Abstract
Proteins in the cupin superfamily have a wide range of biological functions in archaea, bacteria and eukaryotes. Although proteins in the cupin superfamily show very low overall sequence similarity, they all contain two short but partially conserved cupin sequence motifs separated by a less conserved intermotif region that varies both in length and amino acid sequence. Furthermore, these proteins all share a common architecture described as a 6-stranded β-barrel core, and this canonical cupin or “jelly roll” β-barrel is formed with cupin motif 1, the intermotif region, and cupin motif 2 each forming two of the core six β-strands in the folded protein structure. The recently obtained crystal structures of cysteine dioxygenase (CDO), with contains conserved cupin motifs, show that it has the predicted canonical cupin β-barrel fold. Although there had been no reports of CDO activity in prokaryotes, we identified a number of bacterial cupin proteins of unknown function that share low similarity with mammalian CDO and that conserve many residues in the active site pocket of CDO. Putative bacterial CDOs predicted to have CDO activity were shown to have similar substrate specificity and kinetic parameters as eukaryotic CDOs. Information gleaned from crystal structures of mammalian CDO along with sequence information for homologs shown to have CDO activity facilitated the identification of a CDO family fingerprint motif. One key feature of the CDO fingerprint motif is that the canonical metal-binding glutamate residue in cupin motif 1 is replaced by a cysteine (in mammalian CDOs) or by a glycine (bacterial CDOs). The recent report that some putative bacterial CDO homologs are actually 3-mercaptopropionate dioxygenases suggests that the CDO family may include proteins with specificities for other thiol substrates. A paralog of CDO in mammals was also identified and shown to be the other mammalian thiol dioxygenase, cysteamine dioxygenase (ADO). A tentative fingerprint motif for ADOs, or DUF1637 family members, is proposed. In ADOs, the conserved glutamate residue in cupin motif 1 is replaced by either glycine or valine. Both ADOs and CDOs appear to represent unique clades within the cupin superfamily.
Keywords: cysteamine dioxygenase, cysteine dioxygenase, cupin proteins, Domain of Unknown Function 1637 (DUF1637), iron-dependent enzymes, thiols
Introduction
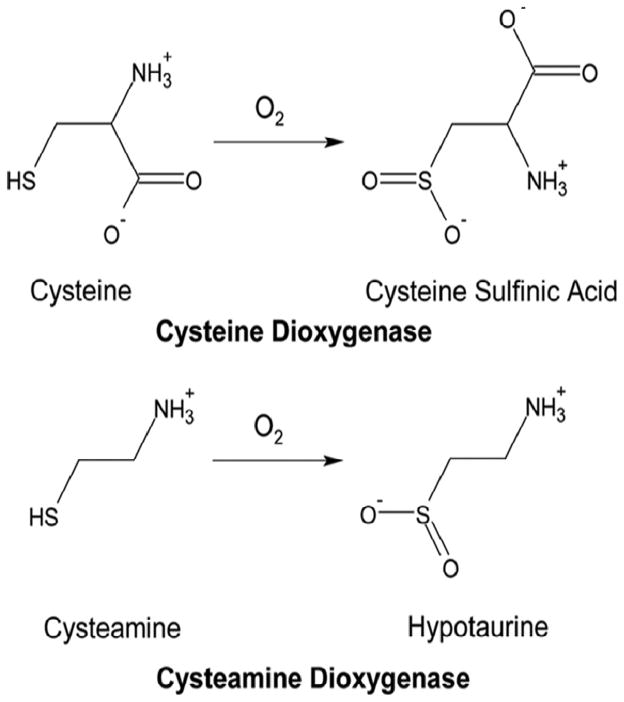
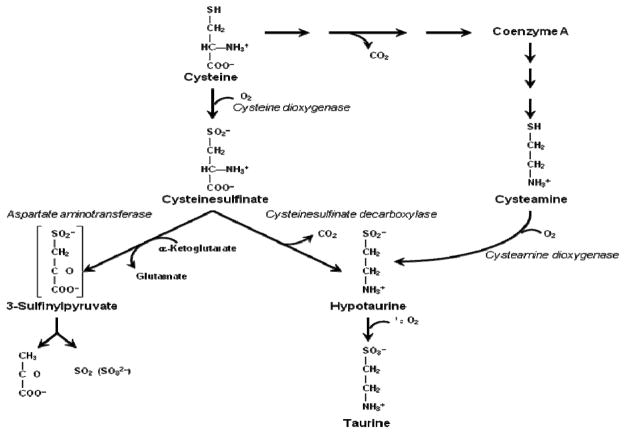
Cysteine metabolism to its sulfoxidation end-products is dependent upon two unique iron-dependent enzymes that are the only known mammalian thiol dioxygenases – enzymes adding molecular oxygen to a sulfhydryl group to form a sulfinic acid (See Fig. 1). These two thiol dioxygenases are cysteine dioxygenase (CDO) and cysteamine dioxygenase (ADO, 2-aminoethanethiol dioxygenase). Both of these thiol dioxygenases are essential for hypotaurine/taurine biosynthesis by the pathways shown in Fig. 2 (Stipanuk 1986, 2004; Stipanuk et al. 2009). CDO uses cysteine and dioxygen (O2) as substrates and converts these substrates to cysteinesulfinic acid, which can subsequently be either (i) decarboxylated to hypotaurine or (ii) transaminated to the putative intermediate 3-sulfinylpyruvate that spontaneously decomposes to pyruvate and sulfite. ADO uses cysteamine as a substrate. A cysteamine moiety (i.e., decarboxylated cysteine) is formed from cysteine in the process of coenzyme A synthesis, and this moiety is subsequently released during coenzyme A degradation. ADO converts cysteamine and O2 to hypotaurine. Hypotaurine is further oxidized to taurine. Hence, functional roles of CDO and ADO in metabolism include removal of thiol substrates (cysteine or cysteamine), thus regulating cysteine or cysteamine levels in body tissues and fluids; production of hypotaurine/taurine from two cysteine metabolites, cysteinesulfinate and cysteamine; and, for CDO, production of inorganic sulfur (sulfite/sulfate).
Fig. 1.
Reactions catalyzed by CDO and ADO.
Fig. 2.
Pathways of cysteine metabolism that require CDO and ADO.
The reactions catalyzed by CDO and ADO are notably different from those catalyzed by other classes of dioxygenases that have been studied: cysteine dioxygenation involves the oxidation of a sulfhydryl group rather than cleavage of a C-C bond or hydroxylation of a carbon atom; and both oxygen atoms from the oxygen molecule are transferred to a single sulfur atom rather than distributed between two carbon atoms. CDO was successfully purified from rat liver by Yamaguchi and coworkers in the 1970s, and they went on to characterize the enzyme and to show that it binds one atom of iron per molecule (Yamaguchi et al. 1978; Sakakibara et al. 1976).
The same group obtained the first cDNA clone for rat CDO (Hosokawa et al. 1990) and deduced its amino acid sequence. McCann et al. (1994) and Hirschberger et al. (2001) subsequently sequenced the cDNA for human and murine CDOs, demonstrating that all three messages encode a 200-amino acid residue protein with identical deduced amino acid sequences for the rat and murine CDOs and a similar sequence for human CDO that differs by only 16 amino acid residues.
CDO is a member of the cupin superfamily
CDO was initially placed in the cupin superfamily because it contains conserved cupin motifs (Dunwell et al. 2000). Proteins in the cupin superfamily or clan have a wide range of biological functions in archaea, bacteria and eukaryotes. The functions of proteins in the cupin superfamily include enzymatic activities, such as decarboxylases, dioxygenases, hydrolases, isomerases, and epimerases, and non-enzymatic functions, such as binding to auxin, transcription factors, and seed storage. Although proteins in the cupin superfamily show very low overall sequence similarity, they all contain two short but partially conserved cupin sequence motifs, Gx5HxHx3-6Ex 6 G (cupin motif 1) and Gx 5-7 PxGx2Hx 3N (cupin motif 2), separated by a less conserved intermotif region that varies both in length (~15 to 50 residues) and amino acid sequence. Despite the very low overall sequence similarity of cupin superfamily members, they all share a common architecture described as a 6-stranded β-barrel core with the functional site of the cupin protein generally located at the center of this conserved barrel (Dunwell et al. 2001).
The canonical cupin or “jelly roll” β-barrel is formed with cupin motif 1, the intermotif region, and cupin motif 2 each forming two of the core six β-strands in the folded protein structure. These core β-strands in cupin family members are denoted as βC-βH, in analogy with the notation first used for viral capsid proteins (Rossman et al. 1983) and subsequently adapted for application to the jelly-roll β-barrels of phaseolin (Lawrence et al. 1994) and canavalin (Ko et al. 2000). Thus, βC and βD, βE and βF, and βG and βH from the motif 1, intermotif, and motif 2 regions, respectively, are folded so that βC and βH, βD and βG, and βE and βF each form antiparallel strands.
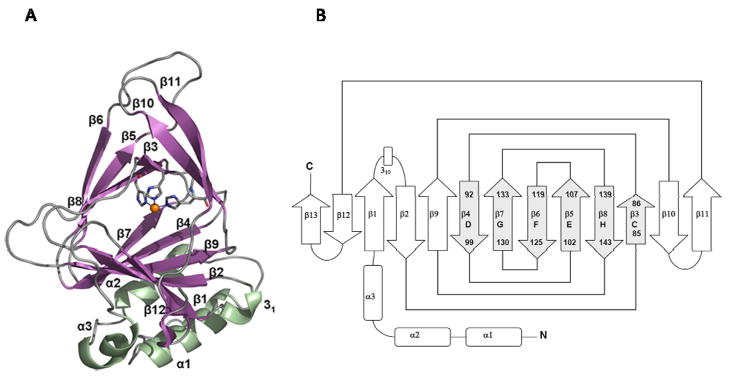
As predicted, this cupin fold was indeed revealed in the first crystal structures of mammalian CDOs. The entire β-sandwich of CDO is composed of seven anti-parallel β-strands (β1, β2, β4, β7, β9, β12, and β13) on the lower side and six anti-parallel β-strands (β3, β5, β6, β8, β10, and β11) on the upper side (McCoy et al. 2006; Simmons et al. 2006b; Ye et al. 2007). As illustrated in the topology diagram in Fig. 3, the 6-stranded β-barrel or cupin core of CDO comprises β-strands 3 and 4 of CDO, which correspond, respectively, to βC and βD of cupin motif 1, β-strands 5 and 6, which correspond, respectively, to βE and βF of the intermotif region, and β-strands 7 and 8, which correspond, respectively, to βG and βH of cupin motif 2).
Fig. 3.
Conserved cupin β barrel structure of CDO. (A) Ribbon illustration of the structure of rat CDO. (B) Topology diagram showing conserved cupin β-strands C-H and their jellyroll topology. Based on Simmons et al. (2006b).
CDO contains a novel Fe2+-center
More surprisingly, the structures of CDO also revealed a novel metal center. Cupins are typically metalloproteins with the metal binding amino acid residues accommodating a variety of metal ions. The identification of metal binding residues in cupin proteins was first accomplished with the structure of germin, a Mn2 +-containing oxalate oxidase in plants. The Mn2 + in germin was coordinated in a tetrad with three histidine residues and one glutamate site (Woo et al. 2000). Subsequently, crystal structures of Mn2+ oxalate decarboxylase from Bacillus subtilis (Tanner et al. 2001; Anand et al. 2002) and Cu2+ quercetin dioxygenase from Aspergillus japonicus (Fusetti et al. 2002) with similar metal coordination suggested that the 3-His-1-Glu metal coordination may be a typical feature of cupin metalloenzymes. In all three cases, the metal coordinating ligands included the two histidine residues and the conserved glutamate residue in cupin motif 1 along with the histidine residue in cupin motif 2 (i.e., the bolded residues in cupin motif 1 and 2 above). Although this 3-His-1-Glu metal coordination pattern is clearly most typical of members of the cupin superfamily, variations on this theme have been reported. For example, in 3-hydroxyanthranilate-3,4-dioxygenase, the first histidine in cupin motif 1 is not used and the glutamate residue in cupin motif 1 instead serves as a bidentate ligand for Fe2+ to preserve the metal-coordinating tetrad ( Zhang et al. 2005). A Mn2 +- containing cupin from Thermotoga maritime contains a fourth histidine in place of glutamate in cupin motif 1, giving rise to 4-His metal coordination (Jaroszewski et al. 2004).
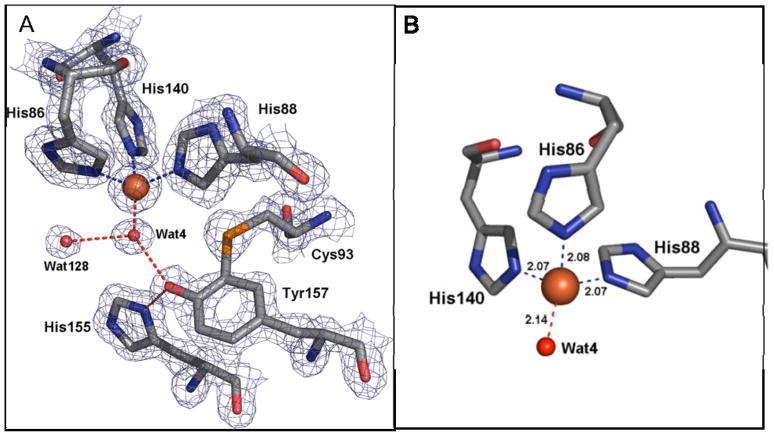
The iron center in CDO, however, was not the 3-His-1-Glu or a variation of this metal-coordinating tetrad. Instead, the crystal structures of CDO revealed coordination of Fe2+ by a facial triad of 3 histidine side chains with no carboxylate available near the iron center to serve as an additional ligand (McCoy et al. 2006; Simmons et al. 2006b; Ye et al. 2007). Although it was already known from sequence data that the conserved glutamate ligand in cupin motif 1 of CDO was replaced by a cysteine residue (i.e., Cys93) in mammalian CDOs, the crystal structure revealed that no other residue replaced the conserved glutamate as a metal-coordinating ligand as might have been predicted. Thus, in contrast to the 2-His-1-carboxylate triad structural motif found in most natural mononuclear non-heme Fe2+-containing proteins of various fold families or the the 3-His-1-Glu metal-coordinating tetrad found in many cupin family proteins, CDO uses a 3-His triad to coordinate Fe2+. The crystal structure of resting CDO most surprising feature of the metallocenter of CDO, because metal-binding centers in the cupin superfamily as well as mononuclear iron enzymes in other protein families typically have penta- or hexa-coordinated metal centers. In fact, to our knowledge, such pure tetrahedral coordination of mononuclear iron has not been seen apart from that found in the Fe(Cys)4 iron centers of rubredoxin-like electron carriers (Chen et al. 2006).
In addition to the Fe2+-coordinating residues (His86, His88, and His140) , the substrate binding pocket of mammalian CDO also contains key conserved residues including Tyr157, the residue that forms the cysteinyltyrosine (Cys-Tyr) cofactor, Tyr58, Arg60, Ser153, and His155 (McCoy et al. 2006; Simmons et al. 2006b; Ye et al. 2007). Also conserved are Leu154 buried in a neighboring aliphatic pocket, and cis-Pro159-Pro160 located in a loop between β9 and β10, Ser83 which H-bonds to the backbone NH of residue 142 (very close to the metal ligand His140), and Phe167 packed behind the main chain containing the metal ligands His86 and His88. In addition, Asp87 is positioned between the iron-coordinating residues His86 and His88, and the Asp87 carboxylate interacts electrostatically with Asp87-N and Thr89-N, maintaining the position of the iron ligands, as well as with the backbone and side chain of His165 in the neighboring β-strand that contributes to the active site.
One might predict that the novel metallocenter of CDO is important for its unique reaction chemistry. The crystal structure of human CDO in complex with its substrate L-cysteine (Ye et al. 2007) and the crystal structure of rat CDO with an Fe2+ bound persulfenate intermediate trapped in its active site pocket (Simmons et al. 2008) have yielded some clues about reaction chemistry. Both the CDO-cysteine and the CDO-cysteine persulfenate structures reveal the coordination of both the S (thiol or persulfenate) and N (α-amino group) atoms with the Fe2+ and the latter revealed that Fe2+ directly activates both the cysteine and O2 substrates. Results of studies done with electron paramagnetic resonance (EPR) were consistent with cysteine binding first to the free enzyme to alter the coordination of the Fe2+ and create the O2 binding site (Pierce et al. 2007). Several catalytic mechanisms for CDO have been predicted, and all mechanisms that preceded the CDO-cysteine persulfenate structure (Simmons et al. 2008) predicted that initial thiol oxidation occurs via sulfur attaching the Fe-distal or terminal oxygen atom (McCoy et al. 2006; Simmons et al. 2006b; Ye et al. 2007; Pierce et al. 2007; Aluri and de Visser 2007). In contrast to these predictions, the CDO-cysteine persulfenate complex suggests the possibility that initial thiol oxidation occurs via sulfur attacking the Fe-proximal oxygen, rather than the distal or terminal oxygen atom, with product formation involving an intramolecular isomerization (Simmons et al. 2008). This evidence along with the electron-rich character of the thiol functional group and the availability of low-energy empty d orbitals on the sulfur suggests that thiol dioxygenases may use a very different reaction mechanism than that used by other dioxygenases that add oxygen to carbon atoms.
Identification of a novel crosslink in eukaryotic CDOs
The crystal structures of mammalian CDO (McCoy et al. 2006; Simmons et al. 2006b; Ye et al. 2007) all revealed a rare Cys-Tyr intramolecular crosslink (between Cys93 and Tyr157), as shown in Fig. 4. Such thioether-containing “cofactors” in the active sites of enzymes are rare and have previously been reported only for galactose oxidase (Ito et al. 1991), glyoxal oxidase (Whittaker et al. 1996, 1999), and NirA, a sulfite reductase (Schnell et al. 2005). The cysteine residue (Cys93) involved in this thioether crosslink is the cysteine residue in CDO that replaces the conserved glutamate residue in cupin motif 1, suggesting that both the lack of a glutamate residue and the Cys-Tyr thioether are important for CDO function.
Fig. 4.
Active site pocket of CDO. (A) Coordination of iron by His86, His88, His140 and a solvent molecule (Wat) and the Cys-Tyr intramolecular crosslink between Cys93-Sγ and Tyr157-Cε2. (B) Tetrahedral geometry of metallocenter in rat CDO. Based on Simmons et al. (2006b).
Two major CDO peaks or bands with a small apparent mass difference are routinely observed when CDO from cells or animal tissues (or recombinant CDO expressed in bacterial systems) is analyzed by chromatographic or electrophoretic methods (Stipanuk et al. 2004; Simmons et al. 2006a; Stipanuk et al. 2002; Dominy et al. 2008); electrophoretic migration yields apparent molecular masses of 23 kDa and 22.5 kDa for the two isoforms. The new information obtained from the crystal structure about the internal Cys-Tyr cross-link allowed us to demonstrate that the apparently smaller “22.5” kDa isoform of CDO was, in fact, the thioether-crosslinked or “mature” enzyme. The knowledge that CDO forms an intramolecular Cys-Tyr crosslink was consistent with the small mass difference as well as with the fact that the “22.5 kDa” isoform band had a slightly higher pI (pI=5.85 versus pI=5.80 for the 23 kDa band). Furthermore, the electrophoretic migration patterns for these two forms of CDO are similar to those reported for galactose oxidase, which also forms a Cys-Tyr crosslink (Whittaker and Whittaker, 2003). When we mutated either Cys93 or Tyr157 of CDO to block Cys-Tyr cross-link formation, the mutant CDO migrated as a single 23 kDa band on SDS-PAGE (Dominy et al. 2008). In addition, 2-D gel electrophoresis to separate the isoforms followed by mass spectrometric studies of trypsin- and chymotrypsin-digests of the two isoforms showed the presence of the predicted unlinked peptides containing Cys93 and Tyr157 in the 23 kDa but not in the “22.5 kDa” band (Dominy et al. 2008). Kleffmann et al. (2009) were subsequently successful in positively identifying the cross-linked peptide. Thus, CDO exists both as a non-thioether-containing immature isoform that migrates at the predicted molecular mass of ~23 kDa and a Cys-Tyr thioether-containing mature isoform that migrates with an apparent mass of ~22.5 kDa.
In galactose oxidase, the protein-derived thioether crosslink, or cofactor, formed spontaneously upon exposure to Cu+1 and O2 and was essential for catalytic activity (Whittaker and Whittaker 2003; Rogers et al. 2000). A recent report indicates that O2 is not strictly required for cofactor formation in galactose oxidase but it is unclear what the final electron acceptor would be under anaerobic conditions (Rogers et al. 2008). Crosslink formation in CDO clearly follows a different pattern. Both the mature and immature isoforms of CDO have catalytic activity (Stipanuk et al. 2004; Simmons et al. 2006a). Additionally, mutation of either the cysteine or tyrosine residue that forms the thioether crosslink in wildtype CDO (i.e., Cys93Ser, Cys93Ala, and Tyr153Phe) yielded active enzyme, albeit with lower activity than wildtype CDO (Dominy et al. 2008; Ye et al. 2007). Having confirmed the correspondence of the presence of the intramolecular crosslink with a shift in CDO’s electrophoretic migration, we had an easy means to “assay” thioether crosslink formation. This, in turn, allowed us to evaluate the requirements for intramolecular thioether synthesis and the effects of thioether cofactor formation to the catalytic activity and stability of the enzyme (Dominy et al. 2008).
Some catalytic activity in the immature CDO was necessary for thioether crosslink formation; inactive mutant forms of CDO (e.g., His86Ala) did not form any of the mature isoform, and mutant forms of CDO with low activity (e.g., Arg60Ala, which has a markedly decreased affinity for cysteine) formed the crosslink more slowly (Dominy et al. 2008). Mutations of nonessential residues (e.g., Tyr58Phe; Ser153Ala) had little effect. Like other amino acid cofactor-containing enzymes, formation of the Cys-Tyr cofactor in CDO required a transition metal cofactor (Fe2+) and O2, but unlike other amino acid cofactor-containing enzymes, biogenesis of the CDO cofactor did not occur immediately upon exposure to metal and O2, and was strictly dependent upon the presence of its specific substrate cysteine. Substrate and substrate turnover were essential for CDO cofactor formation, both in assays of purified CDO and for CDO in intact cells. Even in the presence of Fe2+, O2, and cysteine, cofactor formation was appreciably slower than the rates reported for other amino acid cofactor-containing enzymes and appeared to require hundreds of catalytic turnover cycles to occur.
Although cofactor formation appears to occur as a side reaction, it is clear that CDO does mature in cells in culture and in tissues within the body where both forms are found (Dominy et al. 2008; Stipanuk et al. 2004) and it seems likely that this is of physiological significance.
Although immature CDO that has not formed the Cys-Tyr cofactor possesses appreciable catalytic activity, cofactor formation appears to increase CDO catalytic efficiency by more than 10-fold, as judged by the catalytic efficiency of the mature Cys-Tyr cofactor-containing CDO compared with that of the immature Cys93Ser mutant CDO that cannot form the crosslink (kcat/Km of 3200 M−1s−1 versus 220 M−1s−1) (Dominy et al. 2008). The mature isoform of CDO also appeared to have about double the catalytic half-life as the immature isoform, based on assays performed in vitro.
Overall, the results indicate that cysteine’s ability to modulate internal Cys-Tyr cofactor formation represents a very unusual form of feed-forward activation of enzyme activity. This is a unique finding as substrate-regulation of protein cofactor formation has never before been reported, to our knowledge, as a means of regulating a protein’s normal function.
Identification of Bacterial CDOs
In contrast to the known function of CDO in metazoa and fungi, there had been no experimental evidence for the presence of CDO in prokaryotes. However, a PSI-BLAST search using rat CDO as the query sequence uncovered many bacterial cupin proteins of unknown function that share low similarity with rat CDO (12 to 21% identity). These proteins conserved many of the residues in the active site pocket of CDO, with the notable exception of the Cys93 in the canonical position of the glutamate in cupin motif 1 of mammalian CDO. In these putative bacterial CDOs, this position is highly conserved as a glycine. Because of this difference, combined with the fact that the overall similarity of the proteins is not much higher than the similarity of rat CDO with functionally unrelated cupins such as cupin phosphoglucose isomerase, it was not possible to conclusively classify these proteins as bona fide CDOs based upon sequence data alone. From the PSI-BLAST search data, which yielded 38 non-redundant bacterial sequences encoding putative CDOs, we selected four proteins to characterize (Dominy et al. 2006). First, we selected the YubC protein from Bacillus subtilis, which was the top bacterial hit (e-value = 0.05 vs. rat CDO) and also of interest because it contains a cysteine residue within motif 1, one residue downstream on its relative position in eukaryotic CDO, and thus could conceivably form an internal thioether bond. Then, we selected BC2617 from Bacillus cereus (e-value = 0.83 vs. rat CDO) and two divergent paralogs from Streptomyces coelicolor A3-2 (SCO3035 with e = 0.059 vs. rat CDO and SCO5772 with e = 0.45 vs. rat CDO; 25% identity with each other). BC2617 and SCO3035 contain cysteine residues, but these residues all lie well outside of motif 1 and thus would not be predicted to form an internal thioether. SCO5772 contains no cysteine residues at all and definitely could not form an internal thioether bond. All four of these proteins had previously been annotated as hypothetical cupin proteins of unknown function or as putative cysteine dioxygenase-like proteins. The open reading frames of these putative CDOs were cloned, and the proteins were heterologously expressed and purified to homogeneity by a single-step immobilized metal affinity chromatography procedure. All four of the recombinant proteins converted cysteine to cysteinesulfinate, had kinetic parameters similar to those of rat CDO, and exhibited a high level of substrate specificity as assessed by a lack of inhibition of cysteine dioxygenation by cysteine analogs, suggesting that these four proteins are indeed bona fide CDOs.
Having confirmed that four predicted bacterial CDOs did in fact catalyze the oxidation of cysteine to cysteinesulfinate, we did a BLASTP search using the sequences for the four characterized CDOs as the query sequence (Dominy et al. 2006). Twenty-one putative bacterial homologs (e-value of <1.5) with good conservation of active-site residues were identified. Interestingly, among the 21 putative homologs two proteins (ZP_00991440 from Vibrio splendidus and NP_902852 from Chromobacterium violaceum) differed from the rest, having conservation of the unique cysteine found in motif 1 of eukaryotic CDOs; these two proteins were not originally identified in our initial PSI-BLAST query based on the sequence of rat CDO.
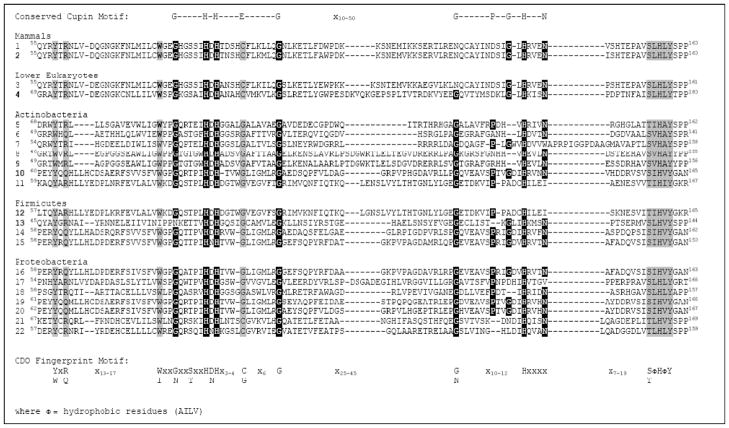
By sequence alignment of eukaryotic and bacterial proteins known to be active cysteine dioxygenases, as well as those of the most closely related homologs identified in the BLAST searches, we derived a consensus CDO family fingerprint (i.e., a set of motifs based on conserved regions of sequence alignments, which may be separated along the sequence although not necessarily separated in 3D space). The CDO protein family fingerprint includes conserved regions on either side of the cupin motifs (see Fig. 5):
where Φ = hydrophobic residues (AILV)
Fig. 5.
Conservation of functional residues within eukaryotic and putative bacterial CDOs. Sequence alignment includes the cupin motifs plus regions on either side of the cupin motifs that are highly conserved in all proteins shown to have CDO activity. Conserved cupin motif residues are highlighted in
 . Conserved CDO “fingerprint motifs” that are not part of the conserved cupin motif are highlighted in
. Conserved CDO “fingerprint motifs” that are not part of the conserved cupin motif are highlighted in
 . Numbered CDO sequences (1–22) are from the following strains or species and for the listed National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) protein sequence accession numbers: 1, Rattus norvegicus, BAA11925; 2, Homo sapiens, BAA12873; 3, Xenopus tropicalis, AAH61333; 4, Histoplasma capsulatum, AAV66535; 5, Frankia sp. strain EAN1pec, YP_001509251; 6, Janibacter sp. strain HTCC2649, ZP_00993719; 7, Mycobacterium tuberculosis, CAA17181; 8, Streptomyces avermitilis[SAV2489], NP_823665; 9, Streptomyces coelicolor A3(2) [SCO3035], NP_627257; 10, Streptomyces coelicolor A3(2) [SCO5772], NP_629897; 11, Bacillus anthracis, ZP_00392898; 12, Bacillus cereus, NP_832375; 13, Bacillus subtilis, NP_390992; 14, Bacillus thuringiensis, ZP_00744219; 15, Azotobacter vinelandii, YP_002800236; 16, Burkholderia ambifaria, YP_77557; 17, Burkholderia cenocepacia, ZP_00461417; 18, Myxococcus xanthus, AAF87926; 19, Polaromonas sp. strain JS666, YP_548490; 20, Ralstonia eutropha JMP134, YP_299237; 21, Vibrio splendidus, ZP_00991440; 22, Chromobacterium violaceum ATCC, NP_902852. The putative CDOs indicated by species/accession numbers shown in bold font have been shown to be functional cysteine dioxygenases with specificity for cysteine
. Numbered CDO sequences (1–22) are from the following strains or species and for the listed National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) protein sequence accession numbers: 1, Rattus norvegicus, BAA11925; 2, Homo sapiens, BAA12873; 3, Xenopus tropicalis, AAH61333; 4, Histoplasma capsulatum, AAV66535; 5, Frankia sp. strain EAN1pec, YP_001509251; 6, Janibacter sp. strain HTCC2649, ZP_00993719; 7, Mycobacterium tuberculosis, CAA17181; 8, Streptomyces avermitilis[SAV2489], NP_823665; 9, Streptomyces coelicolor A3(2) [SCO3035], NP_627257; 10, Streptomyces coelicolor A3(2) [SCO5772], NP_629897; 11, Bacillus anthracis, ZP_00392898; 12, Bacillus cereus, NP_832375; 13, Bacillus subtilis, NP_390992; 14, Bacillus thuringiensis, ZP_00744219; 15, Azotobacter vinelandii, YP_002800236; 16, Burkholderia ambifaria, YP_77557; 17, Burkholderia cenocepacia, ZP_00461417; 18, Myxococcus xanthus, AAF87926; 19, Polaromonas sp. strain JS666, YP_548490; 20, Ralstonia eutropha JMP134, YP_299237; 21, Vibrio splendidus, ZP_00991440; 22, Chromobacterium violaceum ATCC, NP_902852. The putative CDOs indicated by species/accession numbers shown in bold font have been shown to be functional cysteine dioxygenases with specificity for cysteine
Examining this consensus CDO family motif fingerprint confirms the importance of the metal binding ligands, with His 86, His88, and His140 of mammalian CDOs being strictly conserved in bacterial CDOs as well. In addition, the absence of the Glu metal ligand from cupin motif 1 appears to be critical, although the Glu is substituted by Gly rather than Cys in most bacterial CDOs. This suggests that the capacity for CDO to form a Cys-Tyr crosslink (i.e., Cys93-Tyr157 in mammalian CDOs) is not essential, and this is consistent with the structure of the putative CDO from Ralstonia eutropha (Cupriavidus necator) JMP134 (PDB 2GM6), released by the Joint Center for Structural Genomics, which showed a well-conserved active site despite the absence of a cysteinyl-tyrosine bond.
Consistent with all proposed mechanisms for CDO activity, the Tyr157 is strictly conserved, and the Ser153·His155·Tyr157 triad is also conserved in all known CDOs: the sequence motif x7-19(S/T)ΦHΦY appears immediately following cupin motif 2 in CDOs. A small sequence immediately before cupin motif 1 [(Y/W)x(R/Q)x13-17(W/I)x2] contains the Tyr58 and Arg60 that have been proposed to play important roles in the substrate binding pocket. In comparison to residues predicted to be important for CDO activity based on the mammalian CDO structures, the CDO family fingerprint suggests that Thr89, Pro159, Pro160 and Phe167 are not well conserved in bacterial CDOs and, thus, that these residues are not critical.
The importance of the CDO consensus motifs outside the conserved cupin motif regions in predicting which homologs are likely to have CDO activity was illustrated by our ability to predict that two cyanobacterial proteins that were identified in the BLASTP search, which we performed using the four confirmed bacterial CDO sequences as query sequences, would not have CDO activity (Dominy et al. 2006). Both of these proteins shared some of the conserved active-site residues seen in CDOs but were missing other key conserved residues, notably some of those that lie outside of cupin motifs 1 and 2 (i.e., neither had the complete Ser·His·Tyr triad and both lacked the conserved basic residue corresponding to Arg60). Although these cyanobacterial proteins (ZP_00516761 from Crocosphaera watsonii and ZP_00112383 from Nostoc sp. strains PCC73102) were annotated in the National Center for Biotechnology database (www.ncbi.nlm.nih.gov/) as encoding putative CDOs, these do not appear to be true CDOs. We cloned ZP_00112383 from genomic DNA and found the gene product had no CDO activity.
Thus, based on both the differences in CDO sequences and structures, which are quite distinct from other cupins, CDO seems to represent a separate evolutionary clade within the cupin superfamily. Our phylogenetic analyses of the putative bacterial CDO homologs indicate that CDO is distributed among species within the phyla of Actinobacteria, Firmicutes, and Proteobacteria (Dominy et al. 2006). Collectively, these data suggest that a large subset of eubacteria is capable of cysteine sulfoxidation, but the role of this reaction in these organisms remains to be elucidated. It will also be of interest to determine if the two bacterial homologs of CDO that we have listed as putative CDOs and that do contain a conserved Cys93, rather than the Gly substitution commonly observed in bacterial CDOs, are able to form the thioether cofactor.
Other members of the CDO clade
Bruland et al. (2009) recently identified two bacterial CDO homologs that catalyze the oxidation of 3-mercaptopropionate. These investigators were studying the pathway for 3,3-thiodipropionic acid metabolism in a group of proteobacteria that they had enriched from the environment based on the ability of the organisms to use 3,3-thiodipropionic acid as the sole source of carbon and energy for growth. Mutagenesis of isolate Variovorax paradoxus TBEA6 yielded mutants fully or partially impaired in growth on 3,3-thiodipropionic acid. Genotypic characterization of two of these mutants demonstrated the involvement of a putative gene encoding a cysteine dioxygenase homolog in the further catabolism of the 3, 3-thiodipropionic acid cleavage product 3-mercaptopropionic acid. Further characterization of this CDO homolog (ACB72254) showed that it used 3-mercaptopropionate as substrate, converting it to 3-sulfinopropionate, and that the CDO homolog did not use either cysteine or cysteamine as substrate.
One of two putative CDO genes in Ralstonia eutropha H16 was also shown to be a 3-mercaptopropionate dioxygenase (Bruland et al. 2009). Heterologous expression of this putative CDO (YP_841375), which has high sequence similarity to the V. paradoxus mercaptopropionate dioxygenase, in the “putative CDO”-disrupted mutant of V. paradoxus restored its growth on 3,3-thiodipropionic acid. In contrast, another CDO homolog from R. eutropha H16 ( YP_726114), which has much lower sequence similarity to the V. paradoxus mercaptopropionate dioxygenase, did not restore the ability of the V. paradoxus mutants to grow on 3, 3-thiopropionic acid as the sole source for carbon and energy, indicating that this paralog did not have mercaptopropionate dioxygenase activity. In addition, production of 3-sulfinopropionate was detected in cells expressing YP_841375 and grown on 3-mercaptopropionic acid but not in cells expressing the less similar paralog YP_726114. Thus, Bruland et al. (2009) have clearly shown that not all putative CDOs use cysteine as substrate, although the mercaptopropionate dioxygenases carry out a similar reaction with a similar thiol substrate. This raises the question of whether the CDO clade contains a variety of thiol dioxygenases that act on different substrates. This is particularly likely for bacteria (e.g., R. eutropha, Bordetella pertussis, and Verminephrobacter eiseniae) that often encode more than one putative CDO. As shown in Fig. 6, both of the proteins with 3-mercaptopropionate dioxygenase activity possess the CDO family fingerprint, and, thus, they would not have been identified as a potential subclass of CDOs with different substrate specificity on the basis of amino acid sequence. It is, perhaps, notable that the 3-mercaptopropionate dioxygenases from V. paradoxus and R. eutropha both contain a well-conserved cupin motif 2 and have relatively short intermotif regions compared to the CDO fingerprint. In addition, the enzyme from V. paradoxus has an alanine replacing the conserved glutamate in cupin motif 1, whereas that from R. eutropha has the more typical glycine residue. Additional kinetic and structural work should further unfold this interesting story. Interestingly, the putative CDO from R. eutropha JMP134 (YP_299237) whose crystal structure has been deposited (PDB 2GM6) has high sequence similarity (83%) to the mercaptopropionate dioxygenase from R. eutropha H16 (YP_841375).
Fig. 6.
Sequence alignment of CDO “fingerprint motifs” for mercaptopropionate dioxygenases from 1, Variovorax paradoxus , ACB72254 and 2, Ralstonia eutropha H16, YP_841375. Conserved CDO fingerprint motifs and/or cupin motifs are highlighted in
 . An alternate replacement of the glutamate residue in cupin motif 1 (with an alanine, instead of a cysteine or glycine) is shown in
. An alternate replacement of the glutamate residue in cupin motif 1 (with an alanine, instead of a cysteine or glycine) is shown in
 . Both proteins have been demonstrated to be mercaptopropionate dioxygenases instead of cysteine dioxygenases ( Bruland et al. 2009).
. Both proteins have been demonstrated to be mercaptopropionate dioxygenases instead of cysteine dioxygenases ( Bruland et al. 2009).
A number of highly similar homologs of the mercaptopropionate dioxygenases from V. paradoxus and R. eutropha can be found by a BLAST search, particularly in other species within the same Burkholderiales order of betaproteobacteria, which are often found in environmental samples such as waste water or soil. Mercaptopropionate is derived from biological and abiotic reactions and is found in costal sediments. The ability of these organisms to use 3-mercaptopropionate could be beneficial for growth of soil bacteria and also serve to release the sulfur back into the sulfur cycle.
Identification of the Mammalian Cysteamine Dioxygenase
Although many tissues are capable of converting cysteamine to hypotaurine, the lack of identification of the gene for cysteamine dioxygenase and the failure of early efforts to consistently purify a protein with cysteamine dioxygenase activity (Cavallini et al. 1963, 1975; Richerson and Ziegler 1987) had delayed progress in understanding the metabolism of cysteamine, a product of the constitutive degradation of coenzyme A, and the synthesis of taurine, the final product of cysteamine oxidation and the most abundant free amino acid in mammalian tissues. Given that both CDO and cysteamine dioxygenase use a thiol substrate and catalyze a similar thiol dioxygenation reaction, we hypothesized that they may share a phylogenetic connection. A PSI-BLAST search of the murine genome using murine/rat CDO as the query sequence yielded a hypothetical murine paralog of CDO that is encoded by the gene Gm237 (Gene model 237, GI:88984114, protein accession number Q6PDY2) and belongs to the DUF1637 (Domain of Unknown Function 1637) protein family (Dominy et al. 2008). Although its overall identity to CDO is low, it is similar to CDO in that it lacks the highly conserved glutamate residue found in motif 1 of many other metal binding cupins but not in that of CDO. Similarly, a PSI-BLAST search of the human genome using mammalian CDO as the query sequence yielded an unnamed predicted protein product encoded by C10orf22 (chromosome 10 open reading frame 22) that is homologous (85% identity) to that encoded by murine gene Gm237 (i.e., protein Q6PDY2) (Dominy et al. 2008).
When expressed as a recombinant protein, murine protein Q6PDY2 exhibited significant cysteamine dioxygenase activity in vitro (Dominy et al. 2008). The reaction was highly specific for cysteamine; cysteine was not oxidized by the enzyme, and structurally related compounds were not competitive inhibitors of the reaction. When overexpressed in HepG2/C3A cells, protein Q6PDY2 increased the production of hypotaurine from cysteamine. Similarly, when endogenous expression of the human ortholog C10orf22 in HepG2/C3A cells was reduced by RNA-mediated interference, hypotaurine production decreased (Dominy et al. 2008). In addition, purified Q6PDY2 contained one atom of iron per molecule, and mutation of the first conserved metal-binding histidine in cupin motif 1 to an alanine reduced bound iron levels to trace amounts in the recombinant purified protein and also resulted in no detectable catalytic activity (Dominy et al. 2008). Overall, these data suggested that murine Gm237 and human C10orf22 encode the enzyme that is responsible for endogenous cysteamine dioxygenase activity. In response to the publication of this work, the gene/protein data bank sequences for Gm237 and C10orf22 and their orthologs have now been named cysteamine dioxygenases, and the DUF1637 protein family is considered to be made up of putative cysteamine dioxygenases (e.g., see the NCBI Entrez Protein database at www.ncbi.nlm.nih.gov/sites/entrez or the Pfam database at pfam.sanger.ac.uk).
A comparison of mammalian ADO sequences with those of homologs present in other eukaryotes (Fig. 7) indicates that the iron-binding ligand motif of CDO cupin motif 1 is conserved: x6H(D/N)Hx4(G/V)x6G, with the conserved glutamate residue being replaced by either glycine or valine (as compared to cysteine or glycine in CDO). Although the cupin motif 2 residues are less-well conserved among all members of the DUF1637 protein family, all contain what is well-positioned to be the third metal-binding histidine residue and all but one contain the proline six to ten residues earlier (Fig. 7). Although there is not yet proof that the conserved histidine (e.g., His193 in human ADO) is the third iron ligand, the alignment is rather compelling.
Fig. 7.
Conservation of functional residues within eukaryotic ADOs and putative ADOs. Sequence alignment includes the cupin motifs plus regions on either side of the cupin motifs that are highly conserved in all proteins shown to have ADO activity. Conserved cupin motif residues are highlighted in
 . Conserved ADO “fingerprint motifs” that are not part of the conserved cupin motif are highlighted in
. Conserved ADO “fingerprint motifs” that are not part of the conserved cupin motif are highlighted in
 . Numbered CDO sequences (1–15) are from the following strains or species and for the listed NCBI protein sequence accession numbers: 1, Homo sapiens, NP_116193.2; 2,Macaca mulatta, XP_001092839; 3, Mus musculus, Q6PDY2.2; 4, Rattus norvegicus, NP_001101096; 5, Danio rerio, AAH65461 6, Xenopus laevis, AAH82884; 7, Drosophila melanogaster, NP_648176; 8, Schistosoma japonicum, AAW27563; 9, Strongylocentrotus purpuratus, XP_001183066; 10,Dictyostelium discoideum, XP_644380; 11, Leishmania major, CAJ02170; 12, Trypanosoma brucei, AAX78835; 13, Medicago truncatula, ABD28522; 14, Arabidopsis thaliana, NP_191426; 15, Oryza sativa, BAB03364. The putative ADOs indicated by species/accession numbers shown in bold font have been shown to be functional cysteamine dioxygenases with specificity for cysteamine.
. Numbered CDO sequences (1–15) are from the following strains or species and for the listed NCBI protein sequence accession numbers: 1, Homo sapiens, NP_116193.2; 2,Macaca mulatta, XP_001092839; 3, Mus musculus, Q6PDY2.2; 4, Rattus norvegicus, NP_001101096; 5, Danio rerio, AAH65461 6, Xenopus laevis, AAH82884; 7, Drosophila melanogaster, NP_648176; 8, Schistosoma japonicum, AAW27563; 9, Strongylocentrotus purpuratus, XP_001183066; 10,Dictyostelium discoideum, XP_644380; 11, Leishmania major, CAJ02170; 12, Trypanosoma brucei, AAX78835; 13, Medicago truncatula, ABD28522; 14, Arabidopsis thaliana, NP_191426; 15, Oryza sativa, BAB03364. The putative ADOs indicated by species/accession numbers shown in bold font have been shown to be functional cysteamine dioxygenases with specificity for cysteamine.
The nature of the iron-binding site in ADO is undoubtedly different from that in CDO based on the marked difference in the ability of iron to dissociate from the two proteins. Most of the iron was lost from CDO during its purification, whereas essentially all of the bound iron was retained in ADO throughout purification (Dominy et al. 2007; Simmons et al. 2006a, 2006b).
Sequence alignment of members of the DUF1637 family results in the following fingerprint motif for family members and, presumably, for ADOs:
where Φ = hydrophobic residues (LIY)
This fingerprint motif for ADOs differs considerably from that for CDOs. The first residues of cupin motif 1 (just before the first metal-binding histidine) are x3IPx in ADOs and other family members compared to Gx2Sx2. In addition to the extremely weak conservation of cupin motif 2, the particular residues conserved on either side of the cupin regions differ. In DUF1637 family members, the sequence (I/L)x10Fx4 is conserved immediately before cupin motif 1, compared with conservation of (Y/W)x(R/Q)x13-17(W/I)x2 immediately before cupin motif 1 in CDOs. Following cupin motif 2, the sequence x10-13D(I/V)(L/I)xPPYx7-11Cx2(Y/F) is conserved in DUF1637 family members, compared with x10-21(S/T)ΦHΦY in CDOs. The roles of the strictly conserved downstream residues in ADO, especially the tyrosine and cysteine residues, need to be determined. We speculate that the conserved tyrosine in the latter ADO fingerprint is structurally equivalent to the active site tyrosine conserved in the CDO fingerprint. Interestingly, mammalian ADO migrates as two or three bands during electrophoresis, suggesting that it may contain an intramolecular crosslink, like CDO, or undergo other types of posttranslational modification (Dominy et al. 2006).
Further structural studies are needed to shed light on the function of the conserved residues in ADOs or DUF1637 family proteins. Given the likelihood of differences in the catalytic mechanisms of CDO and ADO, further characterization of both catalytic reactions should yield significant insights into the chemistry of thiol oxidation. ADOs, like CDOs, appear to represent a unique clade within the cupin superfamily. Further study of these two thiol dioxygenases and their homologs should clarify the members and functions of the cupin families represented by CDO and by ADO.
Acknowledgments
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases through Public Health Service Grant # DK056649 (to MHS).
Abbreviations
- ADO
cysteamine dioxygenase, or 2-aminoethanethiol dioxygenase
- CDO
cysteine dioxygenase
References
- Aluri S, de Visser SP. The mechanism of cysteine oxygenation by cysteine dioxygenase enzymes. J Am Chem Soc. 2007;129:14846–14847. doi: 10.1021/ja0758178. [DOI] [PubMed] [Google Scholar]
- Anand R, Dorrestein PC, Kinsland C, Begley TP, Ealick SE. Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 A resolution. Biochemistry. 2002;41:7659–7669. doi: 10.1021/bi0200965. [DOI] [PubMed] [Google Scholar]
- Bruland N, Wübbeler JH, Steinbüchel A. 3-mercaptopropionate dioxygenase, a cysteine dioxygenase homologue, catalyzes the initial step of 3-mercaptopropionate catabolism in the 3,3-thiodipropionic acid-degrading bacterium Variovorax paradoxus. J Biol Chem. 2009;284:660–672. doi: 10.1074/jbc.M806762200. [DOI] [PubMed] [Google Scholar]
- Cavallini D, Scandurra R, Demarco C. The enzymatic oxidation of cysteamine to hypotaurine in the presence of sulfide. J Biol Chem. 1963;238:2999–3005. [PubMed] [Google Scholar]
- Cavallini D, Federici G, Ricci G, Dupre S, Antonucci A. The specificity of cysteamine oxygenase. FEBS Lett. 1975;56:348–351. doi: 10.1016/0014-5793(75)81124-0. [DOI] [PubMed] [Google Scholar]
- Chen CJ, Lin YH, Huang YC, Liu MY. Crystal structure of rubredoxin from Desulfovibrio gigas to ultra-high 0.68Å resolution. Biochem Biophys Res Commun. 2006;349:79–90. doi: 10.1016/j.bbrc.2006.07.205. [DOI] [PubMed] [Google Scholar]
- Dominy JE, Jr, Simmons CR, Karplus PA, Gehring AM, Stipanuk MH. Identification and characterization of bacterial cysteine dioxygenases: a new route of cysteine degradation for eubacteria. J Bacteriol. 2006;188:5561–5569. doi: 10.1128/JB.00291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominy JE, Jr, Simmons CR, Hirschberger LL, Hwang J, Coloso RM, Stipanuk MH. Discovery and characterization of a second mammalian thiol dioxygenase, cysteamine dioxygenase. J Biol Chem. 2007;282:25189–198. doi: 10.1074/jbc.M703089200. [DOI] [PubMed] [Google Scholar]
- Dominy JE, Jr, Hwang J, Guo S, Hirschberger LL, Zhang S, Stipanuk MH. Synthesis of amino acid cofactor in cysteine dioxygenase is regulated by substrate and represents a novel post-translational regulation of activity. J Biol Chem. 2008;283:12188–12201. doi: 10.1074/jbc.M800044200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell JM, Khuri S, Gane PJ. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiology and Molecular Biology Reviews. 2000;64:153–179. doi: 10.1128/mmbr.64.1.153-179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell JM, Culham A, Carter CE, Sosa-Aguirre CR, Goodenough PW. Evolution of functional diversity in the cupin superfamily. Trends Biochem Sci. 2001;26:740–746. doi: 10.1016/s0968-0004(01)01981-8. [DOI] [PubMed] [Google Scholar]
- Fusetti F, Schroter KH, Steiner RA, van Noort PI, Pijning T, Rozeboom HJ, Kalk KH, Egmond MR, Dijkstra BW. Crystal structure of the copper-containing quercetin 2,3-dioxygenase from Aspergillus japonicus. Structure (Camb) 2002;10:259–268. doi: 10.1016/s0969-2126(02)00704-9. [DOI] [PubMed] [Google Scholar]
- Hirschberger LL, Daval S, Stover PJ, Stipanuk MH. Murine cysteine dioxygenase gene: structural organization, tissue-specific expression and promoter identification. Gene. 2001;277:153–161. doi: 10.1016/s0378-1119(01)00691-6. [DOI] [PubMed] [Google Scholar]
- Hosokawa Y, Matsumoto A, Oka J, Itakura H, Yamaguchi K. Isolation and characterization of a cDNA for rat liver cysteine dioxygenase. Biochem Biophys Res Commun. 1990;168:473–478. doi: 10.1016/0006-291x(90)92345-z. [DOI] [PubMed] [Google Scholar]
- Ito N, Phillips SE, Stevens C, Ogel ZB, McPherson MJ, Keen JN, Yadav KD, Knowles PF. Novel thioether bond revealed by a 1.7 A crystal structure of galactose oxidase. Nature. 1991;350:87–90. doi: 10.1038/350087a0. [DOI] [PubMed] [Google Scholar]
- Jaroszewski L, Schwarzenbacher R, von Delft F, McMullan D, Brinen LS, Canaves JM, Dai X, Deacon AM, DiDonato M, Elsliger MA, Eshagi S, Floyd R, Godzik A, Grittini C, Grzechnik SK, Hampton E, Levin I, Karlak C, Klock HE, Koesema E, Kovarik JS, Kreusch A, Kuhn P, Lesley SA, McPhillips TM, Miller MD, Morse, Moy K, Ouyang J, Page R, Quijano K, Reyes R, Rezezadeh F, Robb A, Sims E, Spraggon G, Stevens RC, van den Bedem H, Velasquez J, Vincent J, Wang X, West B, Wolf G, Xu Q, Hodgson KO, Wooley J, Wilson IA. Crystal structure of a novel manganese-containing cupin (TM1459) from Thermotoga maritima at 1.65 A resolution. Proteins. 2004;56:611–614. doi: 10.1002/prot.20130. [DOI] [PubMed] [Google Scholar]
- Kleffmann T, Jongkees SA, Fairweather G, Wilbanks SM, Jameson GNJ. Mass-spectrometric characterization of two posttranslational modifications of cysteine dioxygenase. Biol Inorg Chem. 2009;14:913–921. doi: 10.1007/s00775-009-0504-x. [DOI] [PubMed] [Google Scholar]
- Ko TP, Day J, McPherson A. The refined structure of canavalin from jack bean in two crystal forms at 2.1 and 2.0 A resolution. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 4):411–420. doi: 10.1107/s0907444900002237. [DOI] [PubMed] [Google Scholar]
- Lawrence MC, Izard T, Beuchat M, Blagrove RJ, Colman PM. Structure of phaseolin at 2.2 A resolution. Implications for a common vicilin/legumin structure and the genetic engineering of seed storage proteins. J Mol Biol. 1994;238:748–776. doi: 10.1006/jmbi.1994.1333. [DOI] [PubMed] [Google Scholar]
- McCann KP, Akbari MT, Williams AC, Ramsden DB. Human cysteine dioxygenase type I: primary structure derived from base sequencing of cDNA. Biochim Biophys Acta. 1994;1209:107–110. doi: 10.1016/0167-4838(94)90144-9. [DOI] [PubMed] [Google Scholar]
- McCoy JG, Bailey LJ, Bitto E, Bingman CA, Aceti DJ, Fox BG, Phillips GN., Jr Structure and mechanism of mouse cysteine dioxygenase. Proc Natl Acad Sci U S A. 2006;103:3084–3089. doi: 10.1073/pnas.0509262103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BS, Gardner JD, Bailey LJ, Brunold TC, Fox BG. Characterization of the nitrosyl adduct of substrate-bound mouse cysteine dioxygenase by electron paramagnetic resonance: electronic structure of the active site and mechanistic iimplications. Biochemistry. 2007;46:8569–8578. doi: 10.1021/bi700662d. [DOI] [PubMed] [Google Scholar]
- Richerson RB, Ziegler DM. Cysteamine dioxygenase. Methods Enzymol. 1987;143:410–415. doi: 10.1016/0076-6879(87)43071-1. [DOI] [PubMed] [Google Scholar]
- Rogers MS, Baron AJ, McPherson MJ, Knowles PF, Dooley DM. Galactose oxidase pro-sequence cleavage and cofactor assembly are self-processing reactions. J Am Chem Soc. 2000;122:990–991. [Google Scholar]
- Rogers MS, Hurtado-Guerrero R, Firbank SJ, Halcrow MA, Dooley DM, Phillips SE, Knowles PF, McPherson MJ. Cross-link formation of the cysteine 228-tyrosine 272 catalytic cofactor of galactose oxidase does not require dioxygen. Biochemistry. 2008;47:10428–10439. doi: 10.1021/bi8010835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG, Abad-Zapatero C, Murthy MR, Liljas L, Jones TA, Strandberg B. Structural comparisons of some small spherical plant viruses. J Mol Biol. 1983;165:711–736. doi: 10.1016/s0022-2836(83)80276-9. [DOI] [PubMed] [Google Scholar]
- Sakakibara S, Yamaguchi K, Hosokawa Y, Kohashi N, Ueda I. Purification and some properties of rat liver cysteine oxidase (cysteine dioxygenase) Biochim Biophys Acta. 1976;422:273–279. doi: 10.1016/0005-2744(76)90138-8. [DOI] [PubMed] [Google Scholar]
- Schnell R, Sandalova T, Hellman U, Lindqvist Y, Schneider G. Siroheme- and [Fe4-S4]-dependent NirA from Mycobacterium tuberculosis is a sulfite reductase with a covalent Cys-Tyr bond in the active site. J Biol Chem. 2005;280:27319–27328. doi: 10.1074/jbc.M502560200. [DOI] [PubMed] [Google Scholar]
- Simmons CR, Hirschberger LL, Machi MS, Stipanuk MH. Expression, purification, and kinetic characterization of recombinant rat cysteine dioxygenase, a non-heme metalloenzyme necessary for regulation of cellular cysteine levels. Protein Expr Purif. 2006a;47:74–81. doi: 10.1016/j.pep.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Simmons CR, Liu Q, Huang Q, Hao Q, Begley TP, Karplus PA, Stipanuk MH. Crystal structure of mammalian cysteine dioxygenase. A novel mononuclear iron center for cysteine thiol oxidation. J Biol Chem. 2006b;281:18723–18733. doi: 10.1074/jbc.M601555200. [DOI] [PubMed] [Google Scholar]
- Simmons CR, Krishnamoorthy K, Granett SL, Schuller DJ, Dominy JE, Jr, Begley TP, Stipanuk MH, Karplus PA. An Fe2+-bound persulfenate intermediate in cysteine dioxyenase. Biochemistry. 2008;47:11390–11392. doi: 10.1021/bi801546n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipanuk MH. Metabolism of sulfur-containing amino acids. Annu Rev Nutr. 1986;6:179–209. doi: 10.1146/annurev.nu.06.070186.001143. [DOI] [PubMed] [Google Scholar]
- Stipanuk MH. Sulfur amino acid metabolism: pathways for production and removal of homocysteine and cysteine. Annu Rev Nutr. 2004;24:539–577. doi: 10.1146/annurev.nutr.24.012003.132418. [DOI] [PubMed] [Google Scholar]
- Stipanuk MH, Londono M, Lee JI, Hu M, Yu AF. Enzymes and metabolites of cysteine metabolism in nonhepatic tissues of rats show little response to changes in dietary protein or sulfur amino acid levels. J Nutr. 2002;132:3369–3378. doi: 10.1093/jn/132.11.3369. [DOI] [PubMed] [Google Scholar]
- Stipanuk MH, Londono M, Hirschberger LL, Hickey C, Thiel DJ, Wang L. Evidence for expression of a single distinct form of mammalian cysteine dioxygenase. Amino Acids. 2004;26:99–106. doi: 10.1007/s00726-003-0001-4. [DOI] [PubMed] [Google Scholar]
- Stipanuk MH, Ueki I, Dominy JE, Jr, Simmons CR, Hirschberger LL. Cysteine dioxygenase: a robust system for regulation of cellular cysteine levels. Amino Acids. 2009;37:55–63. doi: 10.1007/s00726-008-0202-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner A, Bowater L, Fairhurst SA, Bornemann S. Oxalate decarboxylase requires manganese and dioxygen for activity. Overexpression and characterization of Bacillus subtilis YvrK and YoaN. J Biol Chem. 2001;276:43627–43634. doi: 10.1074/jbc.M107202200. [DOI] [PubMed] [Google Scholar]
- Whittaker MM, Whittaker JW. Cu(I)-dependent biogenesis of the galactose oxidase redox cofactor. J Biol Chem. 2003;278:22090–22101. doi: 10.1074/jbc.M300112200. [DOI] [PubMed] [Google Scholar]
- Whittaker MM, Kersten PJ, Cullen D, Whittaker JW. Identification of catalytic residues in glyoxal oxidase by targeted mutagenesis. J Biol Chem. 1999;274:36226–36232. doi: 10.1074/jbc.274.51.36226. [DOI] [PubMed] [Google Scholar]
- Whittaker MM, Kersten PJ, Nakamura N, Sanders-Loehr J, Schweizer ES, Whittaker JW. Glyoxal oxidase from Phanerochaete chrysosporium is a new radical-copper oxidase. J Biol Chem. 1996;271:681–687. doi: 10.1074/jbc.271.2.681. [DOI] [PubMed] [Google Scholar]
- Woo EJ, Dunwell JM, Goodenough PW, Marvier AC, Pickersgill RW. Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat Struct Biol. 2000;7:1036–1040. doi: 10.1038/80954. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Hosokawa Y, Kohashi N, Kori Y, Sakakibara S, Ueda I. Rat liver cysteine dioxygenase (cysteine oxidase). Further purification, characterization, and analysis of the activation and inactivation. J Biochem (Tokyo) 1978;83:479–491. doi: 10.1093/oxfordjournals.jbchem.a131935. [DOI] [PubMed] [Google Scholar]
- Ye S, Wu X, Wei L, Tang D, Sun P, Bartlam M, Rao Z. An insight into the mechanism of human cysteine dioxygenase. Key roles of the thioether-bonded tyrosine-cysteine cofactor. J Biol Chem. 2007;282:3391–3402. doi: 10.1074/jbc.M609337200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Colabroy KL, Begley TP, Ealick SE. Structural studies on 3-hydroxyanthranilate-3,4-dioxygenase: the catalytic mechanism of a complex oxidation involved in NAD biosynthesis. Biochemistry. 2005;44:7632–7643. doi: 10.1021/bi047353l. [DOI] [PubMed] [Google Scholar]