PDMS) microfluidic channel with a height of 4.5 μm using the stop flow lithography (SFL) technique and setup as previously described.[16] Briefly, the microfluidic channel was placed on an inverted objective microscope (Axiovert 200, Zeiss) and arrays of particles were polymerized cyclically through a 40× microscope objective. The transparency mask used to define the array of particles was ordered from FineLine Imaging (Colorado Springs, CO). The pressure used to drive flow during polymerization was 3psi and the exposure dose of UV light during each cycle of polymerization ranged from 50 to 150 ms depending on the concentration of PEG(700)DA. The particles were collected in phosphate buffered saline (PBS) containing 0.05% v/v Tween 20 and 40% v/v PEG 200. They were then spun down in a benchtop centrifuge and the supernatant fluid was replaced with PBS containing 0.05% Tween 20.
The PEG particles functionalized with a DNA probe were made in a similar fashion with the following changes. Only 18% v/v 3×TE was used and no rhodamine acrylate was added. Additionally, 2% v/v of a 500×10−6 m solution containing an acrylated DNA probe (5′ - Acrydite-Sp18 - ATA GCA GAT CAG CAG CCA GA - 3′, Integrated DNA Technologies) in PBS was added. To test the ability of the DNA-labeled particles to uniquely capture a complimentary strand of DNA, a mixture of inert and functionalized particles (~1 000 particles/μL) was incubated in PBS containing 0.05% v/v Tween 20 and a 25×10−9 m concentration of the target DNA (5′ - Cy3 - TCT GGC TGC TGA TCT GCT AT - 3′, Integrated DNA Technologies) for 30 min at 37 °C. The particles were subsequently washed repeatedly with PBS containing 0.05% v/v Tween 20.
The PEG particles functionalized with antibody against epithelial cell adhesion molecule (EpCAM) were made with the following changes. Acrylic acid (Polysciences) was added to the monomer solution in a concentration of 10% v/v at the expense of some of the PEG 200 and rhodamine acrylate was also omitted. The particles were incubated in a solution containing 10 μg·mL−1 biotinylated anti-EpCAM (R&D Systems Inc.) and 5 mg·mL−1 1-Ethyl-3-[3-dimethylaminopropyl]carbodiimide Hydrochloride (EDC, Sigma–Aldrich) in PBS with 0.05% v/v Tween 20 for 30 min at 37 °C. The particles were subsequently washed repeatedly with PBS containing 0.05% v/v Tween 20. To demonstrate the functionalization of the particles with anti-EpCAM, we incubated a mixture of inert and functionalized particles (≈1 000 particles/μL) in PBS containing 0.05% v/v Tween 20 and a 50 μg·mL−1 concentration of the protein neutravidin labeled with FITC (Pierce Biotechnology) for 30 min at 37 °C and the particles were subsequently washed repeatedly with PBS containing 0.05% v/v Tween 20.
Particle Size Measurement
The size of the particles was measured with images from a microscope mounted camera (KP-M1A, Hitachi) using a 40× objective. The diameter and thickness of the particles were determined by measuring >100 individual colloids. In the case of crosses and S-shapes, the diameter was measured by circumscribing a circle around the particles using image analysis software (ImageJ) and measuring the diameter of the circle. To compare the particle sizes with RBCs, 10 μL of the particle solution (≈1 000 particles/μL) was mixed with 10 μL of whole blood diluted in PBS at a ratio of 1:1 000.
Particle Flow Testing
A 4 μm tall microfluidic channel was molded in PDMS and bonded to a glass coverslip. The channel (shown in Supporting Information (SI) Figure 1) contained entrance and exit regions where access holes were punched, a 960 μm long approach section, five parallel 4 μm constrictions, and a 30 μm long exit section. A liquid filled reservoir containing PBS with 0.5% Tween 20 was inserted into the entrance hole and connected to a pressure source with a regulator (Omega Engineering, 0–15 psig). The channel was filled at 3 psi by opening the pressure valve. The reservoir was then replaced with another containing the particle sample. The driving pressure was set on the regulator for pressures >1 psi or by manometer (custom made) for pressures <1 psi. The frequency of passage and threshold pressure drop were determined by observation with a 20× objective on the inverted microscope. The pressure drop through the constriction was calculated using COMSOL Multiphysics finite element modeling software for a given total pressure drop across the entire channel.
Videos of Particle Passage
The viscosity of the suspending fluid was increased by adding 50% v/v glycerol to the particle suspension and the particles were then filmed while flowing through the constrictions in both brightfield and fluorescence using a microscope mounted camera (KP-M1A, Hitachi) at 30 frames per second (interlaced). The video was de-interlaced using custom routines in MATLAB to achieve an effective 60 frames per second. The videos in the Supplemental Information are slowed down 2×.
Results and Discussion
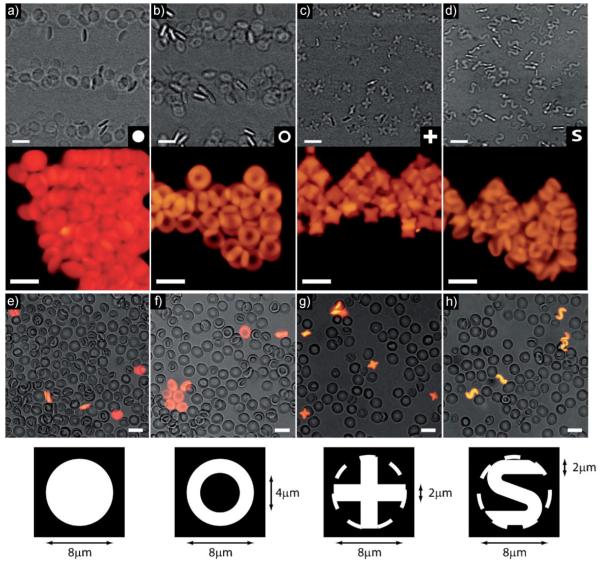
Red blood cells (RBCs) have a biconcave disk shape with a diameter of ≈8 μm and a thickness of ≈2 μm.[17] Using SFL[16] we synthesized a variety of PEG hydrogel colloids with similar sizes. While the shape of RBCs is important biologically, we also demonstrate that other non-biological shapes with the same general size as RBCs can exhibit enhanced properties. Using different photomasks (Figure 1 bottom row) we were able to synthesize monodisperse disks, rings, crosses, and S-shapes in large quantities (>1E6 particles/batch) with a thickness of 2 μm±0.1 μm (Figure 1). The disks and rings were determined to have a diameter of 8 μm±0.2 μm and the mask design was such that an 8 μm diameter circle can just circumscribe the crosses and S-shaped particles. The volumes of the four classes of particles were 100, 75, 56, and 56 μm3 for disks, rings, crosses, and S-shapes, respectively.
Figure 1.
Collections of PEG particles 8 μm in diameter and 2 μm thick with cross sections in the shapes of (a) disks, (b) rings, (c) crosses, and (d) S-shapes. The top row contains brightfield images from video of the particles flowing and the middle row contains fluorescence images of concentrated particles. The bottom row of images contains composite brightfield and fluorescent images of diluted human whole blood (grayscale) and PEG particles (fluorescent) mixed together in PBS illustrating the size of the (e) disks, (f) rings, (g) crosses, and (h) S-shapes with respect to RBCs. The photomask used to define each shape is shown in the bottom row along with relevant dimensions. In the case of crosses and S-shapes, a dashed circle was added to the mask to illustrate their size relative to the disks and rings. In all images, the scale bars are 10 μm.
To tune the deformability of the particles we systematically altered their cross-linking density, a well known method for altering the mechanical properties of bulk and micropatterned hydrogels.[18–20] We have recently shown that in the free radical polymerization which takes place in SFL, the extent of reaction can not be easily altered.[21] However, it has been previously shown for bulk hydrogels synthesized through free radical polymerization that the concentration of reactive species can drastically impact the type of reactions that occur.[22] At low reactive species concentrations, radical termination primarily occurs within a single molecule resulting in a cyclization while at high concentrations, it primarily occurs between two molecules resulting in a crosslink.[22] In the former case, the gel has a more open nano-structure with larger pores than in the latter case where high crosslinking density leads to smaller pores.
The four classes of particles were synthesized using PEG(700)DA concentrations ranging from 10v/v to 40% v/v. There was no observable dependence of the size or shapes of the particles upon the concentration of PEG(700)DA and the particles were approximately the same size as RBCs (Figure 1e–h). It was determined experimentally that below a concentration of ≈7%, intact particles could not be reliably polymerized agreeing well with the gelation point of similar systems.[22]
The mechanical test used to characterize the particles was motivated by the in vivo environment. In circulation, RBCs regularly flow through capillaries with diameters smaller than the nominal size of the cell itself. The smallest of such capillaries is ≈5 μm and thus the RBC must deform to pass through. The pressure difference across the capillary in vivo can be as small as ≈0.2 mmHg.[23] The PEG particles synthesized here were thus tested by flowing through a microfluidic channel containing several 4 μm×4 μm constrictions under a known pressure difference. A detailed description of the flow test system is provided in Supporting Information (SI) Figure 1. For each particle geometry and crosslinking density we determined the critical threshold pressure differential ΔPc, defined as the pressure differential at which 50% of the particles pass through the constrictions. This mechanical test was chosen to highlight the anticipated particle behavior under physiological conditions.
The critical pressure differential is shown in Figure 2 as a function of the concentration of PEG(700)DA for the four different classes of particles. The error bars mark the range of ΔP from 0 to 100% passage. Figure 2 illustrates the importance of both the nano- and micro-structure of the colloids. The most drastic feature is the range of four orders of magnitude in ΔPc over which the data span. In all cases, the particles have the same nominal size (inscribed within an 8 μm×2 μm disk) but their mechanical properties vary over such a large range through tuning the crosslinking density and micro-scale shape. As expected, particles synthesized with a higher concentration of PEG(700)DA are less flexible as the nano-structure of the gel contains smaller pores. A second feature of the data in Figure 2 is the drastic change in ΔPc that occurs near the gelation point. This behavior is consistent with observations in bulk hydrogel materials.[22] In all cases the particles synthesized with 10% PEG(700)DA pass through the constrictions under mild pressure differentials ranging from 2.8 mmHg for disks to 0.05 mmHg for S-shapes, which is smaller than pressure drops observed in human capillaries in both healthy and diseased states.[23–24]
Figure 2.
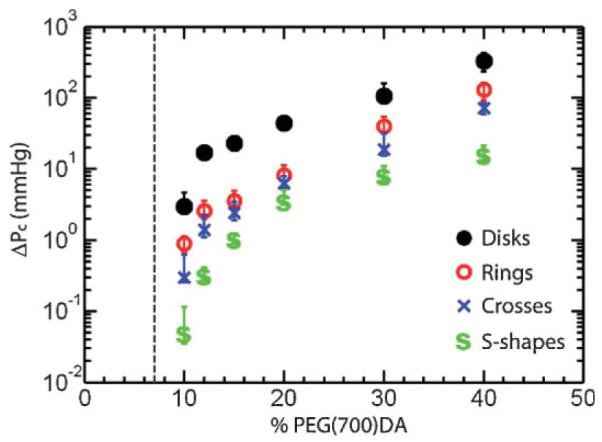
Threshold pressure differential for passage of disks, rings, crosses, and S-shapes through 4 μm×4 μm constrictions as a function of the concentration of PEG(700)DA in the pre-polymer solution. The vertical dashed line is the experimentally determined gelation point below which intact particles could not be synthesized.
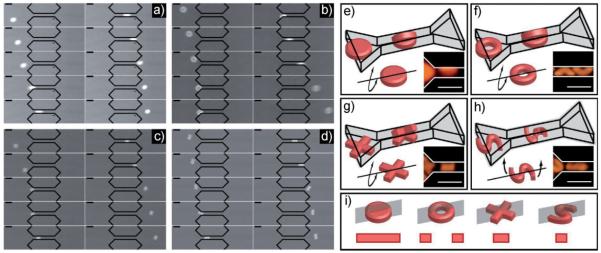
Aside from the dependence upon nano-scale architecture, the shapes also exhibit variable deformability as a function of their micro-structure. Disks appear to be the least flexible of the four followed by rings, crosses, and finally S-shapes. To investigate this varied behavior more closely, we determined their conformations while passing through the constrictions. The mode of passage through a pore offers valuable insight into the behavior of colloidal materials. For instance, in order for flexible molecules such as DNA to pass through a nano-pore, the head of the molecule must enter first with the body threading behind.[25] This has been shown important for biomolecule separations. Here, we observed a rich array of modes of passage including bending, twisting, and orientation effects. For disks, rings, and crosses we observed that the particles prefer to buckle, rather than compress in order to pass through the constriction while S-shapes predominately twist during their passage. Furthermore, crosses and S-shapes exhibit a preferred orientation as they passed through the constriction. The passage of the four types of particles is shown with still-frame image sequences in Figure 3a–d (videos of passage are in SI). The observed length of the deformed particle within the constriction is the same as in the undeformed case before the constriction indicating that the particles are bent rather than stretched. The mode of passage for each shape is further illustrated in Figure 3e–h along with higher magnification images showing that a disk, ring, or cross trapped within the constriction does indeed adopt a folded over shape. In contrast, Figure 3h demonstrates that the two ends of the S-shaped particle twist in opposite directions enabling passage with only minimal bending. For crosses and S-shapes, the particles were seen to orient as illustrated in Figure 3g–h.
Figure 3.
Detailed images and illustrations of particles passing through a 4 μm×4 μm constriction. (a) Image sequence (50 ms between frames) of a PEG disk passing under a pressure difference of 2.8 mmHg. (b) Image sequence (83 ms between frames) of a PEG ring passing under a pressure difference of 0.9 mmHg. (c) Image sequence (133 ms between frames) of a PEG cross passing under a pressure difference of 0.3 mmHg. (d) Image sequence (200 ms between frames) of a PEG S-shape passing under a pressure difference of 0.05 mmHg. Illustrations and experimental images of deformation of the particles within the constriction for (e) disks, (f) rings, (g) crosses, and (h) S-shapes. (i) Illustrations of the plane where deformation takes place in each particle and the cross sectional area of the particle in that plane. The scale bars in all images are 10 μm.
The actual mechanism of passage in this study is quite complicated, driven by hydrodynamic, excluded volume, and surface forces in addition to mechanical deformation of materials with complex shapes. However, a simplified view of the bending and twisting modes helps to explain the trends observed in Figure 2. Along the plane where deformation occurs (Figure 3i) the cross-sectional area of the disk (16 μm2) is larger than the ring (8 μm2) and the cross (5.7 μm2),given its preferred orientation. The torsion of the S-shaped particles occurs within the plane illustrated in Figure 3i which has the smallest area (4 μm2) of the four. It is known that the bending modulus is proportional to the cross-section of the structure in question. If we consider the bulk of the energy penalty for deformation to lie within the plane of deformation we can comment on the trends observed. Under this simplification, the energy required to bend the disks will be largest followed by rings, crosses, and S-shapes.
Another important observation is that the distribution of mass plays a crucial role in determining the behavior of the particles. Crosses and S-shapes have the same volume (56 μm3) but measurably different deformability, important when considering the carrying capacity of these materials for applications such as drug delivery to specific target cells within the blood stream. By distributing the same amount of mass (or volume) into different micro-structures, flexibility can be tuned without affecting the capacity to carry functional molecules.
While the particles in this study are of similar size and shape to RBCs there are important differences which become evident when testing their mechanical properties. RBCs are vesicles with an internal fluid phase contained by a cell membrane. Therefore, when a RBC passes through a constriction, its mode of passage is significantly different and involves a more fluid-like deformation of the membrane.[4] The PEG particles in this study are poroelastic objects with consistent material properties throughout their volumes which governs their deformation during passage through the constriction. As such, their mechanical properties are not comparable to those of RBCs, especially in terms of deformability. Nonetheless, the range of pressure drops at which the PEG particles can pass through the constriction spans the entire physiological range observed in human capillaries.
To emphasize the potential utility of these particles for in vivo applications in the blood stream, we demonstrated the ability to functionalize particles with a variety of biologically relevant chemistries. In Figure 4 we show non-functionalized, non-fluorescent S-shapes mixed with rings containing a 20 base DNA probe (Figure 4a–b) or biotinylated anti-EpCAM (Figure 4c–d) which is frequently overexpressed by epithelial derived cancer cells.[26–27]
Figure 4.
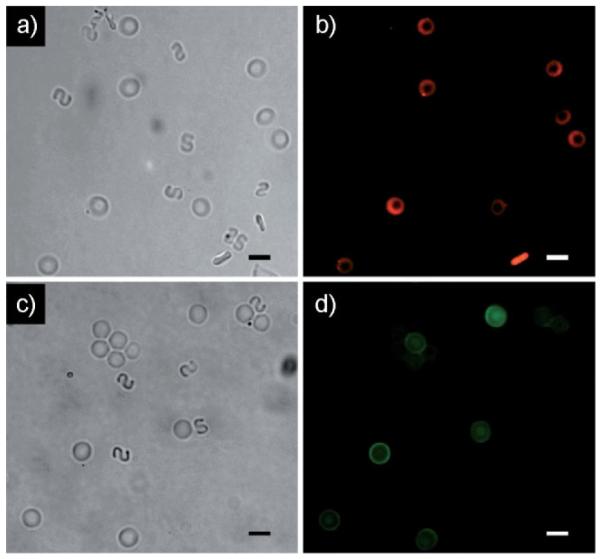
Images of inert S-shaped particles and rings functionalized with a biomolecule. Brightfield (a) and fluorescence (b) images of the particle mixture after incubation with a fluorescent target 20 base DNA strand. In this case the rings were hybridized with a 20 base probe DNA. Brightfield (c) and fluorescence (d) images of the particle mixture after incubation with a fluorescently labeled neutravidin target. In this case the rings were functionalized with a biotinylated antibody relevant in cancer biology. The PEG particles appear oriented randomly due to their Brownian motion. The scale bars in all images are 10 μm.
In the case of rings containing DNA probe, we incubated the particles with a complimentary target 20 base DNA strand labeled with Cy3 fluorescent dye. Fluorescence images of the particle mixture after incubation (Figure 4b) confirm that the target DNA associates only with the functionalized rings indicating that the S-shapes are indeed inert. Likewise, when rings labeled with biotinylated anti-EpCAM are incubated with fluorescein isothiocyanate (FITC) labeled neutravidin in the presence of inert S-shapes, again the FITC labeled protein only interacts with the functionalized particles (Figure 4d). Through this example, we demonstrate the ability to independently synthesize PEG particles with a variety of useful chemistries independent of their size, shape, and mechanical properties.
Conclusion
Using SFL, we synthesized a new class of squishy microparticles with comparable size to RBCs and independently tuned their size/shape, functional chemistry, and mechanical flexibility. Altering the concentration of reactive PEG(700)DA in the monomer solution affected the particle cross-linking density. This variation in nano-structure led to a large difference in the measured mechanical properties when the particles were flowed through narrow microfluidic constrictions. Further tuning the micro-architecture of the particles by changing their shapes expanded the range of flexibilities to four orders of magnitude. The materials of construction for the particles presented here are identical and only their nano- and micro-architecture have been modulated. Finally, we demonstrated that many biologically important functional chemistries can be incorporated into the particles independent of their other properties.
Even with the development of new tools for synthesizing and manipulating polymeric colloids, the range of possibilities for controlling their properties remains largely untapped. This work represents the first systematic demonstration of independent control over size/shape, functional chemistry, and mechanical flexibility of polymeric colloids. The wide range of control demonstrated here will serve as a first step toward new materials that may have impact in many fields including advanced materials, drug delivery, consumer products, and in vivo diagnostics.
Supplementary Material
Acknowledgements
We thank Octavio Hurtado for assistance with microfabrication and David J. Quinn for useful discussions on design of the constriction devices. This work was supported in part by the MIT-MGH Postdoctoral Fellowship in Translational Research and the National Institute of Biomedical Imaging and Bioengineering (BioMEMS Resource Center, P41 EB002503). PSD acknowledges support from the John Simon Guggenheim Foundation and the Rothschild-Yvette Mayent-Institute Curie Fellowship.
Footnotes
Supporting information for this article is available at the bottom of the article’s abstract page, which can be accessed from the journal’s homepage at http://www.mrc-journal.de, or from the author.
Contributor Information
Ramin Haghgooie, BioMEMS Resource Center, Center for Engineering in Medicine and Surgical Services, Massachusetts General Hospital, Shriners Hospital for Children, and Harvard Medical School, 114 16th Street, Room 1239, Charlestown, MA 02129, USA.
Mehmet Toner, BioMEMS Resource Center, Center for Engineering in Medicine and Surgical Services, Massachusetts General Hospital, Shriners Hospital for Children, and Harvard Medical School, 114 16th Street, Room 1239, Charlestown, MA 02129, USA.
Patrick S. Doyle, Massachusetts Institute of Technology, 77 Massachusetts Ave, Room 66-270, Cambridge, MA 02139, USA.
References
- [1].Skalak R, Branemark PI. Science. 1969;164:717. doi: 10.1126/science.164.3880.717. [DOI] [PubMed] [Google Scholar]
- [2].Nash GB, Linderkamp O, Pfafferroth C, Meiselman MJ. Blood Cells, Rheology, and Aging. Springer Verlag; Berlin: 1988. p. 99. [Google Scholar]
- [3].Miller LH, Baruch DI, Marsh K, Doumbo OK. Nature. 2002;415:673. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- [4].Li J, Lykotrafitis G, Dao M, Suresh S. Proc. Nat. Acad. Sci. 2007;104:4937. doi: 10.1073/pnas.0700257104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Discher DE, Eisenberg A. Science. 2002;297:967. doi: 10.1126/science.1074972. [DOI] [PubMed] [Google Scholar]
- [6].Dendukuri D, Pregibon DC, Collins J, Hatton TA, Doyle PS. Nat. Mater. 2006;5:365. doi: 10.1038/nmat1617. [DOI] [PubMed] [Google Scholar]
- [7].Champion JA, Mitragotri S. Proc. Nat. Acad. Sci. 2006;103:4930. doi: 10.1073/pnas.0600997103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Maynor BW, Larue I, Hu Z, Rolland JP, Pandya A, Fu Q, Liu J, Spontak RJ, Sheiko SS, Samulski RJ, Samulski ET, DeSimone JM. Small. 2007:3845. doi: 10.1002/smll.200600507. [DOI] [PubMed] [Google Scholar]
- [9].Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. Nat. Nanotechnol. 2007;2:249. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mitragotri S, Lahann J. Nat. Mater. 2009;8:15. doi: 10.1038/nmat2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gratton SEA, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM. Proc. Nat. Acad. Sci. 2008;105:11613. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Beningo KA, Wang Y. J. Cell Sci. 2002;115:849. doi: 10.1242/jcs.115.4.849. [DOI] [PubMed] [Google Scholar]
- [13].Gratton SEA, Williams SS, Napier ME, Pohlhaus PD, Zhou Z, Wiles KB, Maynor BW, Shen C, Olafsen T, Samulski ET, DeSimone JM. Acc. Chem. Res. 2008;41:1685. doi: 10.1021/ar8000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Borrega R, Cloitre M, Betremieux I, Ernst B, Leibler L. Europhys. Lett. 1999;47:729. [Google Scholar]
- [15].Seth JR, Cloitre M, Bonnecaze RT. J. Rheol. 2006;50:353. [Google Scholar]
- [16].Dendukuri D, Gu SS, Pregibon DC, Hatton TA, Doyle PS. Lab. Chip. 2007;7:818. doi: 10.1039/b703457a. [DOI] [PubMed] [Google Scholar]
- [17].Evans E, Fung YC. Microvasc. Res. 1972;4:335. doi: 10.1016/0026-2862(72)90069-6. [DOI] [PubMed] [Google Scholar]
- [18].Anseth KS, Bowman CN, Brannon-Peppas L. Biomaterials. 1996;17:1647. doi: 10.1016/0142-9612(96)87644-7. [DOI] [PubMed] [Google Scholar]
- [19].Zaari N, Rajagopalan P, Kim SK, Engler AJ, Wong JY. Adv. Mater. 2004;16:2133. [Google Scholar]
- [20].Gray DS, Tien J, Chen CS. J. Biomed. Mater. Res. 2003;66A:605. doi: 10.1002/jbm.a.10585. [DOI] [PubMed] [Google Scholar]
- [21].Dendukuri D, Panda P, Haghgooie R, Kim JM, Hatton TA, Doyle PS. Macromolecules. 2008;41:8547. [Google Scholar]
- [22].Elliott JE, Anseth JW, Bowman CN. Chem. Eng. Sci. 2001;56:3173. [Google Scholar]
- [23].Huang Y, Doerschuk CM, Kamm RD. J. Appl. Physiol. 2001;90:545. doi: 10.1152/jappl.2001.90.2.545. [DOI] [PubMed] [Google Scholar]
- [24].de Graaff JC, Ubbink DT, van der Spruit JA, Lagarde SM, Jacobs MJHM. J. Vasc. Surg. 2003;38:1067. doi: 10.1016/s0741-5214(03)00603-7. [DOI] [PubMed] [Google Scholar]
- [25].Wolterink JK, Barkema GT, Panja D. Phys. Rev. Lett. 2006;96:208301. doi: 10.1103/PhysRevLett.96.208301. [DOI] [PubMed] [Google Scholar]
- [26].Balzar M, Winter MJ, de Boer CJ, Litvinov SV. J. Mol. Med. 1999;77:699. doi: 10.1007/s001099900038. [DOI] [PubMed] [Google Scholar]
- [27].Went PTH, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, Dirnhofer S. Hum. Pathol. 2004;35:122. doi: 10.1016/j.humpath.2003.08.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.