Abstract
Sib matings increase homozygosity and, hence, the frequency of detrimental phenotypes caused by recessive deleterious alleles. However, many species have evolved adaptations that prevent the genetic costs associated with inbreeding. We discovered that the highly invasive longhorn crazy ant, Paratrechina longicornis, has evolved an unusual mode of reproduction whereby sib mating does not result in inbreeding. A population genetic study of P. longicornis revealed dramatic differences in allele frequencies between queens, males and workers. Mother–offspring analyses demonstrated that these allele frequency differences resulted from the fact that the three castes were all produced through different means. Workers developed through normal sexual reproduction between queens and males. However, queens were produced clonally and, thus, were genetically identical to their mothers. In contrast, males never inherited maternal alleles and were genetically identical to their fathers. The outcome of this system is that genetic inbreeding is impossible because queen and male genomes remain completely separate. Moreover, the sexually produced worker offspring retain the same genotype, combining alleles from both the maternal and paternal lineage over generations. Thus, queens may mate with their brothers in the parental nest, yet their offspring are no more homozygous than if the queen mated with a male randomly chosen from the population. The complete segregation of the male and female gene pools allows the queens to circumvent the costs associated with inbreeding and therefore may act as an important pre-adaptation for the crazy ant's tremendous invasive success.
Keywords: ants, parthenogenesis, inbreeding, invasiveness, reproductive strategies
1. Introduction
Inbreeding increases the frequency of detrimental phenotypes caused by recessive deleterious mutations. As a result, inbred families frequently display major abnormalities, such as mutant lethal phenotypes or genetic diseases. Even in the absence of overt major abnormalities, inbreeding depression is detectable by the lower fertility, survival and growth rates of individuals with high inbreeding coefficients (see [1] for review).
In Hymenoptera (ants, bees and wasps), an additional cost associated with inbreeding stems from the unusual mechanism of sex determination, whereby sex is usually determined by a single sex-determining locus (i.e. single-locus complementary sex determinism, sl-CSD). Under sl-CSD in haplodiploid organisms such as Hymenoptera, unfertilized haploid eggs develop into males, and diploid eggs develop into females if heterozygous at the sex-determining locus. However, individuals homozygous at the sex-determining locus develop into diploid males [2–6]. Thus, whenever a queen mates with a male possessing one of her alleles at the sex-determining locus, half of the diploid offspring produced will develop into diploid males. Because diploid males are usually inviable or sterile, inbreeding may result in large costs for the colony [7,8] (but see [9]).
Ants have evolved various adaptations to prevent inbreeding. For example, colonies often produce reproductive individuals of only a single sex, thereby precluding the possibility of mating with relatives. In addition, these individuals depart on highly synchronized mating flights composed of individuals from a large number of colonies, so that matings between related individuals are unlikely [10]. In a few species, however, mating does occur within the nest. Ant species that display nestmate mating are usually characterized by the presence of many queens per colony and a unicolonial mode of social organization whereby individuals mix freely among physically separated nests. Thus, in these cases, the porosity of colony boundaries leads to a lower probability of inbreeding, because queens are likely to mate with males to whom they are not directly related. Interestingly, unicoloniality is also a key attribute responsible for the ecological dominance of several ant species [11–13] and their aptitude to invade new habitats. Indeed, of the 17 land invertebrate species listed among the world's worst invaders [14], five are ant species with documented or inferred unicolonial structures.
The ability of introduced populations to thrive in a new habitat is not trivial, because introduction is generally associated with population genetic bottlenecks. The resulting drop in allelic diversity accompanying bottlenecks increases the level of homozygosity relative to the native population. Therefore, introduction into new habitats may yield consequences similar to those of conventional inbreeding in terms of increasing the frequency of detrimental phenotypes owing to recessive deleterious mutations. For example, all introduced populations of invasive ants studied so far possess fewer alleles than native populations [11,15–19], and introduced populations of the fire ant Solenopsis invicta [8,15] produce high proportions of diploid males.
Recent studies have revealed several cases of unexpected modes of reproduction in ants [20–24]. In these species, queens have been found to use selectively sexual and asexual reproduction to produce new workers and queens, respectively. In this study, we describe the unusual reproduction system of yet another ant species, the longhorn crazy ant Paratrechina longicornis (Latreille 1802). This ant is a highly invasive agricultural and household pest throughout much of the tropics and subtropics, and a pervasive indoor pest in temperate areas [25]. Our genetic analyses of an introduced population show that queens were clonally produced from their mother, males clonally produced from their father and workers produced sexually. We demonstrate that this mode of reproduction allows P. longicornis colonies to maintain heterozygosity over generations, despite population bottlenecks and matings between individuals produced from the same mother.
2. Material and methods
(a). Collections
We collected seven P. longicornis nests in the summer of 2008 and spring of 2009 from a single population in Bangkok, Thailand, near Kasetsart University (GPS: 13.85° N, 100.57° W). The nests were brought back to the laboratory and subdivided to produce new daughter colonies. Specifically, we isolated 21 queens (hereafter referred to as F0 queens) with at least 50 nestmate workers to establish 21 single-queen laboratory colonies. The colonies were kept in climate chambers at a constant temperature of 26°C and 12 L : 12 D cycle. Food (honey, fruits, cockroaches, crickets and mealworms) and water were provided ad libitum. After three months, all the pupae were known to be the offspring of the resident F0 queen. At this time, we collected the queen and 12 worker, gyne (winged virgin queen) and male pupae (F1 offspring) from each colony, if these samples were available. In total, we sampled 89 gyne (mean ± s.d. = 7.4 ± 4.8 from 12 colonies), 139 male (7.7 ± 4.8 from 18 colonies) and 252 worker (12.0 ± 0 from 21 colonies) F1 pupae.
(b). Sib mating
Seven colonies produced more F1 sexual pupae of both sexes than we actually sampled. These F1 gyne and male pupae were left in their mother colony and raised into adult sexuals by nestmate workers. After three weeks, at least one F1 queen shed her wings in each of the seven colonies, suggesting that this queen had mated with an F1 nestmate brother (hereafter the terms brothers, sisters or sibs are defined as individuals produced by the same queen). We isolated one such F1 queen with at least 50 nestmate workers from each colony. We allowed these F1 queens to survive for three more months, at which point we sampled the seven F1 queens, 12 worker pupae and up to 12 gyne and male pupae (depending on availability) from each F1 colony. Overall, we sampled 84 worker (12 ± 0 per colony), 65 gyne (9.3 ± 3 per colony) and 84 male (12 ± 0 per colony) F2 pupae from the seven colonies.
(c). Genotyping
DNA was extracted from whole individual pupae, and from the head and thorax of mated queens, using a Biosprint 96 workstation (Qiagen, CA, USA) following the manufacturer's protocols. Each individual was genotyped at the 13 polymorphic microsatellite loci: Plon-1.B9, Plon-1.H3, Plon-3.C4, Plon-4.E6, Plon-5.C3, Plon-5.F7, Plon-7.H4, Plon-8.A8, Plon-8.F2, Plon-8.G5, Plon-9.A4, Plon-10.B7 and Plon-10.E7 [26]. Amplified fluorescent fragments were visualized using an ABI Prism 3100 automated DNA sequencer. Microsatellite alleles were scored using GeneMapper software v. 4.0 (Applied Biosystems). One of the 13 loci (Plon-7.H4) was found to possess only a single allele and therefore was not considered further.
Genotypes of F0, F1 and F2 individuals could be determined directly, while paternal genotypes were inferred from worker and maternal genotypes. Inference of paternal genotype is generally straightforward in the Hymenoptera, given that males are haploid. In this population, inference was further facilitated by the fact that males and queens had almost no alleles in common (§3). All statistical tests on genotype frequencies were conducted using the software R [27].
3. Results
(a). Population genetic patterns
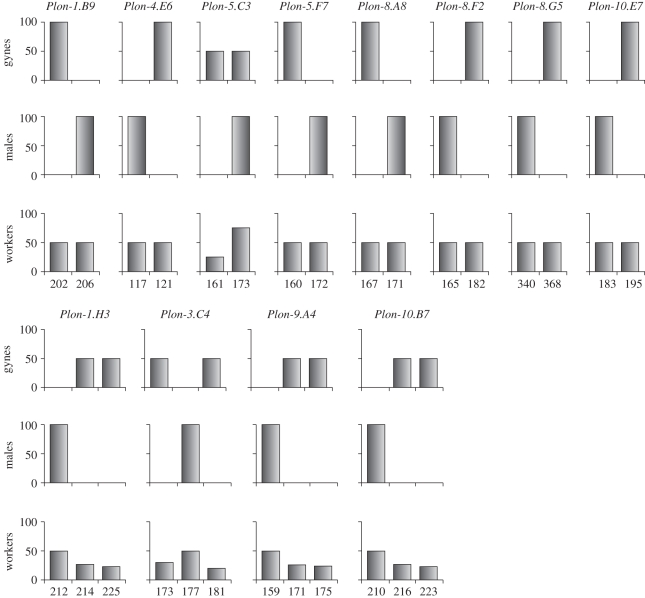
Our population genetic analyses revealed dramatic differences in allele frequencies among queens, males and workers (figure 1). The differences were most extreme between queens and males: for 11 of the 12 polymorphic loci, queens and males had no alleles in common. At all 12 loci, workers exhibited allele frequencies intermediate between those of queens and males.
Figure 1.
Allele frequencies in P. longicornis queens, workers and males sampled from a single population. Frequencies were inferred from 89 gyne, 139 male and 252 worker genotypes.
We suspected that these very unusual genotype frequencies resulted from a combination of sexual and asexual reproduction, whereby workers were produced by normal sexual reproduction, and queens and males were produced asexually. Under this scenario, queens would inherit only maternal alleles and males only paternal alleles (figure 2; §4). To test whether this was indeed the case, we analysed the genotypes of the F0 and F1 individuals produced in the laboratory. Consistent with a generalized system of clonal reproduction of queens and males, we found that all 21 mother (F0) queens had identical genotypes at all 12 loci, and the inferred genotype of the queens' mates were also all identical to each other (but had different alleles than those in queens). Moreover, all 89 F1 queens produced in the laboratory harboured exclusively maternal alleles, and were genetically identical to their mother (F0) queens and, therefore, to each other. The mother–offspring analyses also revealed a complete lack of recombination; all F0 and F1 queens were heterozygotes at five of the 12 loci and homozygous at the remaining seven loci (all queens had the same allele at these seven loci). The complete lack of recombination indicated that thelytokous parthenogenesis (i.e. the production of diploid females from unfertilized eggs) was apomictic (i.e. ameiotic) [28,29].
Figure 2.
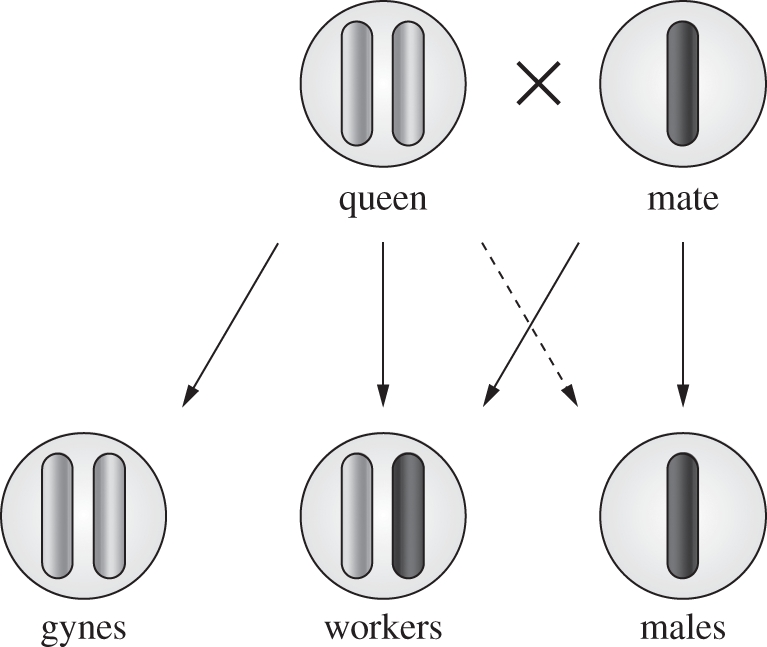
Clonal reproduction in queens and males. The figure summarizes the reproduction system of P. longicornis in the study population. Maternal (light) and paternal (dark) chromosomes are displayed. Contribution to the genome of the offspring is indicated by arrows (dashed arrow represents the mother laying haploid eggs with no actual contribution to the genome).
In stark contrast to the F1 queens, the 139 F1 males never carried any maternal alleles at 11 of the 12 loci studied. The only allele shared between queens and their sons was at the locus Plon-5.C3, where queens were always heterozygous (202/206) and their sons always hemizygous for the 206 allele. Importantly, the genotypes of males were always identical to those of their mother queens’ mates (as inferred by mother–worker analysis) at all 12 loci. Thus, all 139 males were genetic clones of the F0 queens’ mates.
Finally, all but one of the 252 F1 workers had one maternal allele and one paternal allele, demonstrating that workers were sexually produced. The exception was one worker that exhibited exclusively maternal alleles at all loci and was thus parthenogenetically produced (as for new queens).
(b). Sib mating experiments
The young virgin queens raised in the seven laboratory colonies readily mated with their brothers. Parent–offspring analyses between F1 and F2 offspring confirmed that all F2 queens were clonally produced by their mothers and all F2 workers were sexually produced (electronic supplementary material, table S1). The comparison across generations revealed that the level of heterozygosity was similar between queens of both generations as well as between workers of both generations (workers: Ho(F1) = 0.951 ± 0.053, Ho(F2) = 0.946 ± 0.040, p = 0.184, n.s.; gynes: Ho(F1) = 0.417 ± 0.000, Ho(F2) = 0.417 ± 0.000, p = 1.000, n.s.).
4. Discussion
Our genetic analyses revealed an unusual mode of reproduction in an introduced population of the longhorn crazy ant P. longicornis. We found that P. longicornis workers arose from classical sexual reproduction, whereas queens were clones of their mothers and males were clones of their fathers. This latter result was particularly surprising because male ants usually receive no paternal alleles at all. Laboratory mating experiments showed that this mode of reproduction allowed the queens to mate with their brothers inside the nest, and yet maintain heterozygosity in the queen and worker castes over an unlimited number of generations.
Surprisingly, the heterozygosity level of new queens is completely independent of the genealogical link between the mother queen and her mate in this species, as there is no mixing of the paternal and maternal lineages. By contrast, the level of worker heterozygosity depends on the genetic similarity between the maternal and paternal lineages, because workers are sexually produced. But since all queens and all males in our study population had the same genotype, queens were no more genetically similar to their brothers than to any other male in the population, and the level of worker heterozygosity was therefore not affected by whether queens mated with their brothers or other males.
The particular reproductive system of P. longicornis probably provides an important pre-adaptation allowing this species to colonize new habitats. Paratrechina longicornis is arguably the most broadly dispersed of all ant species, distributed widely across the Old World and New World in both the Northern Hemisphere and Southern Hemisphere [25]. The multiple introductions of this species have probably been accompanied by repeated reductions in population size during the founding stages of new populations. However, the complete separation of the maternal and paternal genomes allows such founding events to occur without any cost associated with inbreeding (i.e. mating between related individuals) in workers. Whatever the population size, a high level of heterozygosity can be maintained in workers as a result of the very stark genetic differences between the paternal and maternal lineages. Population bottlenecks only affect the allelic diversity at the population level, without decreasing observed heterozygosity of queens and workers. In the studied population, there was actually no genotypic diversity within either the maternal or paternal clonal lineages, yet the observed heterozygosity in workers was extremely high (across the 12 microsatellite loci, observed heterozygosity was close to 1; electronic supplementary material, table S1).
Recent studies have reported similar reproductive strategies involving clonal reproduction of both sexes in two other ant species: Wasmannia auropunctata and Vollenhovia emeryi [21,22]. On the basis of the results of our study, it is likely that the mating system of these species also translates into reduced effects of bottlenecks and sib mating on the level of heterozygosity of females. Interestingly, W. auropunctata is also an invasive ant, and recent studies have shown that introduced populations often derive from a single queen introduction [30] and are characterized by the presence of a single queen and a single male genotype. The presence of a single queen and a single male genotype in our P. longicornis population is also compatible with the population being initiated by a single mated queen.
Interestingly, our study shows that P. longicornis queens lay male eggs that inherit no maternal genes. There are at least two potential mechanisms that could lead to a male being clonally produced by his father. First, the maternal genome could be eliminated after the egg was fertilized [21]. Indeed, other examples of selective elimination of one parental genome have been reported in species such as the parasitoid wasp Nasonia vitripennis [31,32] and the waterfrog hybridogenetic species complex Rana esculenta [33,34], and as probable in some ants of the genus Formica [35,36]. In these cases, however, it is always the paternal genome that is eliminated and not the maternal one. The alternative possibility is that females sometimes lay anucleated eggs that develop into haploid males when fertilized [37]. Unfortunately, it is not possible to know which eggs will give rise to males and females, and it will thus be very difficult to discriminate between these two potential mechanisms.
In conclusion, this study demonstrates that P. longicornis uses an unusual mode of reproduction acting as a pre-adaptation for situations where inbreeding and bottlenecking occur. Even if populations are initiated by a single female, the clonal reproduction of both males and queens ensures a high level of observed heterozyosity of workers when the founding queen and her mate have different alleles. Moreover, this unusual system of reproduction maintains similar levels of observed heterozygosity for queens and for workers even if matings occur between siblings over several generations. This may be an important factor explaining why this species has been so successful in invading new habitats.
Acknowledgements
We thank Sasithorn Hasin and Mingkwan Nipitwattanaphon for collecting material in Thailand, Vicky Menetrey for her contribution to lab work, and three anonymous reviewers for their useful comments. This work was supported by an EMBO fellowship (M.P.), United States National Science Foundation grant DEB no. 0640690 (M.A.D.G.) and the Swiss National Science Foundation (L.K.).
References
- 1.Charlesworth D., Willlis J. 2009. The genetics of inbreeding depression. Nat. Rev. Genet. 10, 783–796 10.1038/nrg2664 (doi:10.1038/nrg2664) [DOI] [PubMed] [Google Scholar]
- 2.Whiting P. W. 1939. Sex determination and reproductive economy in Habrobracon. Genetics 24, 110–111 [Google Scholar]
- 3.Whiting P. W. 1943. Multiple alleles in complementary sex determination of Habrobracon. Genetics 28, 365–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butcher R. D. J., Whitfield W. G. F., Hubbard S. F. 2000. Single-locus complementary sex determination in Diadegma chrysostictos (Gmelin) (Hymenoptera: Ichneumonidae). J. Hered. 91, 104–111 10.1093/jhered/91.2.104 (doi:10.1093/jhered/91.2.104) [DOI] [PubMed] [Google Scholar]
- 5.Beye M., Hasselmann M., Fondrk M. K., Page R. E., Omholt S. W. 2003. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell 114, 419–429 10.1016/S0092-8674(03)00606-8 (doi:10.1016/S0092-8674(03)00606-8) [DOI] [PubMed] [Google Scholar]
- 6.Van Wilgenburg E., Driessen G., Beukeboom L. W. 2006. Single locus sex determination in Hymenoptera: an unintelligent design? Front. Zool. 3, 1–15 10.1186/1742-9994-3-1 (doi:10.1186/1742-9994-3-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross K. G., Fletcher D. J. C. 1986. Diploid male production—a significant colony mortality factor in the fire ant Solenopsis invicta (Hymenoptera: Formicidae). Behav. Ecol. Sociobiol. 19, 283–291 10.1007/BF00300643 (doi:10.1007/BF00300643) [DOI] [Google Scholar]
- 8.Krieger M. J. B., Ross K. G., Chang C. W. Y., Keller L. 1999. Frequency and origin of triploidy in the fire ant. Solenopsis invicta. Heredity 82, 142–150 10.1038/sj.hdy.6884600 (doi:10.1038/sj.hdy.6884600) [DOI] [Google Scholar]
- 9.Cowan D. P., Stahlhut J. K. 2004. Functionally reproductive diploid and haploid males in an inbreeding hymenopteran with complementary sex determination. Proc. Natl Acad. Sci. USA 101, 10 374–10 379 10.1073/pnas.0402481101 (doi:10.1073/pnas.0402481101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook J. M., Crozier R. H. 1995. Sex determination and population biology of the Hymenoptera. Trends Ecol. Evol. 10, 281–286 10.1016/0169-5347(95)90011-X (doi:10.1016/0169-5347(95)90011-X) [DOI] [PubMed] [Google Scholar]
- 11.Tsutsui N. D., Suarez A. V., Holway D. A., Case T. J. 2000. Reduced genetic variation and the success of an invasive species. Proc. Natl Acad. Sci. USA 97, 5948–5953 10.1073/pnas.100110397 (doi:10.1073/pnas.100110397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Dowd D. J., Green P. T., Lake P. S. 2003. Invasional ‘meltdown’ on an oceanic island. Ecol. Lett. 6, 812–817 10.1046/j.1461-0248.2003.00512.x (doi:10.1046/j.1461-0248.2003.00512.x) [DOI] [Google Scholar]
- 13.Wilson E. O. 2005. Early ant plagues in the New World. Nature 433, 32. 10.1038/433032a (doi:10.1038/433032a) [DOI] [PubMed] [Google Scholar]
- 14.Invasive Species Specialist Group. 2010. 100 of the world's worst invasive alien species. See http://www.issg.org .
- 15.Ross K. G., Vargo E. L., Keller L., Trager J. C. 1993. Effect of a founder event on variation in the genetic sex-determining system of the fire ant Solenopsis invicta. Genetics 135, 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van der Hammen T., Pedersen J. S., Boomsma J. J. 2002. Convergent development of low-relatedness supercolonies in Myrmica ants. Heredity 89, 83–89 10.1038/sj.hdy.6800098 (doi:10.1038/sj.hdy.6800098) [DOI] [PubMed] [Google Scholar]
- 17.Foucaud J., Orivel J., Fournier D., Delabie J. H. C., Loiseau A., Le Breton J., Cerdans P., Estoup A. 2009. Reproductive system, social organization, human disturbance and ecological dominance in native populations of the little fire ant, Wasmannia auropunctata. Mol. Ecol. 18, 5059–5073 10.1111/j.1365-294X.2009.04440.x (doi:10.1111/j.1365-294X.2009.04440.x) [DOI] [PubMed] [Google Scholar]
- 18.Helanterä H., Queller D. C., Carrillo J., Strassmann J. 2009. Unicolonial ants—where do they come from, what are they and where are they going? Trends Ecol. Evol. 24, 341–349 10.1016/j.tree.2009.01.013 (doi:10.1016/j.tree.2009.01.013) [DOI] [PubMed] [Google Scholar]
- 19.Vogel V., Pedersen J. S., Giraud T., Krieger M. J. B., Keller L. 2010. The worldwide expansion of the Argentine ant. Divers. Distrib. 16, 170–186 10.1111/j.1472-4642.2009.00630.x (doi:10.1111/j.1472-4642.2009.00630.x) [DOI] [Google Scholar]
- 20.Pearcy M., Aron S., Doums C., Keller L. 2004. Conditional use of sex and parthenogenesis for worker and queen production in ants. Science 306, 1780–1783 10.1126/science.1105453 (doi:10.1126/science.1105453) [DOI] [PubMed] [Google Scholar]
- 21.Fournier D., Estoup A., Orivel J., Foucaud J., Jourdan H., Le Breton J., Keller L. 2005. Clonal reproduction by males and females in the little fire ant. Nature 435, 1230–1234 10.1038/nature03705 (doi:10.1038/nature03705) [DOI] [PubMed] [Google Scholar]
- 22.Ohkawara K., Nakayama M., Satoh A., Trindl A., Heinze J. 2006. Clonal reproduction and genetic caste differences in a queen-polymorphic ant, Vollenhovia emeryi. Biol. Lett. 2, 359–363 10.1098/rsbl.2006.0491 (doi:10.1098/rsbl.2006.0491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drescher J., Blüthgen N., Feldhaar H. 2007. Population structure and intraspecific aggression in the invasive ant species Anoplolepis gracilipes in Malaysian Borneo. Mol. Ecol. 16, 1453–1465 10.1111/j.1365-294X.2007.03260.x (doi:10.1111/j.1365-294X.2007.03260.x) [DOI] [PubMed] [Google Scholar]
- 24.Goodisman M. A. D., Kovacs J. L., Hunt B. G. 2008. Functional genetics and genomics in ants (Hymenoptera: Formicidae): the interplay of genes and social life. Myrmecol. News 11, 107–117 [Google Scholar]
- 25.Wetterer J. K. 2008. Worldwide spread of the longhorn crazy ant, Paratrechina longicornis (Hymenoptera: Formicidea). Myrmecol. News 11, 137–149 [Google Scholar]
- 26.Molecular Ecology Resources Primer Development Consortium et al. 2011. Permanent genetic resources added to Molecular Ecology Resources database 1 August 2010–30 September 2010. Mol. Ecol. Res. 11, 219–222 [DOI] [PubMed] [Google Scholar]
- 27.Ihaka R., Gentleman R. 1996. R: a language for data analysis and graphics. J. Comp. Graph. Stat. 5, 299–314 10.2307/1390807 (doi:10.2307/1390807) [DOI] [Google Scholar]
- 28.Suomalainen E., Saura A., Lokki J. 1987. Cytology and evolution in parthenogenesis. Boca Raton, FL: CRC Press [Google Scholar]
- 29.Pearcy M., Hardy O., Aron S. 2006. Thelytokous parthenogenesis and its consequences on inbreeding in an ant. Heredity 96, 377–382 10.1038/sj.hdy.6800813 (doi:10.1038/sj.hdy.6800813) [DOI] [PubMed] [Google Scholar]
- 30.Mikheyev A. S., Bresson S., Conant P. 2009. Single-queen introductions characterize regional and local invasions by the facultatively clonal little fire ant Wasmannia auropunctata. Mol. Ecol. 18, 2937–2944 10.1111/j.1365-294X.2009.04213.x (doi:10.1111/j.1365-294X.2009.04213.x) [DOI] [PubMed] [Google Scholar]
- 31.Werren J. H., Nur U., Eickbush D. 1987. An extrachromosomal factor causing loss of paternal chromosomes. Nature 327, 75–76 10.1038/327075a0 (doi:10.1038/327075a0) [DOI] [PubMed] [Google Scholar]
- 32.Nur U., Werren J. H., Eickbush D. G., Burke W. D., Eickbush T. H. 1988. A ‘selfish’ B chromosome that enhances its transmission by eliminating the paternal genome. Science 240, 512–514 10.1126/science.3358129 (doi:10.1126/science.3358129) [DOI] [PubMed] [Google Scholar]
- 33.Tunner H. G. 1970. Das Serumeiweitßbild einheimischer Wasserfrösche und der Hybridcharakter yon R. esculenta. Ferh. dr. zool. Ges. 64, 352–358 [Google Scholar]
- 34.Graf J.-D., Müller W. P. 1979. Experimental gynogenesis provides evidence of hybridogenetic reproduction in the Rana esculenta complex. Experientia 35, 1574–1576 10.1007/BF01953200 (doi:10.1007/BF01953200) [DOI] [PubMed] [Google Scholar]
- 35.Kulmuni J., Seifert B., Pamilo P. 2010. Segregation distortion causes large-scale differences between male and female genomes in hybrid ants. Proc. Natl Acad. Sci. USA 107, 7371–7376 10.1073/pnas.0912409107 (doi:10.1073/pnas.0912409107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keller L. 2010. Genetics: biased transmission of genomes according to parents of origin. Curr. Biol. 20, 601–602 10.1016/j.cub.2010.05.048 (doi:10.1016/j.cub.2010.05.048) [DOI] [PubMed] [Google Scholar]
- 37.Foucaud J., Estoup A., Loiseau A., Rey O., Orivel J. 2010. Thelytokous parthenogenesis, male clonality and genetic caste determination in the little fire ant: new evidence and insights from the lab. Heredity 105, 205–212 10.1038/hdy.2009.169 (doi:10.1038/hdy.2009.169) [DOI] [PubMed] [Google Scholar]