Abstract
The T cells scan the surface of antigen-presenting cells with their T cell receptors (TCR) in order to find and respond to specific peptide-major histocompatibility complexes (pMHC). Since mainly self-peptides are expressed on antigen-presenting cells, the T cells must utilize sensitive mechanisms in order to quickly discriminate between self and nonself-peptides. A range of different models have been proposed to account for this process. Due to the experimental inconsistency of how T cells respond to altered peptides it has been difficult to validate the competing models. Recent models, based on the kinetic proofreading model, propose that a short life-time of the TCR/pMHC complexes may be compensated by fast rebinding of the individual molecules. Hence, both the on- and off-rate involved in the interaction between pMHCs and TCRs will determine the fate of the T cell discrimination. I here briefly review some of the proposed models on T cell discrimination and scanning, and discuss the significance of determining self-peptide kinetics to validate the different models.
Key words: mathematical, model, scanning, antigen, TCR, discrimination
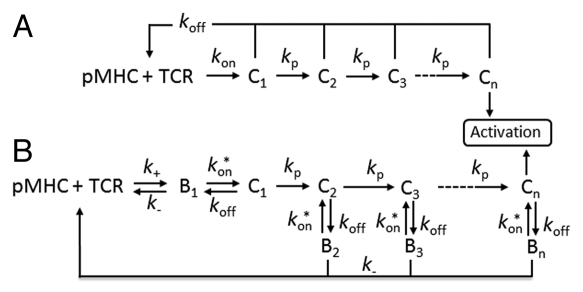
McKeithan proposed the kinetic proofreading (KP) model 16 years ago in order to explain how T cells discriminate between nonself and self-peptides.1 Since then, numerous studies have used this model as an underlying framework for more detailed models and to fit kinetic data of altered peptides to T cell activation. The original KP-model proposes that a T cell receptor (TCR) must be bound to a peptide-major histocompatibility complex (pMHC) long enough to complete a series of biochemical modification steps in order to induce T cell activation (Fig. 1A). The series of modifications can be thought of as the phosphorylation steps initiated by the TCR engagement, which are rapidly dephosphorylated if the TCR/pMHC complex dissociates. Thus, only the life-time/dissociation rate of the TCR/pMHC complex determines if a T cell should respond to a peptide or not. The KP-model has been refined and extended to include feedback mechanisms during the intracellular signalling events, which generate an even sharper discrimination between different pMHCs based on their mean life-time to the TCRs.2,3 The life-time threshold to initiate T cell activation has been predicted to be at least 3 seconds.2 Complexes between self-pMHCs and TCRs have a much shorter life-time than TCR/agonist-pMHC complexes, which is a consequence of the negative selection of high affinity self-antigens in the thymus.4,5 The KP-model has been challenged by the “equilibrium affinity (Kd) model”, where T cell activation has been correlated with the Kd values of different peptides.6,7 Hence, this model states, in contrast to the KP-model, that the association rate is also involved in the TCR discrimination where fast on-rates increase the number of engaged TCRs.
Figure 1.
Kinetic proofreading models. (A) The original KP-model proposed by McKeithan.1 A TCR and a pMHC associate at rate kon. The TCR/pMHC complex (C1) must undergo n modification steps at rate kp to generate an active TCR that initiates the activation signals. At each step, the complex may dissociate at rate koff, leading to a complete unmodified form of the TCR. (B) Rebinding in the KP-model assumes that the interacting molecules must first diffuse into close proximity (B1) before they can form complex. The interacting molecules (B) can either associate at the intrinsic rate kon* or diffuse apart at rate k. A bound TCR (C) undergoing the KP steps that dissociates may either rebind and proceed with the subsequent proofreading steps or revert to its unmodified form if the molecules diffuse apart.
A recent theoretical study has, however, recently found support for both models. By refining the original KP-model, Dushek et al.8 proposed a model in which fast on-rates allowed individual TCRs to rebind to the same pMHC before diffusing apart (Fig. 1B). The rebinding is assumed to be fast enough to sustain the TCR in its current proofreading step, which allows the TCR to proceed towards the subsequent activation step without reverting to its unmodified form. Hence, a short life-time of the TCR/pMHC complex can be compensated by a fast on-rate. In addition, two different research groups have recently investigated the theory of rebinding experimentally. Aleksic et al. and Govern et al. validated such “rebinding models” against the KP and the Kd-models. They found that the “rebinding model” correlated best with the experimental data in which T cell activation was measured as a function of the TCR/pMHC binding properties. Although the models based on the KP-model seem to explain T cell discrimination, we still know little about the signalling properties when a TCR reaches the final kinetic proofreading step. It is not clear whether optimal T cell activation requires TCRs arriving at the final proofreading step continuously, where individual TCRs only signal a short period of time, or if TCRs signal as long as they are bound to a pMHC at their active state. The serial triggering model was proposed based on the observation that a few specific pMHCs could trigger many TCRs.11–13 Calculations based on a specific agonist peptide showed that each individual pMHC could potentially trigger five TCRs within a period of 100 seconds.14 By combining the serial triggering model with the KP-model, Coombs et al.15 found an optimal life-time of TCR/pMHC complexes. If the life-time was too short, the TCR failed to reach its active proofreading step, whereas a too long life-time prevented efficient serial engagement of the TCRs. This prediction is supported by experiments in which T cell activation is optimal for intermediate life-times of TCR/pMHC complexes.16,17 However, several other experimental systems do not provide support for this prediction7,9,10 and the importance of serial triggering is thus considered controversial. Experiments in which the T cell response is correlated with increasing life-time of the TCR/pMHC complexes indicate that an active TCR generates continuous signals as long as it is bound to a pMHC.
The observation that just a few pMHCs can trigger a T cell raises another question: how can a T cell find agonist peptides in an ocean of expressed self-pMHCs? T cells are mainly exposed to abundant self-peptides, expressed by MHCs on antigen-presenting cells (APC), during their search for nonself-peptides.18,19 Since the T cell response needs to be fast, T cells must utilize efficient search mechanisms to find nonself-peptides. A T cell only spends a few minutes on each APC,20 which suggests that the sampling of pMHCs, by its TCRs, must be a very fast process in order to sample a large fraction of the expressed pMHCs. We have recently attempted to shed light on this process by developing a mathematical framework for calculating the speed at which a T cell samples pMHCs.21 The model revealed a number of critical parameters that determine the accuracy and speed of the sampling process. For example, there is a tradeoff between the speed at which a T cell moves on the surface of the APC and the fraction of unique sampled pMHC molecules during the scanning process. The calculations showed that a single TCR generally samples 1–10 pMHC per second. Therefore, a T cell can theoretically scan 800–8,000 pMHCs per second on the APC (assuming that the contact area contains 800 TCRs and more than 2,500 pMHCs). Hence, these calculations indicate that T cells scan the APCs at an extremely high rate and provide a possible explanation to finding a needle in the haystack. In addition, simulations based on the recently proposed “rebinding model” also predicted that the sampling rate of unsampled pMHCs was optimal for a certain range of on-rates. Fast on-rates favor TCR rebinding to the same pMHC, which consequently impair efficient sampling of new unsampled pMHCs. However, the predictions are highly dependent on the binding kinetics of self-peptides. Almost all kinetic studies focus on altered peptides, whereas very few are dedicated to natural self-peptides. Therefore, in order to understand the T cell discrimination between self and nonself, based on the binding kinetics, more details are needed concerning the kinetics of peripheral T cells interacting with self-peptides and the abundance of distinct self-peptides that are possibly shared among different TCRs.19 If a kinetic interval of peripheral TCR/self-pMHC complexes could be established, the different models on T cell discrimination could be validated from the view of self-peptides and more accurate calculations on T cell scanning can be performed.
References
- 1.McKeithan TW. Kinetic proofreading in T-cell receptor signal transduction. Proc Natl Acad Sci USA. 1995;92:5042–5046. doi: 10.1073/pnas.92.11.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan-Bonnet G. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science. 2008;321:1081–1084. doi: 10.1126/science.1158013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wylie DC, Das J, Chakraborty AK. Sensitivity of T cells to antigen and antagonism emerges from differential regulation of the same molecular signaling module. Proc Natl Acad Sci USA. 2007;104:5533–5538. doi: 10.1073/pnas.0611482104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, et al. Thymic selection threshold defined by compartmentalization of Ras/MAPK signaling. Nature. 2006;444:724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- 5.Crites TJ, Varma R. On the issue of peptide recognition in T cell development. Self/Nonself. 2010;1:55–61. doi: 10.4161/self.1.1.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian S, Maile R, Collins EJ, Frelinger JA. CD8+ T cell activation is governed by TCR-peptide/MHC affinity, not dissociation rate. J Immunol. 2007;179:2952–2960. doi: 10.4049/jimmunol.179.5.2952. [DOI] [PubMed] [Google Scholar]
- 7.Stone JD, Chervin AS, Kranz DM. T-cell receptor binding affinities and kinetics: impact on T-cell activity and specificity. Immunology. 2009;126:165–176. doi: 10.1111/j.1365-2567.2008.03015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dushek O, Das R, Coombs D. A role for rebinding in rapid and reliable T cell responses to antigen. PLoS Comput Biol. 2009;5:1000578. doi: 10.1371/journal.pcbi.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aleksic M, Dushek O, Zhang H, Shenderov E, Chen JL, Cerundolo V, et al. Dependence of T cell antigen recognition on T cell receptor-peptide MHC confinement time. Immunity. 2010;32:163–174. doi: 10.1016/j.immuni.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Govern CC, Paczosa MK, Chakraborty AK, Huseby ES. Fast on-rates allow short dwell time ligands to activate T cells. Proc Natl Acad Sci USA. 2010;107:8724–8729. doi: 10.1073/pnas.1000966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct observation of ligand recognition by T cells. Nature. 2002;419:845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- 12.Ma Z, Sharp KA, Janmey PA, Finkel TH. Surface-anchored monomeric agonist pMHCs alone trigger TCR with high sensitivity. PLoS Biol. 2008;6:43. doi: 10.1371/journal.pbio.0060043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valitutti S, Müller S, Cella M, Padovan E, Lanzavecchia A. Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature. 1995;375:148–151. doi: 10.1038/375148a0. [DOI] [PubMed] [Google Scholar]
- 14.Wofsy C, Coombs D, Goldstein B. Calculations show substantial serial engagement of T cell receptors. Biophys J. 2001;80:606–612. doi: 10.1016/S0006-3495(01)76041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coombs D, Kalergis AM, Nathenson SG, Wofsy C, Goldstein B. Activated TCRs remain marked for internalization after dissociation from pMHC. Nat Immunol. 2002;3:926–931. doi: 10.1038/ni838. [DOI] [PubMed] [Google Scholar]
- 16.Kalergis AM, Boucheron N, Doucey MA, Palmieri E, Goyarts EC, Vegh Z, et al. Efficient T cell activation requires an optimal dwell-time of interaction between the TCR and the pMHC complex. Nat Immunol. 2001;2:229–234. doi: 10.1038/85286. [DOI] [PubMed] [Google Scholar]
- 17.Cemerski S, Das J, Locasale J, Arnold P, Giurisato E, Markiewicz MA, et al. The stimulatory potency of T cell antigens is influenced by the formation of the immunological synapse. Immunity. 2007;26:345–355. doi: 10.1016/j.immuni.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Germain RN, Stefanová I. The dynamics of T cell receptor signaling: complex orchestration and the key roles of tempo and cooperation. Annu Rev Immunol. 1999;17:467–522. doi: 10.1146/annurev.immunol.17.1.467. [DOI] [PubMed] [Google Scholar]
- 19.Mahajan VS, Leskov IB, Chen JZ. Homeostasis of T cell diversity. Cell Mol Immunol. 2005;2:1–10. [PubMed] [Google Scholar]
- 20.Miller MJ, Hejazi AS, Wei SH, Cahalan MD, Parker I. T cell repertoire scanning is promoted by dynamic dendritic cell behavior and random T cell motility in the lymph node. Proc Natl Acad Sci USA. 2004;101:998–1003. doi: 10.1073/pnas.0306407101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansson A. A mathematical framework for analyzing T cell receptor scanning of peptides. Biophys J. 2010;99:2717–2725. doi: 10.1016/j.bpj.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]