Abstract
Intercontinental dispersal via land bridge connections has been important in the biogeographic history of many Holarctic plant and animal groups. Likewise, some groups appear to have accomplished trans-oceanic dispersal via rafting. Dibamid lizards are a clade of poorly known fossorial, essentially limbless species traditionally split into two geographically disjunct genera: Dibamus comprises approximately 20 Southeast Asian species, many of which have very limited geographical distributions, and the monotypic genus Anelytropsis occupies a small area of northeastern Mexico. Although no formal phylogeny of the group exists, a sister–taxon relationship between the two genera has been assumed based on biogeographic considerations. We used DNA sequence data from one mitochondrial and six nuclear protein-coding genes to construct a phylogeny of Dibamidae and to estimate divergence times within the group. Surprisingly, sampled Dibamus species form two deeply divergent, morphologically conserved and geographically concordant clades, one of which is the sister taxon of Anelytropsis papillosus. Our analyses indicate Palaearctic to Nearctic Beringian dispersal in the Late Palaeocene to Eocene. Alternatively, a trans-Pacific rafting scenario would extend the upper limit on dispersal to the Late Cretaceous. Either scenario constitutes a remarkable long-distance dispersal in what would seem an unlikely candidate.
Keywords: Beringia, dispersal, phylogeny, biogeography, Dibamidae
1. Introduction
Faunal and floral interchange between the Palaearctic (PA) and Nearctic (NA) has played a major role in shaping the current Holarctic biodiversity patterns, and this interchange has been facilitated at various times by both trans-Atlantic [1–3] and trans-Beringian [4,5] land bridges. For many groups, this complexity has meant that the current spatial patterns are congruent with more than one historical explanation. Temporal data are needed to detect instances of pseudocongruence [6], and palaeoclimatological inferences can further narrow the list of plausible histories by matching climatic conditions with known physiological tolerances of particular taxa [7].
The Beringian land bridge has formed an important dispersal corridor between the eastern PA and NA for many vertebrate groups, including fishes, birds, mammals, reptiles and amphibians (e.g. [8–10]). Though this bridge first formed around 100 Ma in the mid-Cretaceous [10], for warm-adapted taxa this route was probably viable only during the period of increased global temperatures that produced and maintained the boreotropical forest belt throughout the Eocene (i.e. about 55–35 Myr ago; [11]). On the Atlantic side, the Thulean land bridge provided a similar opportunity for the interchange of warm-adapted taxa (albeit much shorter, owing to the breaking of the bridge around 50 Ma; [1]).
The fossorial and basically limbless lizard family Dibamidae (males have small flaps near the cloaca; [12]) has long been an enigma to both systematists and biogeographers. Although some early morphological studies (e.g. [13]) hinted that dibamids were not closely related to any extant group of squamate reptiles (lizards and snakes), most morphological studies favoured hypotheses relating dibamids to various squamate clades (e.g. gekkonids, scincids; reviewed by Greer [12]). Only recently have molecular phylogenetic studies confirmed that dibamids are an early-diverging group within the Squamata (e.g. [14–16]), and that the two dibamid genera do form a clade [17]. Equally intriguing is the geographical distribution of the group, which consists of tropical Southeast Asia (Indochina, the Malay Peninsula and islands stretching across to western New Guinea) and a relatively small area over 14 000 km away in northeastern Mexico [12]. The Asian populations comprise approximately 20 named species of the genus Dibamus [18], and include several island endemics (e.g. [18–20]) and mainland species with very restricted, geographically proximate (i.e. microendemic) distributions (e.g. [21,22]). The morphologically very similar Mexican populations comprise the monotypic genus Anelytropsis (A. papillosus; [12]).
Dibamids apparently do not fossilize well, as there are no known fossil forms [23]. Given this fact and the great age of the stem clade (potentially originating in the Late Triassic; [16]), it is plausible that dibamids once had a wide Pangean distribution, and that Dibamus and Anelytropsis represent deeply divergent clades with highly disjunct, relictual distributions. Assuming no overseas dispersal, under this scenario, the divergence between these clades should predate the formation of the Turgai seaway that split the PA around 160 Ma in the Middle Jurassic [24].
Beringian dispersal in either direction could also account for the current distributions. Because all extant dibamids are found in tropical (Southeast Asia) or subtropical (eastern Mexico) climates, it seems likely that ancestral taxa would share similar requirements, thus limiting the possibility of dispersal to the warmer Eocene interval at 55–35 Myr ago. Alternatively, convincing examples of trans-Pacific overseas dispersal do exist [25,26]. Although no absolute upper time limit exists for an oceanic rafting event, support for this route would be bolstered if shallow trans-Pacific genetic divergences make Beringian dispersal untenable.
Opportunities for trans-Atlantic (Thulean) land bridge dispersal were probably limited by climatological conditions to a small window around 50–55 Myr ago (i.e. after the climate warmed, but before the Thulean bridge submerged). Because the Turgai Sea (which splits the PA) did not close until the Oligocene, any now-extinct European populations that may have supplied migrants to the New World would have been last connected by land to Asian populations before the opening of the Turgai Sea (160 Ma; [24]). This would in turn predict a Jurassic-age genetic divergence between the New World and Asia. Thulean-bridge dispersal (or trans-Atlantic rafting for that matter) in the other direction would require an additional Oligocene or later (i.e. post-closure of the Turgai Sea) dispersal to Asia from the now-extinct European populations. This scenario would predict relatively shallow divergences among Asian populations relative to PA–NA divergences.
To test for topological and temporal agreement with these hypotheses, and to discover the direction of dispersal along the inferred most likely route, we conducted a molecular phylogenetic study of Dibamidae using both mitochondrial and nuclear DNA sequence data.
2. Material and methods
(a). Taxon and gene sampling
Taxon sampling included seven putative species (eight individuals) of Dibamus that roughly span the known distribution of the genus from southeastern Indochina to western New Guinea, and one individual of A. papillosus from Tamaulipas, Mexico. To provide calibration nodes and sufficiently dense taxon sampling for robust divergence-dating analyses, we also included as outgroups the tuatara (Sphenodon punctatus) and 63 other squamate taxa (see electronic supplementary material). We sequenced all individuals for the mitochondrial protein-coding gene, ND2, and six nuclear protein-coding genes (BDNF, CMOS, DNAH3, NKTR, RAG1 and R35) for a total alignment of 5864 base pairs (bp). See electronic supplementary material, tables for primer sequences and GenBank numbers.
(b). DNA amplification, sequence editing and phylogenetic analyses
Genomic DNA was extracted and amplified using standard protocols [15]. Sequence contigs were assembled using Sequencher v. 4.1.2 (Gene Codes Corporation). The Clustal algorithm [27] implemented in the program DAMBE [28] was used to align sequences by their amino acid translations, and alignments were adjusted by eye using MacClade v. 4.03 [29]. Regions deemed ambiguously aligned by this process were excluded from all analyses. Partitioned unrooted maximum-likelihood (ML) analyses were performed on the full data using RAxML v. 7.0.4 [30], and partitioned unrooted Bayesian (UB) analyses using MrBayes v. 3.1.2 [31]. Gaps were treated as a separate (binary) character partition in the MrBayes analyses, and as missing data in the ML analyses. For ML analyses, data were partitioned by gene and codon position (scheme favoured through Bayes-factor testing; data not shown), and a GTR + Γ model was used for each partition. For UB analyses, the same data partitions were used, but evolutionary models for each partition were determined using the Akaike information criteria as implemented in MrModeltest [32]. Two separate MrBayes runs were conducted; the average standard deviation of split frequencies (≤0.01 used as a cut-off) and the potential scale reduction factor ( ≤0.1 deviation from 1.0 accepted) statistics from MrBayes [31] were used to evaluate topological and branch-length (BL) convergence, respectively. To help spot potential problems with overestimation of BLs owing to the influence of BL priors [33], we also examined relative partition rates to see that they made biological sense (i.e. second codon positions slowest, followed by first positions, and then the more quickly-evolving third positions). All RAxML and MrBayes analyses were performed via the CIPRES 2 portal at the San Diego Supercomputer Centre [34].
Preliminary full-data analyses suggested a potential problem with rooting the dibamid portion of the tree using the standard outgroup method (see figure 1 and §3). Molecular clocks can help infer the root of a clade when appropriate close outgroups are unavailable [35]. Given the distant relationship of Sphenodon (the closest living relatives of squamates) and the relatively basal placement of dibamids in the squamate phylogeny, we therefore performed a series of dibamids-only analyses using (i) all positions of all genes, (ii) the nuclear data as a whole, and (iii) the data from individual genes. For each of these data subsets, we performed partitioned Bayesian and unpartitioned ML phylogenetic analyses under unrooted and strict-clock models (using MrBayes and RAxML, respectively; the available ML programs do not allow strict-clock partitioned analyses). We evaluated relative support for these two models in a Bayesian framework by comparing the harmonic means of the log-likelihoods of the respective analyses using Bayes factors [36,37], and for the analogous ML analyses we used likelihood-ratio testing (LRT; [38]) implemented in PAUP* v. 4.0b10 [39]. We also performed a partitioned relaxed-clock Bayesian (RCB) analysis on the dibamids-only dataset (all character partitions included) using BEAST v. 1.4.8 [40]. For this analysis, no calibrations were applied, and branch rates following an uncorrelated lognormal distribution, with a mean of 1.0. This both demonstrated the effect of a relaxed clock on the rooting position, and also allowed another evaluation of the fit of our data to a strict clock. If the 95 per cent highest probability density (HPD) of the coefficient of variation of rates from such an analysis includes zero, it suggests that a strict clock may be supported by the data [41].
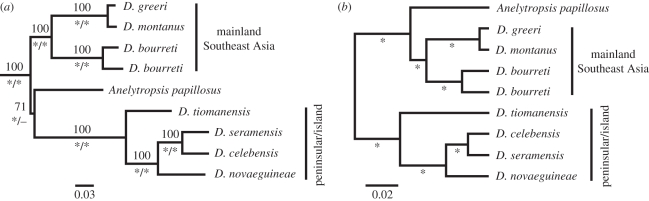
Figure 1.
Results of phylogenetic analyses. (a) Maximum-likelihood (ML) phylogram using Sphenodon and representatives of all major squamate clades (not shown) to root dibamids. Numbers above branches are ML bootstraps and asterisks below branches signify unrooted/relaxed-clock Bayesian posterior probabilities greater than 0.95 (topologies were concordant across analyses). Note the marked BL asymmetry across the root resulting from outgroup rooting. (b) Tree rooted without outgroups using a Bayesian relaxed-clock method. Asterisks below branches signify Bayesian posterior probabilities of 1.0.
(c). Divergence dating and biogeographic testing
We performed an RCB analysis using BEAST v. 1.4.8 [40] on the full 72-terminal (8 dibamids + 63 other squamates + Sphenodon) dataset to estimate divergence times within dibamids. The same partitioning scheme and evolutionary models used for the UB (i.e. MrBayes) analyses was applied, except that mtDNA third-codon positions were excluded owing to poor convergence in analyses using the full data. Fossils or geological data suitable for constraining nodes within Dibamidae are not available; therefore, we used several external calibration points for our analyses. The dibamid root was constrained to match that recovered in the dibamids-only Bayesian and ML analyses performed under relaxed and strict clocks, respectively.
Biogeographic analyses were conducted in an ML framework using ancestral state reconstruction in Mesquite [42] and dispersal-extinction-cladogenesis (DEC) analysis in Lagrange [43]. See electronic supplementary material for details of these analyses.
3. Results
Full-taxa ML, UB and RCB analyses all gave an unexpected topology in which Anelytropsis is not the sister taxon of a monophyletic Dibamus, but is instead the sister taxon of one of the two deeply divergent, geographically concordant Dibamus clades (figures 1a and 2). Although all these analyses recover the peninsular/island clade of Dibamus as the sister taxon of Anelytropsis, marked BL asymmetry across the dibamid root is evident, implying substantially different rates of molecular evolution in the two major clades. In light of this asymmetry, and given the very long branches connecting dibamids to the clade containing all remaining squamates and the outgroup Sphenodon, we considered the possibility that the standard outgroup method had incorrectly rooted this clade.
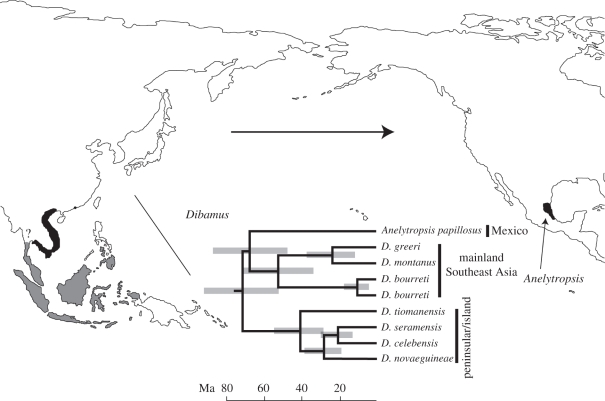
Figure 2.
Bayesian relaxed-clock chronogram from analyses using all outgroups (not shown) and topology constrained to place Anelytropsis as sister to the mainland Dibamus clade, as in figure 1b (see text). Grey bars represent 95% HPDs for node ages. Approximate distribution of peninsular/island Dibamus in grey, distributions of mainland Dibamus and Anelytropsis in black. Question mark denotes uncertainty in the exact geographical limits of the two Dibamus clades. Arrow indicates most likely direction of dispersal.
Sampling only within dibamids, a series of paired Bayesian strict-clock/non-clock analyses using various datasets (all positions, nuclear-only and individual genes) all yielded similar results. In each case, a strict clock placed Anelytropsis as the sister taxon of the mainland Dibamus clade (i.e. changed the position of the root), a result supported by high posterior probabilities in all but the CMOS analysis (table 1). Moreover, although the non-clock model was favoured for three datasets (all positions, combined nuclear data and R35-only), a strict clock was favoured for the other six individual-gene datasets (table 1). Additional ML-based analyses using LRT to compare models found similar results (see the electronic supplementary material, table S1).
Table 1.
Results of Bayes factor comparisons of strict-clock versus non-clock models for various character subsets of the Dibamids-only dataset. (Interpretation based on recommendations of Kass & Raftery [37].)
| partition | Bayes factor | interpretation | Bayes PPa |
|---|---|---|---|
| all positions | 53.52 | very strongly non-clock | 1.0 |
| nuclear only | 85.28 | very strongly non-clock | 1.0 |
| ND2 | −4.72 | moderately favours clock | 1.0 |
| BDNF | −1.94 | weakly favours clock | 0.99 |
| CMOS | −2.84 | moderately favours clock | 0.65 |
| DNAH3 | −5.86 | moderately favours clock | 0.98 |
| NKTR | −8.1 | strongly favours clock | 1.0 |
| R35 | 3.96 | moderately favours non-clock | 1.0 |
| RAG1 | −0.9 | weakly favours clock | 1.0 |
aPosterior probabilities of the Anelytropsis/mainland Dibamus clade recovered in all strict-clock analyses.
RCB analysis of the dibamids-only all-positions dataset also recovered the alternate rooting (i.e. Anelytropsis + mainland Dibamus clade) with maximal support (figure 1b). The coefficient of variation of rates from this analysis has a mean of 0.257, and the lower end of its 95 per cent HPD reaches 0.125, suggesting a relaxed clock as more appropriate than a strict clock for the all-positions dataset. In summary, our results strongly suggest that outgroup rooting is inappropriate in this case, and we therefore favour the clock-based alternate rooting. Regardless of which rooting is used, we note that Anelytropsis is still nested within Dibamus, and the striking fact remains that the genetic divergence between the two Asian lineages is on par with their respective divergences from the Mexican Anelytropsis.
Fossil-calibrated RCB analyses including all outgroups, and constrained to conform to our favoured dibamid rooting, found divergences within each of the two Dibamus clades dating back to the Eocene. The split between Anelytropsis and mainland Dibamus dated to 69 Ma (95% HPD of 49–90 Myr ago), and the age of crown-clade dibamids as a whole was 72 Ma (95% HPD of 54–95 Myr ago; figure 2).
Both ML-based ancestral state reconstruction using Mesquite [42] and DEC analysis using Lagrange [43] favoured an Asian distribution for the most recent common ancestor of mainland Dibamus and Anelytropsis, and thus Old World to New World dispersal (79% and 78% probabilities, respectively; see the electronic supplementary material for discussion of some limitations of these two methods for our system).
4. Discussion
The Late Cretaceous divergences among the major dibamid clades are incompatible with Pangean vicariance. Thus, our results reveal a counterintuitive evolutionary history for one of the most enigmatic of squamate groups, one combining striking microendemicity with intercontinental dispersal. This is noteworthy, because most of the approximately 24 independently evolved lineages of burrowing, snake-like squamate groups have evolved geographically in situ [44]. Of these burrowing lineages (excluding snakes), only dibamids and amphisbaenians [45] have exhibited long-distance (intercontinental) dispersal. Although the isolated distribution of Anelytropsis in Mexico makes deep divergence from other dibamids intuitively reasonable, the even deeper split between two geographically adjacent and non-overlapping Dibamus clades was quite unexpected. Such an ancient divergence between morphologically similar congeners (no morphological characters diagnose these clades; [20]) provides a striking example of morphological stasis, one apparently predating other recently documented cases [46–48]. Further taxon sampling is necessary to know the full extent of geographical concordance within clades, but our results (along with the substantial number of coastal island endemics and very restricted mainland ranges; see the electronic supplementary material) thus suggest a remarkably low propensity for even relatively local dispersal throughout the Tertiary.
Our formal biogeographic analyses support a scenario of dispersal from Asia to North America. An Atlantic crossing via the Thulean bridge would require Jurassic PA–NA divergences (assuming the Turgai Sea formed a barrier to dispersal) as well as extinction across most of Asia and all of Europe. Thus, overland dispersal via the more geographically proximate Bering land bridge is clearly a more plausible alternative. Although our analysis places the origin of the Anelytropsis lineage between 90 and 49 Myr ago, the boreotropical belt of forest that would allow Beringian passage of warm-adapted taxa is thought to have formed only after global temperatures increased during the Palaeocene and culminated in the thermal maximum around the Palaeocene–Eocene boundary at approximately 55 Ma ([7], but see [49] for evidence of possible Cretaceous lizard faunal interchange between Asia and North America). The terminal Eocene cooling event (approx. 35 Ma) that followed once again excluded warm-adapted taxa from Beringia [10]. Several reptile groups, including colubrid [50] and viperid [51] snakes, skinks [9,52] and anguids [9,53] apparently crossed Beringia to the New World after the climate cooled, in the Oligoene or even Miocene. However, all of these taxa contain extant members occupying more northern, temperate regions than those occupied by extant dibamids. Combining these lines of evidence, we suggest that the Anelytropsis lineage probably dispersed across Beringia in the Late Palaeocene or Eocene.
An alternative to overland dispersal is trans-Pacific rafting. After the widespread acceptance of plate tectonics theory in the 1970s, oceanic dispersal hypotheses fell out of favour with most biogeographers. However, recent studies have demonstrated the biogeographic importance of this phenomenon for many groups, including taxa not generally thought to be good candidates for survival in an oceanic environment (reviewed in de Queiroz [54]). For example, the Fijian iguanas (Brachylophus) almost certainly originated via trans-Pacific rafting from the New World, where all their closest relatives (both extant and fossil) are found [25]. Although the dispersal was in the opposite direction, this might seem to provide a precedent for dibamids. However, the case for overseas dispersal in Brachylophus is strengthened by the fact that, unlike dibamids, they are completely absent (including fossils, which should be more likely from these large-bodied lizards) from Australasia and the Asian mainland.
Vidal et al. [45] concluded that members of the limbless, fossorial, squamate group Amphisbaenia dispersed to the New World twice by trans-Atlantic rafting. However, this inference relied on molecular divergence dates in the Eocene, long after the separation of Africa and South America. Overland dibamid dispersal is not problematic in this regard. Moreover, present-day trans-Pacific rafting would require a voyage three times as long as trans-Atlantic rafting, and this ratio would have been even greater in the Eocene. For these reasons, we favour the well-established Beringian route. If fossil dibamids are eventually discovered in the northern PA or NA, this position will obviously be bolstered. Alternatively, if further taxon sampling reveals an Asia–North America split much more recent than our current estimate, then the rafting hypothesis should be more strongly considered.
Advances in divergence dating using DNA sequence data have led to amazing and sometimes counterintuitive biogeographic discoveries in recent years. Our study provides a striking example of a long-distance dispersal event in a taxon that otherwise appears to have exceptionally low vagility. Additionally, this study reveals that the morphologically conserved and geographically restricted genus Dibamus harbours surprisingly ancient phylogenetic diversity, both at the deepest and more nested hierarchical levels. Further sampling of geographical regions and species may provide clues to the geological and/or ecological forces that led to such an unexpected biogeographic pattern, and will be crucial to taxonomic revision of this fascinating group.
Acknowledgements
We thank P. Wisniewski for laboratory assistance. For generous loans of Dibamus tissues and specimens, we thank D. Cannatella, D. Frost, L. Grismer, M. Kearney, T. LaDuc, J. McGuire, R. Murphy and C. Spencer. We thank A. Peralta-Garcia and P. Scott for their assistance in collecting, and O. Flores-Villela for collecting permits. We thank the editor and two anonymous reviewers for constructive suggestions that improved the paper. This project was funded by NSF grant EF0334967 to T.W.R.
References
- 1.McKenna M. C. 1983. Cenozoic paleogeography of North Atlantic land bridges. In Structure and development of the Greenland–Scotland bridge: new concepts and methods (eds Bott M. H. P., Saxov S., Talwani M., Thiede J.), pp. 351–395 New York, NY: Plenum [Google Scholar]
- 2.Tiffney B. H. 1985. The Eocene North Atlantic land bridge: its importance in tertiary and modern phytogeography of the Northern Hemisphere. J. Arnold Arbor. 66, 243–273 [Google Scholar]
- 3.Tiffney B. H. 2008. Phylogeography, fossils, and Northern Hemisphere biogeography: the role of physiological uniformitarianism. Ann. Mo. Bot. Gard. 95, 135–143 10.3417/2006199 (doi:10.3417/2006199) [DOI] [Google Scholar]
- 4.Tiffney B. H. 1985. Perspectives on the origin of the floristic similarity between eastern Asia and eastern North America. J. Arnold Arbor. 66, 73–94 [Google Scholar]
- 5.Wolfe J. A. 1972. An interpretation of Alaskan Tertiary floras. In Floristics and paleofloristics of Asia and Eastern North America (ed. Graham A.), pp. 201–233 Amsterdam, The Netherlands: Elsevier [Google Scholar]
- 6.Cunningham C. W., Collins T. M. 1994. Developing model systems for molecular biogeography: vicariance and interchange in marine invertebrates. In Molecular ecology and evolution, approaches and application (eds Schierwater B., Streit B., Wagner G. P., DeSalle R.), pp. 405–433 Basel, Switzerland: Birkhauser Verlag [Google Scholar]
- 7.Wolfe J. A. 1987. Late Cretaceous–Cenozoic history of deciduousness and the terminal Cretaceous event. Paleobiology 13, 215–226 [Google Scholar]
- 8.Keogh J. S. 1998. Molecular phylogeny of elapid snakes and a consideration of their biogeographic history. Biol. J. Linn. Soc. 63, 177–203 10.1111/j.1095-8312.1998.tb01513.x (doi:10.1111/j.1095-8312.1998.tb01513.x) [DOI] [Google Scholar]
- 9.Macey J. R., Schulte J. A., Strasburg J. L., Brisson J. A., Larson A., Ananjeva N. B., Wang Y., Parham J. F., Papenfuss T. J. 2006. Assembly of the eastern North American herpetofauna: new evidence from lizards and frogs. Biol. Lett. 2, 388–392 10.1098/rsbl.2006.0473 (doi:10.1098/rsbl.2006.0473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanmartin I., Enghoff H., Ronquist F. 2001. Patterns of animal dispersal, vicariance and diversification in the Holarctic. Biol. J. Linn. Soc. 73, 345–390 10.1006/bijl.2001.0542 (doi:10.1006/bijl.2001.0542) [DOI] [Google Scholar]
- 11.Wolfe J. A. 1975. Some aspects of plant geography of the Northern Hemisphere during the Late Cretaceous and Tertiary. Ann. Mo. Bot. Gard. 62, 264–279 10.2307/2395198 (doi:10.2307/2395198) [DOI] [Google Scholar]
- 12.Greer A. L. 1985. The relationships of the lizard genera Anelytropsis and Dibamus. J. Herpetol. 19, 116–156 10.2307/1564427 (doi:10.2307/1564427) [DOI] [Google Scholar]
- 13.Gasc J. P. 1968. Contribution à ľosteologie et la myologie de Dibamus novaeguineae Gray (Sauria, Reptilia). Discussion systématique. Ann. Sci. Nat. Zool. 10, 127–250 [Google Scholar]
- 14.Wiens J. J., Kuczynski C. A., Townsend T. M., Reeder T. W., Mulcahy D. G., Sites J. W., Jr 2010. Combining phylogenomics and fossils in higher-level squamate phylogeny: molecular data change the placement of fossil taxa. Syst. Biol. 59, 674–688 10.1093/sysbio/syq048 (doi:10.1093/sysbio/syq048) [DOI] [PubMed] [Google Scholar]
- 15.Townsend T. M., Larson A., Louis E., Macey J. R. 2004. Molecular phylogenetics of Squamata: the position of snakes, amphisbaenians, and dibamids, and the root of the squamate tree. Syst. Biol. 53, 735–757 10.1080/10635150490522340 (doi:10.1080/10635150490522340) [DOI] [PubMed] [Google Scholar]
- 16.Vidal N., Hedges S. B. 2005. The phylogeny of squamate reptiles (lizards, snakes, and amphisbaenians) inferred from nine nuclear protein-coding genes. C. R. Biol. 328, 1000–1008 [DOI] [PubMed] [Google Scholar]
- 17.Zaldivar-Riveron A., de Oca A. N. M., Manriquez-Moran N., Reeder T. W. 2008. Phylogenetic affinities of the rare and enigmatic limb-reduced Anelytropsis (Reptilia: Squamata) as inferred with mitochondrial 16S rRNA sequence data. J. Herpetol. 42, 303–311 10.1670/06-2273.1 (doi:10.1670/06-2273.1) [DOI] [Google Scholar]
- 18.Das I., Lim K. K. P. 2009. A new species of Dibamus (Squamata: Dibamidae) from Pulau Simeuleu, Mentawai Archipelago, Indonesia. Zootaxa 2088, 15–23 [Google Scholar]
- 19.Das I., Lim K. K. P. 2005. New species of Dibamus (Squamata: Dibamidae) from Pulau Nias, Indonesia. J. Herpetol. 39, 113–117 10.1670/0022-1511(2005)039[0113:NSODSD]2.0.CO;2 (doi:10.1670/0022-1511(2005)039[0113:NSODSD]2.0.CO;2) [DOI] [Google Scholar]
- 20.Honda M., Ota H., Hikida T., Darevsky I. S. 2001. A new species of the worm-like lizard, Dibamus Dumeril & Bibron 1839 (Squamata Dibamidae), from Vietnam. Trop. Zool. 14, 119–125 [Google Scholar]
- 21.Darevsky I. S. 1992. Two new species of worm-like lizard Dibamus (Sauria: Dibamidae), with remarks on the distribution and ecology of Dibamus in Vietnam. Asiat. Herpetol. Res. 4, 1–12 [Google Scholar]
- 22.Das I., Lim K. K. P. 2003. Two new species of Dibamus (Squamata: Dibamidae) from Borneo. Raffles Bull. Zool. 51, 137–141 [Google Scholar]
- 23.Estes R. 1983. The fossil record and early distribution of lizards. In Advances in herpetology and evolutionary biology: essays in honor of Ernest E Williams (eds Rhodin A., Miyata K.), pp. 365–398 Cambridge, MA: Museum of Comparative Zoology, Harvard University [Google Scholar]
- 24.Briggs J. C. 1995. Global biogeography. Amsterdam, The Netherlands: Elsevier Science [Google Scholar]
- 25.Keogh J. S., Edwards D. L., Fisher R. N., Harlow P. S. 2008. Molecular and morphological analysis of the critically endangered Fijian iguanas reveals cryptic diversity and a complex biogeographic history. Phil. Trans. R. Soc. B 363, 3413–3426 10.1098/rstb.2008.0120 (doi:10.1098/rstb.2008.0120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanmartin I., Wanntorp L., Winkworth R. C. 2007. West Wind Drift revisited: testing for directional dispersal in the Southern Hemisphere using event-based tree fitting. J. Biogeogr. 34, 398–416 10.1111/j.1365-2699.2006.01655.x (doi:10.1111/j.1365-2699.2006.01655.x) [DOI] [Google Scholar]
- 27.Thompson J. D., Higgins D. G., Gibson T. J. 1994. Clustal-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 10.1093/nar/22.22.4673 (doi:10.1093/nar/22.22.4673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia X., Xie Z. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 92, 371–373 10.1093/jhered/92.4.371 (doi:10.1093/jhered/92.4.371) [DOI] [PubMed] [Google Scholar]
- 29.Maddison D. R., Maddison W. P. 2000. MacClade 4: analysis of phylogeny and character evolution, v. 4.03 Sunderland, MA: Sinauer; [DOI] [PubMed] [Google Scholar]
- 30.Stamatakis A., Hoover P., Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771 10.1080/10635150802429642 (doi:10.1080/10635150802429642) [DOI] [PubMed] [Google Scholar]
- 31.Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 10.1093/bioinformatics/btg180 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 32.Nylander J. A. A. 2004. MrModeltest v. 2. Program distributed by the author. Uppsala, Sweden: Evolutionary Biology Centre, Uppsala University [Google Scholar]
- 33.Marshall D. C. 2010. Cryptic failure of partitioned Bayesian phylogenetic analyses: lost in the land of long trees. Syst. Biol. 59, 108–117 10.1093/sysbio/syp080 (doi:10.1093/sysbio/syp080) [DOI] [PubMed] [Google Scholar]
- 34.Miller M., Holder M., Vos R., Midford P., Liebowitz T., Chan L., Hoover P., Warnow T. 2009. The CIPRES Portals, CIPRES 2009-08-04. See http://www.phyloorg/sub_sections/portal (accessed 4 August 2009; archived by Webcite(r) at http://www.webcitationorg/5imQlJeQa) [Google Scholar]
- 35.Huelsenbeck J. P., Bollback J. P., Levine A. M. 2002. Inferring the root of a phylogenetic tree. Syst. Biol. 51, 32–43 10.1080/106351502753475862 (doi:10.1080/106351502753475862) [DOI] [PubMed] [Google Scholar]
- 36.Brandley M. C., Schmitz A., Reeder T. W. 2005. Partitioned Bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Syst. Biol. 54, 373–390 10.1080/10635150590946808 (doi:10.1080/10635150590946808) [DOI] [PubMed] [Google Scholar]
- 37.Kass R. E., Raftery A. E. 1995. Bayes factors (review). J. Am. Stat. Assoc. 90, 773–795 10.2307/2291091 (doi:10.2307/2291091) [DOI] [Google Scholar]
- 38.Kishino H., Hasegawa M. 1989. Evaluation of the maximum-likelihood estimate of the evolutionary tree topologies from DNA-sequence data, and the branching order in Hominoidea. J. Mol. Evol. 29, 170–179 10.1007/BF02100115 (doi:10.1007/BF02100115) [DOI] [PubMed] [Google Scholar]
- 39.Swofford D. L. 2002. PAUP* phylogenetic analysis using parsimony (*and other methods), v. 4.0b10 Sunderland, MA: Sinauer Associates [Google Scholar]
- 40.Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. 10.1186/1471-2148-7-214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drummond A. J., Ho S. Y. W., Phillips M. J., Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88. 10.1371/journal.pbio.0040088 (doi:10.1371/journal.pbio.0040088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maddison W. P., Maddison D. R. 2006. Mesquite: a modular system for evolutionary analysis, v. 1.1 See http://mesquiteproject.org [Google Scholar]
- 43.Ree R. H., Smith S. A. 2008. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 57, 4–14 10.1080/10635150701883881 (doi:10.1080/10635150701883881) [DOI] [PubMed] [Google Scholar]
- 44.Wiens J. J., Brandley M. C., Reeder T. W. 2006. Why does a trait evolve multiple times within a clade? Repeated evolution of snakelike body form in squamate reptiles. Evolution 60, 123–141 [PubMed] [Google Scholar]
- 45.Vidal N., Azvolinsky A., Cruaud C., Hedges S. B. 2008. Origin of tropical American burrowing reptiles by transatlantic rafting. Biol. Lett. 4, 115–118 10.1098/rsbl.2007.0531 (doi:10.1098/rsbl.2007.0531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adalsteinsson S. A., Branch W. R., Trape S., Vitt L. J., Hedges S. B. 2009. Molecular phylogeny, classification, and biogeography of snakes of the Family Leptotyphlopidae (Reptilia, Squamata). Zootaxa 2244, 1–50 [Google Scholar]
- 47.Vidal N., et al. 2010. Blindsnake evolutionary tree reveals long history on Gondwana. Biol. Lett. 6, 558–561 10.1098/rsbl.2010.0220 (doi:10.1098/rsbl.2010.0220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lavoué S., Miya M., Arnegard M. E., McIntyre P. B., Mamonekene V., Nishida M. 2011 Remarkable morphological stasis in an extant vertebrate despite tens of millions of years of divergence. Proc. R. Soc. B 278, 1003–1008 10.1098/rspb.2010.1639 (doi:10.1098/rspb.2010.1639) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nydam R. L., Cifelli R. L. 2002. Lizards from the Lower Cretaceous (Aptian–Albian) Antlers and Cloverly formations. J. Vert. Paleontol. 22, 286–298 10.1671/0272-4634(2002)022[0286:LFTLCA]2.0.CO;2 (doi:10.1671/0272-4634(2002)022[0286:LFTLCA]2.0.CO;2) [DOI] [Google Scholar]
- 50.Burbrink F. T., Lawson R. 2007. How and when did Old World ratsnakes disperse into the New World? Mol. Phylogenet. Evol. 43, 173–189 10.1016/j.ympev.2006.09.009 (doi:10.1016/j.ympev.2006.09.009) [DOI] [PubMed] [Google Scholar]
- 51.Wuster W., Peppin L., Pook C. E., Walker D. E. 2008. A nesting of vipers: phylogeny and historical biogeography of the Viperidae (Squamata: Serpentes) (review). Mol. Phylogenet. Evol. 49, 445–459 10.1016/j.ympev.2008.08.019 (doi:10.1016/j.ympev.2008.08.019) [DOI] [PubMed] [Google Scholar]
- 52.Brandley M. C., Wang Y. Z., Guo X. G., de Oca A. N. M., Ortiz M. F., Hikida T., Ota H. 2010. Bermuda as an evolutionary life raft for an ancient lineage of endangered lizards. PLoS ONE 5, 4. 10.1371/journal.pone.0011375 (doi:10.1371/journal.pone.0011375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brandley M. C., Huelsenbeck J. P., Wiens J. J. 2008. Rates and patterns in the evolution of snake-like body form in squamate reptiles: evidence for repeated re-evolution of lost digits and long-term persistence of intermediate bodyforms. Evolution 62, 2042–2064 10.1111/j.1558-5646.2008.00430.x (doi:10.1111/j.1558-5646.2008.00430.x) [DOI] [PubMed] [Google Scholar]
- 54.de Queiroz A. 2005. The resurrection of oceanic dispersal in historical biogeography. Trends Ecol. Evol. 20, 68–73 10.1016/j.tree.2004.11.006 (doi:10.1016/j.tree.2004.11.006) [DOI] [PubMed] [Google Scholar]