Abstract
Within the global pharmaceutical and biotech industries, there is significant interest in identifying in vitro screening systems that are more human-relevant—i.e., that offer greater utility in predicting subcellular and cellular physiological responses in humans in vivo—and that thereby allow investigators to reduce the incidence of costly late-stage failures during pharmaceutical clinical trials, as well as to reduce the use of animals in drug testing. Currently incumbent in vitro screening methods, such as culturing human hepatocytes in suspension, while useful, are limited by a lack of long term cellular function. In order to address this limitation, we have established an integrated, microfluidic, in vitro platform that combines the patented HμREL® microdevice with a hepatic coculture system. In the present report, we use this platform to study clearance and metabolite generation of a battery of molecular entities. The results show that the flow-based coculture system is capable of clearing, with improved resolution and predictive value, compounds with high, medium, and low clearance values. In addition, when coculture is coupled with flow, higher metabolite production rates are obtained than in static systems.
Keywords: Microfluidics, Human hepatocyte, Coculture, Hepatic clearance, Metabolism
1. Introduction
Collectively, the pharmaceutical and biotechnology industries have long been interested in identifying in vitro screening systems that are more human-relevant–i.e., that offer greater utility in predicting subcellular and cellular physiological response in humans in vivo. An improved screening system would allow investigators to reduce the incidence of costly late-stage failures during pharmaceutical clinical trials, as well as to reduce the use of animals in drug testing. While static in vitro cell-based assays are used quite frequently within the pharmaceutical industry, they are limited by the fact that they do not adequately mimic the complexity of the physiological environment and thus may not accurately give effect to relevant human parameters. In addition, because human hepatocyte cultures rapidly lose function over time, they are of limited use in cases where compounds clear slowly, or where metabolites are generated over extended periods of time. While animal testing replicates some complex inter-cellular and inter-tissue effects, animal studies are expensive, labor-intensive and, in certain instances, not relevant to the human physiological response. Thus, a need exists for the development of stable and effective human cell-based in vitro methods that can improve the prediction of in vivo drug disposition.
Many primary hepatocyte based culture methods are limited by their lack of long term in vitro function. Most hepatospecific functions are typically lost in the first days of culture. Methods to stabilize in vitro adult primary hepatocyte function have been developed, and include techniques which use specific extracellular matrices (ECM), which spatially orient hepatocytes between layers of extracellular matrix, or which involve coculture with nonparenchymal cells [1]. For example, it has been shown that when primary hepatocytes are cultured with nonparenchymal cells, there is a marked increase in hepatocyte function as compared to hepatocytes cultured alone [2,3]. In general, systems yielding the most promising results are based upon the promotion of homotypic hepatocyte interactions induced by the coculture [2].
Another consideration is the format used for culturing hepatocytes. The vast majority of current in vitro screening assays utilize cells cultured under static conditions. In this format, continuous flow of culture media over the isolated hepatocytes, mimicking the physiologic state, is absent. A fairly large number of prior studies have demonstrated the beneficial effects of flow on hepatocyte function [4–13]. For example, a small-scale bioreactor incorporating rat hepatocytes was shown to provide superior performance in drug metabolism studies [14]. More recent studies have explored a number of adherent mammalian cells in microbioreactors, and in some cases have incorporated two chamber microfluidic designs seeded with HepG2 cells and L2 lung cells or 3T3 adipocytes in series. A number of studies have incorporated primary hepatocytes into microfluidic arrays to create a more physiologically relevant model of liver metabolism [15–17].
In a prior report, we described a set of clearance and metabolism studies using a microfluidic cell culture analog (CCA) system seeded with primary human hepatocytes alone [18]. In the present report, we use an improved, more stable coculture system within the microdevice to study clearance and metabolite generation of a battery of molecular entities. The results show that the flow-based coculture system is capable of clearing, with resolution and improved predictive value, compounds with high, medium, and low clearance values. In addition, when coculture is coupled with flow, higher metabolite production rates are obtained than in static systems.
2. Materials and methods
2.1. Materials and reagents
All chemicals were obtained from Sigma-Aldrich (St. Louis, MO) unless specified otherwise. Sildenafil was obtained from American Custom Chemicals Corporation (San Diego, CA). Methanol and all other organic solvents were purchased from Fisher Scientific (Waltham, MA). Cryopreserved human hepatocytes were obtained from Celsis in vitro technologies (Baltimore, MD). Rat tail collagen type I, BD Biocoat™ collagen I 96-well microplates were obtained from BD Biosciences (Franklin Lakes, NJ). Polystyrene HμREL® biochips, Hurel PlatinumHeps™ media and the polycarbonate housing sets were obtained from HμREL® Corporation (Beverly Hills, CA)
2.2. Preparation of polystyrene HμREL® biochips
The biochips were sterilized by soaking in 70% isopropanol for an hour followed by rinsing with sterile distilled water. The biochips were then dried and subsequently treated with air plasma using a high frequency generator (Electro-Technic Products Inc.) for 3–5 s to modify the surface properties of the biochips to be more hydrophilic, and to increase the collagen-coating efficiency. The “liver chamber” of the biochips was coated with rat tail type I collagen (BD Biosciences) and the biochips were stored aseptically at 4 °C until use. The biochips were rinsed with phosphate buffered saline (PBS) (Gibco-Invitrogen), three times before cell seeding.
2.3. Cell culture
2.3.1. Preparation of hepatocyte/nonparenchymal cell cocultures
Cryopreserved human hepatocytes were removed from liquid nitrogen and thawed quickly in a water bath at 37 °C. Human hepatocytes were transferred to a 50 mL conical tube containing 20 mL warm Hurel PlatinumHeps™ medium, 9 mL Percoll (Sigma), 1mL 10× PBS (Gibco-Invitogen) and centrifuged at 500 × g (Beckman Coulter, TJ-25) for 5 min at room temperature. After removing the supernatant, the cells were re-suspended in Hurel PlatinumHeps™ medium with a cell density of 4 × 106 cells/mL for seeding. The cell viability and number were determined using trypan blue exclusion (70–95% viable).
Nonparenchymal cells (passages 10–20) were passed in a CO2 incubator at 37 °C until used for experimental plating. On plating day cells were detached from the plate surface using standard trypsinization, suspended in 15 mL DMEM medium (Gibco-Invitogen) and centrifuged at 1000 × g for 7 min at room temperature to obtain a cell pellet. After removing the supernatant, the cells were re-suspended in Hurel PlatinumHeps™ medium with a cell density of 4 × 106 cells/mL for seeding. The cell viability and density were determined using trypan blue exclusion (75–95% viable).
2.3.2. Static culture in 96-well microplates
For the monoculture conditions human hepatocytes suspended in Hurel PlatinumHeps™ medium were confluently seeded in BD Biocoat™ collagen I 96-well microplates. For the coculture condition, human hepatocytes and nonparenchymal cells were premixed and confluently seeded. The cells were allowed to attach in a CO2 incubator at 37 °C for 4 h before 100 μL media is replaced. Media was replaced every 48 h for duration of experiments. On the experimental day, drug solutions were prepared and pre-warmed to 37 °C. The cell containing plates were incubated with 100 μL of respective drug solution for 24 h in a CO2 incubator at 37 °C on an orbital shaker with a shaking speed of 670 rpm. Compounds used in the clearance studies (buspirone, imipramine, sildenafil, timolol, nifedipine, diclofenac, indomethacin, carabamazepine, antipyrine) were dosed at a starting concentration of 1 μM. Compounds used in metabolite generation studies (midazolam, bupropion, detromethorphan, phenacetin, warfarin, 7-hydroxy coumarin, and omeprazole) were dosed at a starting concentration of 25 °M. Supernatant aliquots of 5 °L were taken at pre-determined time points (0, 1, 2, 4, 24 h) and added to 100 °L methanol containing 10 ng/mL loperamide as the internal standard. Samples were stored at −20 °C until analyzed by LC/MS/MS.
2.3.3. Flow-based culture
Monocultures and cocultures suspended in the plating medium were confluently seeded in the “liver chamber” of the HμREL® biochips. The cells were allowed to attach to the biochips in a CO2 incubator at 37 °C for 4 h before assembling the biochips to the HμREL® housing sets and applying the flow of culture medium. Once the biochips were enclosed in the housing sets and connected to the tubing and the pump, the microfluidic device was transferred to a humidified CO2 incubator at 37 °C for 10 min to equilibrate the system. The exposure of the cells to the drugs was initiated by replacing the reservoirs to drug containing culture medium. Compound concentrations for the clearance and metabolite studies are the same as described for the static culture system. Aliquots of 5 μL were taken from the reservoirs at pre-determined time points add to 100 μL methanol containing 10 ng/mL loperamide (internal standard). Samples were stored at −20 °C until analyzed by LC–MS/MS.
2.4. LC–MS/MS method
Samples were centrifuged at 1000 × g for 10 min, and an aliquot (10 μL) of the supernatant was analyzed by LC–MS/MS. The LC–MS/MS system comprised a Shimadzu LC-10ADvp pump (Shimadzu, Columbia, MD), HTS PAL CTC autosampler (Leap Technologies, Carboro, NC), and an API 4000 mass spectrometer with a Turbo Ion Spray probe (Applied Biosystems/MDS SCIEX, Ontario, Canada). The separation of compounds was achieved using a reversed phased stationary phase (Synergi Hydro, Phenomenex). The mobile phase was a gradient with 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) with a flow rate of 0.8 mL/min. The initial composition of the mobile phase was 5% of B for 0.1 min, followed by a linear gradient to 90% of B over 1.1 min, held at 90% B for 0.2 min, and back to 5% B in 0.1 min. Most samples, with the exception of diclofenac, indomethacin, 7-hydroxy coumarin, 7-hydroxy coumarin sulfate, and 7-hydroxy coumarin glucuronide were detected using multiple reaction monitoring (MRM) in positive ion mode. The three exceptions were detected using MRM in negative ion mode. The area ratio of the analytes to the internal standard was calculated using the Analyst® software v. 1.4.1 (Applied Biosystems).
2.5. Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
RNA isolation was carried out using Promega SV Total Isolation Kit according to manufacturer instructions for seeded cells. RNA was purified and concentration was quantified using Molecular Devices Analyst GT spectrophotometer. TaqMan Reverse Transcription Reagents (Applied Biosystems) were used to transcribe RNA. Reactions were initiated using 1 μg RNA and transcribed in a Applied Biosystems GeneAmp PCR System 9700. PCR was initiated using 15 μL TaqMan Universal PCR Master Mix (2×), 1.5 μL Taqman gene expression assay and 3 μL target cDNA in a total volume of 30 μL on Applied Biosystem 7900HT Fast Real Time PCR System in a 96 well optical plate format. Gene transcription was evaluated using the [delta][delta]Ct method normalized to β-actin, a cytoskeletal microfilament, and to RNA from freshly thawed human hepatocytes as well as nonparenchymal cells.
2.6. Bile canaliculi analysis
To detect functional bile canaliculi we incubated hepatocytes with 2 μg/mL of 5(6)-carboxy-2′,7′-dichlorofluorescein diacetate (CDFDA) for 10 min, washed with phenol red-free media, and imaged using fluorescence microscopy.
2.7. Data analysis
2.7.1. Calculation of the intrinsic clearance for the static culture system
The in vitro human hepatocyte intrinsic clearances (CLint) are calculated from the substrate concentration profile in the hepatocyte incubation medium for static the based culture systems using previously described method [18]. In the static conditions, the well-defined and widely used well-stirred model [19] is used to scale up the in vitro intrinsic clearance to the estimated hepatic clearance (CLH), assuming the drug is totally unbound in the serum-free culture medium. A human hepatic blood flow of 20.7 mL/min/kg and a scaling factor (2.3) from in vitro to in vivo [20].
2.7.2. Calculation of the intrinsic clearance for the flow culture system
For the flow conditions, the liver chamber in the system is the sole eliminating compartment, which is connected to a non-eliminating compartment, the reservoir. The clearance of the flow system by hepatocytes cultured in the device can be calculated from previously described equations [18]. In short, a clearance rate is first established from the substrate concentration profile. This in turn is then used to calculate an extraction ratio. Finally, the predicted human hepatic clearance for cells under flow can be obtained by up-scaling the extraction ratio by multiplying by the human liver flow rate.
2.8. Statistical analysis
Statistical calculations were performed using Microsoft Excel (Redmond, WA). Each data point represents the mean of at least three experiments (each with three biological replicates), and the error bars represent the standard deviation of the mean. Statistical significance was determined using the Student's t-test for unpaired data.
3. Results
3.1. Optimization of static coculture
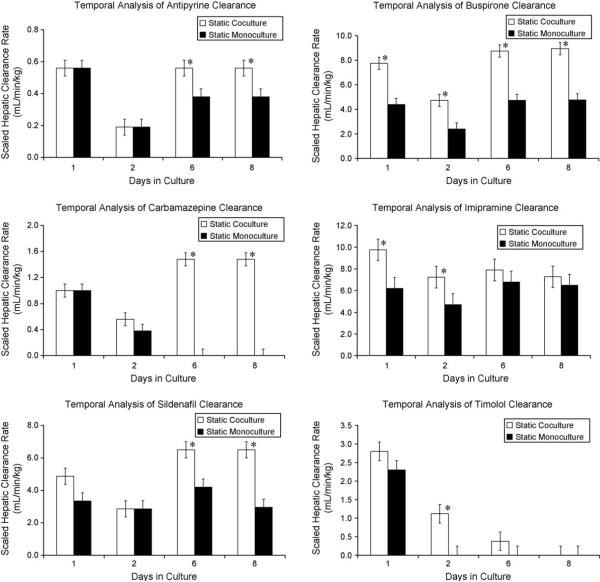
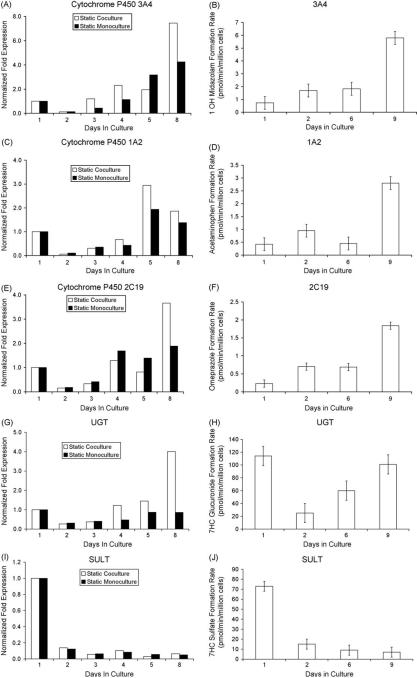
In the current report, we describe an integrated microfluidic platform that combines a previously described HuREL microfluidics device [18] with a hepatic coculture system. As the first step in these studies, we used the coculture system in a static environment to measure the clearance of nine compounds (described in Section 2) over a period of 10 days of culture, with media changes occurring prior to the addition of the drug every the other day. A representative set of results is shown in Fig. 1 which compares the clearance of six compounds in monoculture and coculture formats. In virtually all cases, the coculture outperformed the monoculture, and in general maintained activity over longer periods of time. In a second set of studies, we assessed CYP and phase II enzyme gene expression in both formats, and also measured metabolic formation rates for a variety of metabolites. As shown in Fig. 2 messenger RNA for CYP 3A4, 1A2, 2C19, and the phase II enzyme UGT were all expressed at significant levels throughout the period of analysis. In addition to monitoring gene expression, we also measured metabolite formation of key compounds on multiple days throughout the culture period. As can be seen in Fig. 2(B, D, F, H, J) the temporal trend in metabolite formation rates parallels that of mRNA expression.
Fig. 1.
Metabolic clearance for representative compounds under static culture conditions. Each data point represents the clearance of the parent compound in terms of (mL/min/kg). Clearance rates were evaluated at 1, 2 and 6 days after initial cell seeding. Each time point includes clearance rates of the static coculture (white) and static monoculture (black). The data were presented as a mean ± standard deviation with at least three replicates. Asterisks (*) indicate a statistically significant difference (p < 0.05) as determined by a Student's t-test.
Fig. 2.
RNA expression and metabolite generation under static culture conditions. (A, C, E, G, I) demonstrate the temporal RNA fold expression for the static coculture (white) and static monoculture (black) systems, determined using quantitative PCR methods. (B, D, F, H, J) demonstrate the corresponding enzymatic function in the static coculture system. The data were presented as a mean ± standard deviation with at least three replicates.
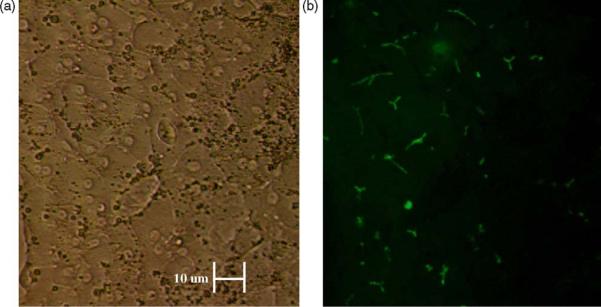
As a final step of coculture characterization, we probed whether the cocultured hepatocytes expressed bile canaliculi. This was accomplished by staining static cultures with 5(6)-carboxy-2′,7′- dichlorofluorescein diacetate as described in Section 2, at days 1–10. Positive staining for bile canaliculi was observed as early as day 4, and was maintained through day 10. A representative image is presented in Fig. 5, from the coculture at day 6 (Fig. 3).
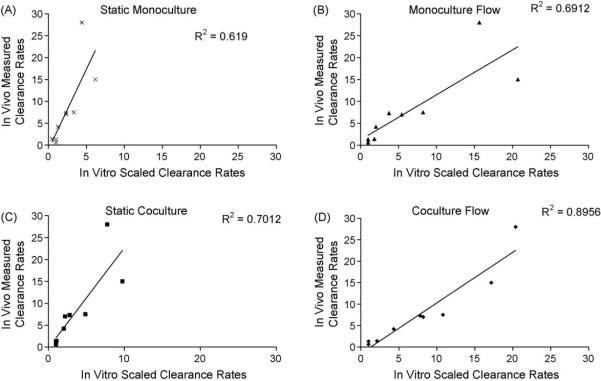
Fig. 5.
In vitro in vivo correlation at day 1 for nine benchmarked compounds. The IVIVC calculations were generated based on scaled clearance data for nine compounds for each of the four culture conditions: (a) Static monoculture; (b) Monoculture flow; (c) Static coculture; (d) Coculture flow. To scale the static systems, an in vitro intrinsic clearance rate was first determined and then scaled to an in vivo value using a well stirred model. For the flow systems, and intrinsic clearance rate was determined, used to calculate an extraction ratio, and then scaled to an in vivo value by multiplying by the liver flow rate. The predicted in vivo values were then compared to published clinical data to obtain the R-squared values.
Fig. 3.
Functional polarization in static coculture. (a) Phase micrograph of primary human hepatocytes in static cocultures following 6 days of culture. (b) Fluorescence image of phase three transporter activity in primary human hepatocytes in cocultures (day 6). CDFDA is internalized by hepatocytes, cleaved by intracellular esterases and excreted into bile canaliculi as fluorescent CDF by active MRP2. The scale bar represents a distance of 10 um.
3.2. Performance of optimized culture under flow
After demonstrating superior performance in our coculture system, we then plated the coculture cell suspension within the first chamber of the HμREL® chip, termed the hepatocyte chamber. After cell seeding, the chip was mounted within the device housing, and flow was applied. The first set of experiments conducted with the combined coculture and flow platform was an evaluation of the clearance of compounds used in the prior analysis of the static coculture system. Fig. 4 shows results from three compounds each representing a high, moderate, or slow clearing compound.
Fig. 4.
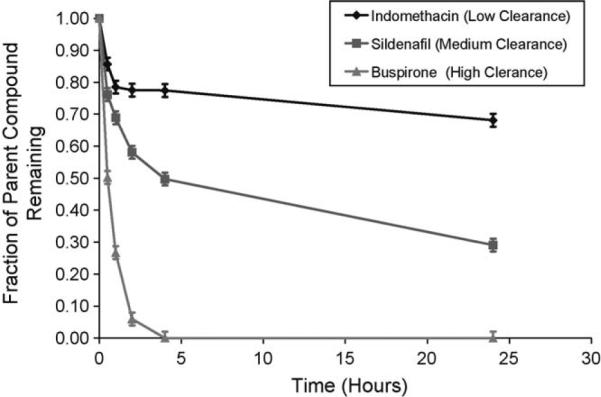
Metabolic profiles of model compounds by human hepatocytes cocultured under flow. Fraction of remaining indomethacin (◆, low clearance), sildenafil (■, medium clearance), and buspirone (▲, low clearance) with initial concentration of 1 μM was plotted as a function of time. The data were presented as a mean ± standard deviation with at least three replicates.
We next evaluated whether the clearance data obtained using the integrated coculture and flow platform, would yield superior in vivo clearance prediction. To accomplish this, we analyzed the clearance of nine compounds by human hepatocytes cultured under four conditions: flow based culture in the presence and absence of nonparenchymal cells, and static culture in the presence and absence of the nonparenchymal cells. The intrinsic rates determined for the static system were scaled, as previously described [18]. Extraction ratios for the flow system were calculated and scaled to in vivo values, also as previously described [18]. As can be seen from regression of the data (Fig. 5), a R-squared coefficient of 0.9 is obtained for the coculture system under flow, whereas poorer correlations are obtained for the monoculture flow and static coculture systems (0.7), and for the static monoculture system (0.6).
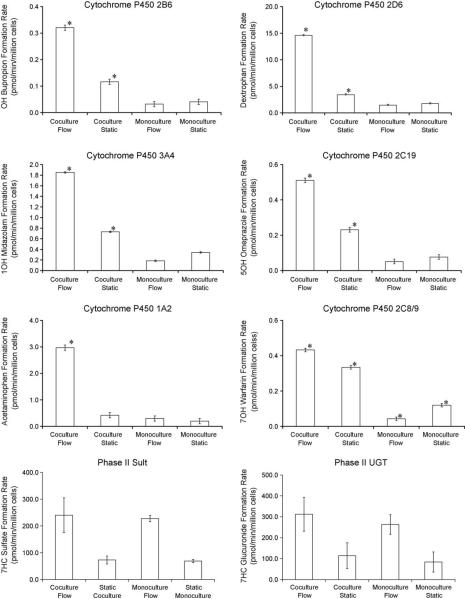
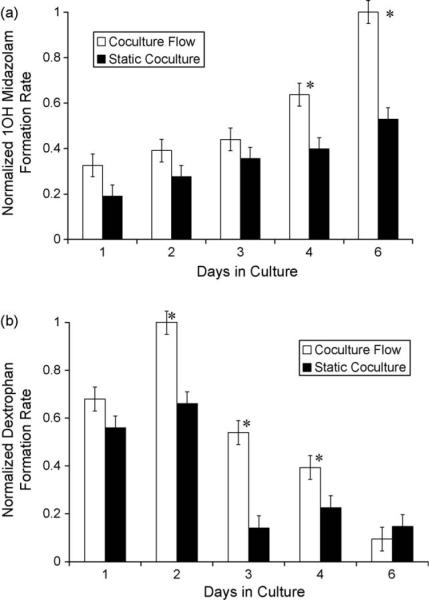
In addition to assaying hepatic clearance, we also evaluated metabolite generation in the integrated system, in a manner similar to the multi day static experiment. As can be seen in Fig. 6, the coculture system in conjunction with flow outperformed the static cultures as well as monoculture under flow, except in the case of phase II enzymes (SULT, UGT) at day 1. Furthermore, when extending the analysis to multiple days, in this case in the metabolism of midazolam (Fig. 7), it appears that the increased performance obtained from adding flow to the coculture system is also maintained.
Fig. 6.
Metabolite formation rates on day 1 of culture. The concentrations of the metabolites were monitored over time, and used to calculate metabolite formation rates. Each plot represents a different enzyme. The data were presented as a mean ± standard deviation with at least three replicates. Asterisks (*) indicate a statistically significant difference (p < 0.05) from other culture conditions as determined by a Student's t-test.
Fig. 7.
Midazolam (a) and dextromethorphan (b) metabolism under flow (white) and in static culture (black). The concentrations of the metabolites were monitored over time, and used to calculate metabolite formation rates. The formation rates were then scaled with respect to the highest formation rate observed over the 6 day study, for each metabolite independently. The data were presented as a mean ± standard deviation with at least three replicates. Asterisks (*) indicate a statistically significant difference (p < 0.05) as determined by a Student's t-test.
4. Discussion
A major effort in drug discovery is focused on evaluating the hepatic metabolism and pharmacokinetics of new molecular entities. However, with currently incumbent in vitro screening systems, drug discovery and preclinical development are at times limited in their predictive capabilities [21,22]. One retrospective study of a variety of randomly selected investigational drugs estimated that a single 10% improvement in preclinical screens could reduce total cost of drug development by over $100 million per approved drug [23]. The development of rapid and predictive preclinical screens requires the engineering of new microenvironments which more completely recapitulate tissue microenvironments. With respect to this goal we have developed a robust culture system that provides an improved coculture method for use in conjunction with the previously described Hurel microfluidics system [18].
It is important to note that when using cells from human subjects, initial levels of metabolic competency vary from donor to donor. This is unavoidable and drives the need for evaluating culture systems such as the one described herein with multiple donor lots of varying initial phase I and II enzyme expression levels. It may also be beneficial to combine a number of lots into one multi-lot culture which may be more representative of the overall population. However, irrespective of initial enzymatic levels, maintenance of those levels is the focus of our studies. Many prior studies have shown that long-term hepatocyte viability and function are maintained during coculture with fibroblasts or endothelial cells [24–26]. It is thought that this synergistic interaction is mediated through a combination of cell-cell contacts, secreted extracellular matrix (ECM) as well as soluble factors. In our studies as well, we demonstrated the benefit of coculture with a nonparenchymal cell type with respect to the maintenance of metabolic competency for the majority of Cytochrome P450 enzymes (3A4, 1A2, 2C19, 2C9), as well as the phase II enzyme UGT. Furthermore, we also demonstrated that bile canaliculi appear as early as day 4 after plating. Future experiments will address the maintenance of other phase I and II enzymes involved in drug metabolism as well as the use of substrates that demonstrate Michaelis-Menten kinetics at clinical concentrations.
Following optimization of the coculture system, we tested its functional capacity within the Hurel microfluidics device [18]. There is a body of evidence suggesting that fluid flow applied to various culture systems can result in better function via a variety of means including: (1) increased mass transport [27], (2) maintenance of better enzymatic activities for long term culture of primary hepatocytes [1,15] and of cell lines [28], (3) better preservation of the viability and morphology of liver tissue slices [29], and (4) better metabolism of xenobiotics by hepatocytes [16,30–32]. In our studies, we have shown that adding the element of fluid flow to the coculture system provides resolution with regard to clearance of compounds with different clearance rates, a result which is often difficult to obtain in static culture systems [33]. As an illustrative plot, Fig. 4 shows the discrimination of three different categories of compound clearance, namely buspirone, a high clearance compound with an in vivo clearance value of 28 mL/min/kg, sildenafil, a medium clearance compound with an in vivo clearance value of 7.5 mL/min/kg, and indomethacin, a low clearance compound with an in vivo clearance value of 1.4 mL/min/kg. When this analysis was then applied across all four culture conditions (i.e. coculture versus monoculture, and static verses flow) to determine the degree of in vitro in vivo correlation for nine compounds, we found that a greater correlation was obtained for the flow systems with an R-squared value of 0.69 for the monoculture flow system, 0.89 for the coculture flow system, as compared to 0.61, and 0.70 for the static monoculture and static coculture systems, respectively.
We also conducted a comparison of the four conditions mentioned above with respect to metabolite formation. Again the system with coculture and fluid flow showed an increased metabolite formation rate as compared to either of the static systems or the monoculture flow system. Furthermore, this increased formation was maintained over multiple days. Although this study addressed the formation of primary metabolites, it is understood that in drug development full metabolite profiles must be generated and that these results (i.e. larger quantities of metabolites generated with an equivalent number of cells) will become more important as secondary and tertiary metabolite generation and identification takes a more prominent role in drug evaluation [34,35].
As to the reason for our observations of increased functional capacity under flow, we can offer two initial hypotheses. The first hypothesis is that the addition of flow has a direct effect on the cells leading to an up-regulation of key functional genes [36,37]. While this is plausible and has been demonstrated for other culture systems [38], we conducted PCR analysis on the cells under flow at multiple time points (data not shown), and observed no major shift in the gene profile for cells under flow, as compared to cells in the static condition. The second hypothesis is that the fluid flow simply increases mass transport within the system. The benefit in the flow system may be two-fold, with a thinner boundary layer forming than in the static system leading to higher clearance for transport limited reactions or additional enhanced transport because of the shallower overall fluid volume above the cells as compared to the situation with the conventional 96 well plate in the static culture embodiment. In addition, increased mass transport may potentially remove unwanted by-products [39,40]. A third possibility involves shear stress induced cellular uptake [41]. We are currently performing additional detailed experiments and analysis in order to better characterize the mechanism of enhanced function.
In summary, we have developed a microfluidic, hepatic cell-based assay platform that combines the attributes of coculture and flow, and which has yielded both superior metabolite generation and superior IVIVC prediction, when compared to traditional culture approaches. Current and future studies are focused on developing an understanding of the mechanisms underlying the improved function observed, and on developing additional capabilities of this system.
References
- [1].Zeilinger K, Sauer IM, Pless G, Strobel C, Rudzitis J, Wang A, et al. Three-dimensional co-culture of primary human liver cells in bioreactors for in vitro drug studies: effects of the initial cell quality on the long-term maintenance of hepatocyte-specific functions. Altern Lab Anim. 2002;30:525–38. doi: 10.1177/026119290203000506. [DOI] [PubMed] [Google Scholar]
- [2].Bhatia SN, Balis UJ, Yarmush ML, Toner M. Microfabrication of hepatocyte/fibroblast co-cultures: role of homotypic cell interactions. Biotechnol Prog. 1998;14:378–87. doi: 10.1021/bp980036j. [DOI] [PubMed] [Google Scholar]
- [3].Allen JW, Khetani SR, Bhatia SN. In vitro zonation and toxicity in a hepatocyte bioreactor. Toxicol Sci. 2005;84:110–9. doi: 10.1093/toxsci/kfi052. [DOI] [PubMed] [Google Scholar]
- [4].Rotem A, Toner M, Tompkins RG, Yarmush ML. Oxygen uptake rates in cultured rat hepatocytes. Biotechnol Bioeng. 1992;40:1286–91. doi: 10.1002/bit.260401020. [DOI] [PubMed] [Google Scholar]
- [5].Borel Rinkes IH, Toner M, Tompkins RG, Yarmush ML. An extracorporeal microscopy perfusion chamber for on-line studies of environmental effects on cultured hepatocytes. J Biomech Eng. 1994;116:135–9. doi: 10.1115/1.2895711. [DOI] [PubMed] [Google Scholar]
- [6].Stevens EJ, Ryan CM, Friedberg JS, Barnhill RL, Yarmush ML, Tompkins RG. A quantitative model of invasive pseudomonas infection in burn injury. J Burn Care Rehabil. 1994;15:232–5. doi: 10.1097/00004630-199405000-00005. [DOI] [PubMed] [Google Scholar]
- [7].Matthew HW, Sternberg J, Stefanovich P, Morgan JR, Toner M, Tompkins RG, et al. Effects of plasma exposure on cultured hepatocytes: implications for bioartificial liver support. Biotechnol Bioeng. 1996;51:100–11. doi: 10.1002/(SICI)1097-0290(19960705)51:1<100::AID-BIT12>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- [8].Stefanovich P, Matthew HW, Toner M, Tompkins RG, Yarmush ML. Extracorporeal plasma perfusion of cultured hepatocytes: effect of intermittent perfusion on hepatocyte function and morphology. J Surg Res. 1996;66:57–63. doi: 10.1006/jsre.1996.0372. [DOI] [PubMed] [Google Scholar]
- [9].Ledezma GA, Folch A, Bhatia SN, Balis UJ, Yarmush ML, Toner M. Numerical model of fluid flow and oxygen transport in a radial-flow microchannel containing hepatocytes. J Biomech Eng. 1999;121:58–64. doi: 10.1115/1.2798043. [DOI] [PubMed] [Google Scholar]
- [10].Tilles AW, Baskaran H, Roy P, Yarmush ML, Toner M. Effects of oxygenation and flow on the viability and function of rat hepatocytes cocultured in a micro-channel flat-plate bioreactor. Biotechnol Bioeng. 2001;73:379–89. doi: 10.1002/bit.1071. [DOI] [PubMed] [Google Scholar]
- [11].Shito M, Kim NH, Baskaran H, Tilles AW, Tompkins RG, Yarmush ML, et al. In vitro and in vivo evaluation of albumin synthesis rate of porcine hepatocytes in a flat-plate bioreactor. Artif Organs. 2001;25:571–8. doi: 10.1046/j.1525-1594.2001.025007571.x. [DOI] [PubMed] [Google Scholar]
- [12].Roy P, Baskaran H, Tilles AW, Yarmush ML, Toner M. Analysis of oxygen transport to hepatocytes in a flat-plate microchannel bioreactor. Ann Biomed Eng. 2001;29:947–55. doi: 10.1114/1.1415524. [DOI] [PubMed] [Google Scholar]
- [13].Roy P, Washizu J, Tilles AW, Yarmush ML, Toner M. Effect of flow on the detoxification function of rat hepatocytes in a bioartificial liver reactor. Cell Transplant. 2001;10:609–14. [PubMed] [Google Scholar]
- [14].Bader A, Fruhauf N, Zech K, Haverich A, Borlak JT. Development of a small-scale bioreactor for drug metabolism studies maintaining hepatospecific functions. Xenobiotica. 1998;28:815–25. doi: 10.1080/004982598239074. [DOI] [PubMed] [Google Scholar]
- [15].Schmitmeier S, Langsch A, Jasmund I, Bader A. Development and characterization of a small-scale bioreactor based on a bioartificial hepatic culture model for predictive pharmacological In vitro screenings. Biotechnol Bioeng. 2006;95:1198–206. doi: 10.1002/bit.21089. [DOI] [PubMed] [Google Scholar]
- [16].Kane BJ, Zinner MJ, Yarmush ML, Toner M. Liver-specific functional studies in a microfluidic array of primary mammalian hepatocytes. Anal Chem. 2006;78:4291–8. doi: 10.1021/ac051856v. [DOI] [PubMed] [Google Scholar]
- [17].Sivaraman A, Leach JK, Townsend S, Iida T, Hogan BJ, Stolz DB, et al. A microscale In vitro physiological model of the liver: predictive screens for drug metabolism and enzyme induction. Curr Drug Metab. 2005;6:569–91. doi: 10.2174/138920005774832632. [DOI] [PubMed] [Google Scholar]
- [18].Chao P, Maguire TJ, Novik E, Cheng KC, Yarmush ML. Evaluation of a microfluidic based cell culture platform with primary human hepatocytes for the prediction of hepatic clearance in human. Biochem Pharmacol. 2009;78(6):625–32. doi: 10.1016/j.bcp.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pang KS, Rowland M. Hepatic clearance of drugs. I. Theoretical considerations of a “well-stirred” model and a “parallel tube” model. Influence of hepatic blood flow, plasma and blood cell binding, and the hepatocellular enzymatic activity on hepatic drug clearance. J Pharmacokinet Biopharm. 1977;5:625–53. doi: 10.1007/BF01059688. [DOI] [PubMed] [Google Scholar]
- [20].Li C, Liu T, Cui X, Uss AS, Cheng KC. Development of In vitro pharmacokinetic screens using Caco-2, human hepatocyte, and Caco-2/human hepatocyte hybrid systems for the prediction of oral bioavailability in humans. J Biomol Screen. 2007;12:1084–91. doi: 10.1177/1087057107308892. [DOI] [PubMed] [Google Scholar]
- [21].Chiba M, Ishii Y, Sugiyama Y. Prediction of Hepatic Clearance in Human From In vitro Data for Successful Drug Development. AAPS J. 2009 doi: 10.1208/s12248-009-9103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Houston JB, Galetin A. Methods for predicting in vivo pharmacokinetics using data from In vitro assays. Curr Drug Metab. 2008;9:940–51. doi: 10.2174/138920008786485164. [DOI] [PubMed] [Google Scholar]
- [23].Cavero I. Optimizing the preclinical/clinical interface: an Informa Life Sciences conference; London, UK. 12–13 December, 2006; Expert Opin Drug Saf 2007;6:217-24. [DOI] [PubMed] [Google Scholar]
- [24].van Poll D, Sokmensuer C, Ahmad N, Tilles AW, Berthiaume F, Toner M, et al. Elevated hepatocyte-specific functions in fetal rat hepatocytes co-cultured with adult rat hepatocytes. Tissue Eng. 2006;12:2965–73. doi: 10.1089/ten.2006.12.2965. [DOI] [PubMed] [Google Scholar]
- [25].Cho CH, Park J, Nagrath D, Tilles AW, Berthiaume F, Toner M, et al. Oxygen uptake rates and liver-specific functions of hepatocyte and 3T3 fibroblast co-cultures. Biotechnol Bioeng. 2007;97:188–99. doi: 10.1002/bit.21225. [DOI] [PubMed] [Google Scholar]
- [26].Washizu J, Berthiaume F, Mokuno Y, Tompkins RG, Toner M, Yarmush ML. Long-term maintenance of cytochrome P450 activities by rat hepatocyte/3T3 cell co-cultures in heparinized human plasma. Tissue Eng. 2001;7:691–703. doi: 10.1089/107632701753337654. [DOI] [PubMed] [Google Scholar]
- [27].Hansen C, Quake SR. Microfluidics in structural biology: smaller, faster em leader better. Curr Opin Struct Biol. 2003;13:538–44. doi: 10.1016/j.sbi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- [28].Gebhardt R, Lippert C, Schneider A, Doehmer J. Improved Determination of drug metabolism by perifusion of recombinant V79 cells carrying human CYP3A4. Toxicol In vitro. 1999;13:639–43. doi: 10.1016/s0887-2333(99)00020-x. [DOI] [PubMed] [Google Scholar]
- [29].Schumacher K, Khong YM, Chang S, Ni J, Sun W, Yu H. Perfusion culture improves the maintenance of cultured liver tissue slices. Tissue Eng. 2007;13:197–205. doi: 10.1089/ten.2006.0046. [DOI] [PubMed] [Google Scholar]
- [30].Viravaidya K, Shuler ML. Incorporation of 3T3-L1 cells to mimic bioaccumulation in a microscale cell culture analog device for toxicity studies. Biotechnol Prog. 2004;20:590–7. doi: 10.1021/bp034238d. [DOI] [PubMed] [Google Scholar]
- [31].Viravaidya K, Sin A, Shuler ML. Development of a microscale cell culture analog to probe naphthalene toxicity. Biotechnol Prog. 2004;20:316–23. doi: 10.1021/bp0341996. [DOI] [PubMed] [Google Scholar]
- [32].Gebhardt R, Wegner H, Alber J. Perifusion of co-cultured hepatocytes: optimization of studies on drug metabolism and cytotoxicity In vitro. Cell Biol Toxicol. 1996;12:57–68. doi: 10.1007/BF00143356. [DOI] [PubMed] [Google Scholar]
- [33].Obach RS. Prediction of human clearance of twenty-nine drugs from hepatic microsomal intrinsic clearance data: an examination of In vitro half-life approach and nonspecific binding to microsomes. Drug Metab Dispos. 1999;27:1350–9. [PubMed] [Google Scholar]
- [34].Leclercq L, Cuyckens F, Mannens GS, de Vries R, Timmerman P, Evans DC. Which human metabolites have we MIST? Retrospective analysis, practical aspects, and perspectives for metabolite identification and quantification in pharmaceutical development. Chem Res Toxicol. 2009;22:280–93. doi: 10.1021/tx800432c. [DOI] [PubMed] [Google Scholar]
- [35].Tolonen A, Turpeinen M, Pelkonen O. Liquid chromatography-mass spectrometry in In vitro drug metabolite screening. Drug Discov Today. 2009;14:120–33. doi: 10.1016/j.drudis.2008.11.002. [DOI] [PubMed] [Google Scholar]
- [36].Jekir MG, Donahue HJ. Gap junctions and osteoblast-like cell gene expression in response to fluid flow. J Biomech Eng. 2009;131:011005. doi: 10.1115/1.3005201. [DOI] [PubMed] [Google Scholar]
- [37].Abkarian M, Faivre M, Horton R, Smistrup K, Best-Popescu CA, Stone HA. Cellular-scale hydrodynamics. Biomed Mater. 2008;3:034011. doi: 10.1088/1748-6041/3/3/034011. [DOI] [PubMed] [Google Scholar]
- [38].Papachristou DJ, Papachroni KK, Basdra EK, Papavassiliou AG. Signaling networks and transcription factors regulating mechanotransduction in bone. Bioessays. 2009 doi: 10.1002/bies.200800223. [DOI] [PubMed] [Google Scholar]
- [39].Korin N, Bransky A, Khoury M, Dinnar U, Levenberg S. Design of well and groove microchannel bioreactors for cell culture. Biotechnol Bioeng. 2009;102:1222–30. doi: 10.1002/bit.22153. [DOI] [PubMed] [Google Scholar]
- [40].Cimetta E, Figallo E, Cannizzaro C, Elvassore N, Vunjak-Novakovic G. Micro-bioreactor arrays for controlling cellular environments: design principles for human embryonic stem cell applications. Methods. 2009;47:81–9. doi: 10.1016/j.ymeth.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hallow DM, Seeger RA, Kamaev PP, Prado GR, LaPlaca MC, Prausnitz MR. Shear-induced intracellular loading of cells with molecules by controlled micro-fluidics. Biotechnol Bioeng. 2008;99:846–54. doi: 10.1002/bit.21651. [DOI] [PMC free article] [PubMed] [Google Scholar]