Abstract
Ever since the days of Ehrlich and the birth of humoral immunity, self-reactivity or ‘horror autotoxicus’ as referred to by Paul Ehrlich, has been of great concern. For instance, in patients with the autoimmune disease systemic lupus erythematosus (SLE), anti-nuclear and anti-DNA antibodies have been recognized for many years. Despite this, the exact mechanism as to how the immune system fails to protect the individual and allows these autoantibodies to develop in this and other systemic autoimmune diseases remains uncertain. So how can we explain their presence? Evidence suggests that B cells expressing autoreactive antibodies do not normally arise but rather undergo negative selection as they develop. In light of this, it might seem contradictory that not all autoreactive B cell clones are eliminated, although this may not even be the intention since autoantibodies are also found in healthy individuals and may even protect from autoimmunity. Here, we will discuss autoantibodies, in particular those recognizing DNA, with regard to their reactivity and their potentially pathogenic or protective properties.
Key words: anti-DNA antibodies, anti-nuclear antibodies, anti-nucleosome antibodies, autoantibodies, systemic autoimmunity, B cells, B cell receptors, CDR3, natural antibodies, SLE
Antibodies
Antibodies are soluble molecules present in biological fluids such as blood, colostrum, saliva, cerebrospinal fluids and the intestinal lumen. By definition they are mediators of humoral immune responses and protect against infections.
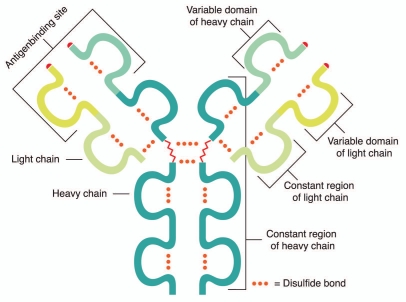
An antibody consists of an identical pair of immunoglobulin (Ig) heavy chains (HC) and light chains (LC), each containing a variable and constant region (Fig. 1). The variable region contains hypervariable sub-regions, referred to as complementarity determining regions (CDR1-3) (Fig. 2), which are responsible for the actual antigen contact. The HC constant region, on the other hand, determines the variety of antibody effector functions.
Figure 1.
General structure of an IgG antibody. This consists of an identical pair of Ig HCs and LCs each containing a variable and constant region.
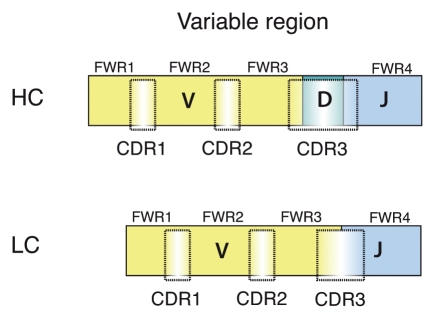
Figure 2.
Schematic picture of HC and LC CDRs and FWRs. The HC and LC variable regions are encoded by the recombined V(D)J gene segments. The relative positions of the CDR1-3 and the FWR1-4 are indicated.
The enormous diversity of the primary antibody repertoire observed in the mature B cell pool is the result of a somatic process mediated by the recombination-activating-genes 1 and 2 (RAG). In this, random recombination of germline-encoded VH (variable), D (diversity) and JH (joining) gene segments generates the HC variable region and VL, JL that of the LC (Fig. 2).1 The reaction itself also contributes to the diversity by the imprecise joining of the gene segments, which results in the antibody HCs showing the greatest variability at the VH-D-JH junction (junctional diversity). Furthermore, the enzyme N-nucleotidyl-transferase (TdT) adds non-templated N-nucleotides to the VH-D and D-JH junctions, thereby further increasing the junctional diversity. The TdT enzyme is mainly active in the bone marrow, hence increasing the adult primary antibody repertoire over that of the fetal repertoire. The respective V gene segment encodes the CDR1 and -2. The VH-D-JH junction encodes the HC-CDR3, which is a major determinant of the antibody-antigen interaction, in some cases even dominant.2
Although the primary antibody repertoire of B cells is very diverse, it has the capacity to increase even further through a process termed somatic hypermutation (SHM), which may lead to an increase in the affinity of the antibody for the antigen, i.e., affinity maturation. In addition, antibodies may change effector function through a process termed class switch recombination (CSR), resulting in a switch from for instance IgM to IgG. Both SHM and CSR are processes that take place in more mature B cells during an immune response.
B Cell Development
During fetal life, B cell development takes place in the liver whilst in adults, in the bone marrow. Early in B cell development the Ig HC locus is first DJ and subsequently VDJ recombined.3,4 After functional recombination, the synthesized µHC assembles with surrogate LC, an invariant LC that is composed of the VpreB and λ5 polypeptides, and expressed as a pre-B cell receptor (pre-BCR) on pre-B cells.5–9 Subsequently, surrogate LC is silenced and the Ig κ and/or λ LC loci undergo VJ recombination. Upon bona fide LC synthesis, these assemble with the existing HCs and form a membrane-anchored antibody, a BCR, in fact, IgM. These, immature, B cells thereafter migrate to secondary lymphoid organs, e.g., spleen where they develop into mature B cells.
Mature B cells can be divided into B1 and B2 subsets. In mice, the B1 subset can be further subdivided into CD5+ B1-a and CD5- B1-b cells.10–12 In humans, because the degree of CD5 expression varies there is still no good marker for B1 B cells.13 Also B2 B cells can be further subdivided, into follicular (FO) and marginal zone (MZ) B cells, in the spleen found in follicles and outside of the marginal sinus, respectively, whereas in lymph nodes the vast majority of B cells are of the FO type.
Mono-, Oligo- and Polyreactive Antibodies
There are, as of yet, no clear way to group antibodies based on common structure, e.g., V gene usage but their reactivity can be defined based on antigen binding patterns, e.g., mono-, oligo- or polyreactive as well as their affinity for antigens. In general, a polyreactive antibody binds several unrelated antigens with low affinity whereas a monoreactive antibody binds one antigen with high affinity, with the antibodies between the two extremes representing those that are oligoreactive.14
It has been estimated that within the pool of mature naïve human B cells (IgM+CD27-CD10−) approximately 5% express polyreactive BCRs, here defined as reactivity to more than one of the following antigens ssDNA, dsDNA, insulin or LPS15 whereas the frequency within IgM+ memory B cells (IgM+CD27+CD10−) is around 1%,16 indicating that these are more specific in their reactivity, as might be expected of a memory population. Surprisingly, therefore, the proportion among IgG+ memory B cells (IgG+CD27+CD10−) is almost 25%.17 As one would expect that IgG+ memory B cells from healthy individuals are mostly directed against non-self antigens and would show a more restricted specificity with high affinity towards the antigens that elicit them, these results suggest that a simple inverse correlation between polyreactivity and affinity is not observed for IgG antibodies.
Natural Antibodies
Natural antibodies are present in newborn humans as well as germ-free mice suggesting that their repertoire is independent of external antigenic contact. These antibodies are characterized by their use of germline-encoded genes and lack of somatic mutations in the V region.14,18,19 Natural antibodies are of the IgM, IgG and IgA isotypes, though the vast majority is IgM. Most bind antigens with low-affinity and the majority bind to several unrelated antigens, i.e., are polyreactive. The B1 subset is usually considered to produce natural antibodies although it has been shown that low-affinity, germline encoded, polyreactive antibodies are not always of B1 origin and can be found in Peyer's patches, lamina propria, MZ of the spleen and in the thymus.20 Natural antibodies play a major role in the primary line of defense against infections and are considered to be a part of the innate immunity.
Because natural antibodies are polyreactive and recognize a large number of antigens, which includes both exogenous (bacteria, virus and fungi) and self-antigens (nucleic acids, phospholipids, erythrocytes, serum proteins, cellular components, insulin or thyroglobulin), the pool of natural antibodies contains autoantibodies, termed natural autoantibodies.21 In addition to the role of the natural antibody repertoire against certain bacterial infections, it has been proposed that natural autoantibodies serve other functions.22–24 For instance, oxidation-specific epitopes are found to be a target of natural IgM antibodies, indicating a role for clearance and neutralization of oxidized lipids. In fact, natural autoantibodies have been found to decrease atherosclerosis, possibly by preventing the accumulation of oxidized low-density lipoprotein particles in the artery wall.25 In addition, natural autoantibodies can also participate in the removal of apoptotic cells and may under normal circumstances play a role in inducing and/or maintaining immunological tolerance.26
Autoantibodies
The recombination of the V(D)J gene segments is random and therefore, this process generates antibodies that recognize foreign antigens and those that recognize self-(auto) antigens. Some autoantigens are found in all cells, such as DNA or chromatin, whereas others are only found in one cell type, e.g., thyroglobulin in the cells of the thyroid gland or the acetylcholine receptor at the postsynaptic side of the neuromuscular junction. Although there are autoreactivity among natural antibodies, most of us would think of autoantibodies as those present in autoimmune diseases, mostly mono- or oligoreactive, e.g., anti-DNA and anti-nuclear antibodies (ANAs) in SLE, anti-phospholipid (cardiolipin) in antiphospholipid antibody syndrome. These autoantibodies can be IgM or class switched and some carry somatic mutations that may result in affinity maturation.
Pathogenic Autoantibodies
Some autoantibodies are considered pathogenic and could be one of the plausible factors responsible for the onset of autoimmune disease, followed by injury to the tissue for which they bear specific reactivity. However, autoantibodies may not by default be pathogenic but can be defined as such according to certain criteria. Based on Koch's postulate for linking a specific disease to a given microbe, half a century ago Witebsky et al. proposed postulates to define autoimmunity.27 Due to and based on the progress of knowledge in the field, the postulates of Witebsky were revised and Bona proposed the following criteria to define pathogenic autoantibodies:28 (1) An autoantigen is to be identified and its ability to induce autoantibodies and autoimmune disease needs to be demonstrated in animals; (2) Isolation of pathogenic autoantibodies from affected organs is required; (3) Passive transfer of autoantibodies should induce lesions or symptoms of autoimmune disease; (4) Occurrence of autoimmune disease in mice expressing V genes encoding pathogenic autoantibodies. One could therefore ask whether there are autoantibodies that fulfill the criteria proposed by Bona and, according to these, could be defined as pathogenic? We will initially consider autoantibodies in SLE, which is a chronic, systemic autoimmune disease that can affect the blood and blood vessels, joints, skin, lungs, heart, kidneys and nervous system. Glomerulonephritis is one of the most severe clinical manifestations in SLE, affecting about 30% of patients and is in part caused by autoantibodies binding to the glomeruli resulting in inflammation. Below we will discuss autoantibodies in light of the four postulates, whether they fulfill these and, based on this knowledge, current thoughts around their development.
Testing the First Two of the Postulates
The presence of anti-nuclear antibodies (ANA) and anti-DNA antibodies in SLE patients was first described in 1957,29–32 and it has since been found that the vast majority (>95%) of SLE patients present with serum ANAs. As the targets of these autoantibodies are nuclear antigens, e.g., DNA, chromatin, nucleoproteins, we conclude that the antigens that these autoantibodies recognize have been defined.
To fulfill the first of Bona's criterion, the defined antigen(s) should give rise to autoantibodies and disease when injected. However, injection of DNA into healthy animals does not generate anti-DNA antibodies, as DNA itself is not very immunogenic. Even so, immunization of complexes of an immunogenic (nonself) peptide together with tightly bound mammalian DNA induces anti-dsDNA autoantibodies with characteristics similar to anti-dsDNA antibodies from lupus prone mice.33 Likewise, the transgenic expression of polyoma virus T antigen results in anti-DNA autoantibodies.34 In the latter two cases, the tight association of DNA with an immunogenic DNA-binding protein, presumably providing peptides for T cell reactivity and T cell help for autoreactive B cells, leads to the generation of anti-DNA autoantibodies. Furthermore, injection of apoptotic cells, representing a source of nuclear antigens, leads to the production of ANAs including anti-DNA antibodies.35 Even though these effects are only transient, it also results in IgG deposition in renal glomeruli. Taken together, we conclude that these autoantibodies fulfill the first postulate.
Because autoantibodies, e.g., anti-DNA antibodies, have been isolated from glomerular eluates in patients with active lupus nephritis,36,37 the second postulate is also fulfilled.
The Third Postulate
Regarding the third postulate, it has been found that passive transfer of SLE autoantibodies partially induces features of the autoimmune disease. For instance, administration of a panel of anti-DNA antibodies showed that some, but not all, induce nephritis in non-autoimmune mice38–42 and thus, fulfilling also the third postulate.
In this context, we will also consider Goodpasture's disease (GP), which is characterized by circulating autoantibodies against glomerular basement membrane (GBM) and a linear deposition of IgG along the glomerular and pulmonary alveolar basement membranes.43 The anti-GMB antibodies can be directly pathogenic as shown by the injection of antibodies, eluted from the kidneys of GP patients, causing acute glomerulonephritis in monkeys.44 However, a recent report has discovered such autoantibodies also in healthy individuals, although at very low levels.45 Thus, an antibody can fulfill the first three postulates and consequently be considered pathogenic but may not always cause disease. This is also supported by the observation that around 25% of healthy relatives of SLE patients present with serum ANAs. This would imply that, in some cases, pathogenicity of autoantibodies might depend on correct exposure of the autoantigen.
Cross-Reactivity of Anti-DNA Autoantibodies
Autoantibodies might be pathogenic in various ways, for instance anti-DNA antibodies, rather than directly binding DNA may interact with nucleosomes (e.g., DNA and histones). In fact, it has been reported that anti-DNA antibodies do react also with chromatin/nucleosomes although anti-nucleosome antibodies appear not to bind to DNA.46 Alternatively, anti-DNA antibodies could also be pathogenic by cross-reacting with other antigens present in kidneys and/or the skin. In support of this, anti-DNA antibodies have been reported to cross-react with α-actinin,47,48 heparan sulfate (HS),49 collagen IV and laminin.50,51 However, more recent work shed light on this and suggest that at least some of the reported cross-reactivity can be explained by an indirect binding via nucleosomes.52 It appears that when anti-DNA antibodies cross-reactive to HS proteoglycans (HSPG) are treated with DNase I, the binding to HSPG is abolished. More detailed analysis revealed that the anti-DNA antibodies bound nucleosomes, derived from the dying hybridoma cells and the binding to HSPG was mediated by these nucleosomes.53 This is also supported by recent data demonstrating that nucleosomes show high affinity to antigens, e.g., laminin and collagen IV51 and may thus explain some of the observed crossreactivities.
In the 1970's, electron dense deposits (EDDs) in GBMs were described in SLE patients and lupus prone mice.54–56 Recently, it was found that EDDs in nephritic mice (NZBxNZW)F1 (NZB/W) contain extracellular nucleosomes and that anti-DNA antibodies co-localize with such EDDs.57 Furthermore, the anti-DNA antibodies were clearly separated from and did not co-localize with other material in the GBM, e.g., laminin, which renders additional support for the importance of the nucleosome as a target antigen in SLE. Moreover, the presence of nucleosome-containing EDDs in nephritic but not non-nephritic kidneys indicates an increase in apoptosis or an inability to clean up apoptotic material in the former, which is also supported by the presence of apoptotic cell products in the circulation in SLE patients and mice.58
Autoantibody Sequences
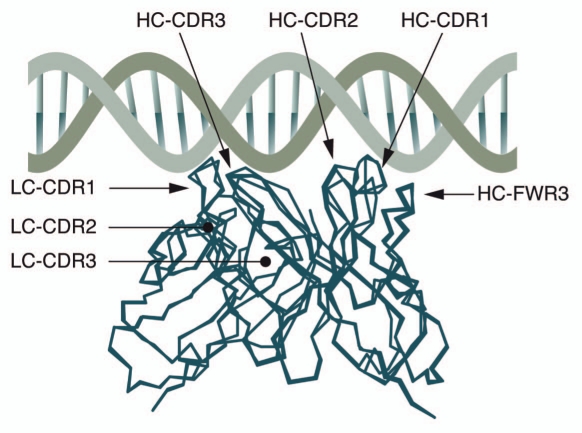
In terms of the fourth of Bona's postulates, we first need to consider the V genes encoding autoantibodies. Extensive sequence analyses of antibodies recognizing different autoantigens have not revealed any definite common structural feature, with the exception of anti-DNA antibodies. Here, it has been shown that arginine residues in the HC-CDRs are strong mediators of DNA binding.59–62 By generating and analyzing a large collection of hybridomas from autoimmune NZB/W mice, enrichment of arginines in the CDR3 region was found and later also a correlation between the positions of these arginines and their specificity for DNA.59,61 Reverting somatic mutations in prototypic anti-DNA antibodies supported the importance of arginines for DNA binding.63 In addition, by putting together the, by that time, over 250 mouse and a few human anti-DNA sequences described in the literature, evidence was presented to suggest structural and genetic selection of anti-DNA antibodies.64 Together with data from other groups,61,65 a molecular model of how the anti-DNA antibodies may react with DNA was introduced, stressing the importance of the antibody HC in the binding of DNA. According to the proposed model, the HC-CDR1 and HC-CDR2 extend into the major groove of DNA and the HC-CDR3 straddles one of the phosphate backbones. The stretches between the CDRs are termed the framework regions (FWRs) and the loop of the HC-FWR3 is positioned to contribute to contacts within the minor groove whilst the LC binds mostly at LC-CDR1, which reaches into the minor groove (Fig. 3). The model emphasizes not only the importance of certain amino acids but also their position. Apart from the contribution of arginine, asparagine and lysine to DNA binding, the authors pin point several positions in the HC-CDR1 and 2, HC-FWR3 and LC-CDR1 where these amino acids play a crucial role but still it is in the HC-CDR3 where they seem to matter the most, as the HC-CDR3 of most spontaneous anti-DNA antibodies contain at least one arginine. Although arginine in HC-CDR3 is not a feature that is generally connected to affinity for autoantigens, it has also been found of importance in antiphospholipid antibodies.66–68 The arginines in HC-CDR3 can arise in several ways, e.g., nucleotide additions, D-D fusions as well as the D gene segment in an unusual reading frame (RF), i.e., in RF3. Arginines can also be introduced into all CDRs by SHM.69,70 In addition to the importance of the arginines for DNA binding, the sequence analyses of IgM and IgG anti-DNA antibodies clearly suggest that they are clonally related and, therefore, derive from an antigendriven response rather than from a polyclonal activation.71
Figure 3.
Molecular model of the interaction between an anti-DNA antibody and DNA. According to the proposed model, the HC-CDR1 and HC-CDR2 extend into the major groove of DNA whereas the HC-CDR3 straddles one of the phosphate backbones. The loop of the HC-FWR3 is positioned such that it contributes to contacts within the minor groove. The LC binds primarily through CDR1, which reaches into the minor groove. Adapted from reference 64.
Pathogenic Anti-DNA Antibodies and the Fourth Postulate
Returning to the fourth postulate, several autoantibody-encoding V genes have been tested to determine whether they do indeed induce autoimmune disease. The results of these studies are not a simple yes- or no-answer and, therefore, we will go through some of these in some detail. We will start with the well-characterized 3H9 antibody, an IgM anti-DNA antibody produced by a hybridoma established from MRL/lpr mice,60 mice that spontaneously develop SLE-like disease, including serum ANAs and anti-DNA antibodies. Passive transfer of the 3H9 hybridoma induces symptoms of autoimmune disease, e.g., binding to glomeruli.39 Furthermore, analysis of the 3H9 HC sequence shows that it is typical of anti-DNA antibodies in that the CDR3 contains one arginine, the CDR2 show evidence of SHM resulting in an additional arginine and, supporting the importance of arginines, mutation of the CDR3 arginine completely abrogates DNA binding.63
The 3H9 HC, when combined with a variety of LCs, binds to ss- and dsDNA with high affinity whereas together with a particular LC, Vk8, it binds only ssDNA. Establishment of a transgenic (TG) mouse line expressing the 3H9 antibody HC, resulted in the development of B cells, although reduced in numbers due to deletion.72 Crossing the 3H9 TG with another TG line that expresses the Vk8 LC also results in reduced B cell numbers due to deletion. Despite this, in both the single 3H9 and double 3H9/Vk8 mice, autoreactive B cells that recognize DNA develop. However, the BCRs expressed by these B cells only bind to ss- but not to dsDNA. This, of course, would be expected in the double TGs but not in the single TGs. Moreover, despite the presence of autoreactive, in fact, anti-ssDNA-reactive B cells, these cells seem unable to develop into plasma cells since anti-DNA antibodies are not detectable in the serum. An inability of the autoreactive B cells to differentiate into plasma cells in vivo is supported by the fact that hybridomas established from these B cells are able to secrete anti-ssDNA antibodies. Because none of the hybridomas produced anti-dsDNA antibodies, this supports that such B cells do not develop. It was later shown that negative selection takes place at the pre-B to B cell transition in the bone marrow.72,73 Thus, these data suggest that B cells expressing BCRs with high affinity for dsDNA undergo negative selection by deletion and, as discussed below, receptor editing whereas those that express BCRs with high affinity to ssDNA are functionally inactivated and do not differentiate into autoantibody secreting plasma cells. The analysis of transgenic mice carrying V-genes from other anti-DNA antibodies derived from diseased NZB/W lupus mice revealed similar results in a normal genetic background.74 As expression of these V genes did not result in autoimmune disease in a normal mouse background, the fourth postulate is evidently not fulfilled. Rather, these experiments, together with other transgenic models for B cell tolerance have provided detailed insight into tolerance induction, mechanisms that prevent the development of cells expressing ‘pathogenic’ autoantibodies in normal individuals.
BCR Checkpoints
Although the above discussion was based on the fourth postulate and anti-DNA antibodies, extensive research based on additional TG mice expressing autoantibodies that are not linked to disease, e.g., those recognizing MHC (3–83) and hen egg lysozyme (HEL), demonstrate that negative selection is not confined to B cells expressing disease linked autoantibodies. However, in the non-disease models, selection only takes place upon introduction of the autoantigen, e.g., the appropriate MHC background or TG expression of soluble/membrane bound HEL. This implies that in the anti-DNA TG mice the autoantigen is present, a possible source of which may be apoptotic cells. Nevertheless, these and many other experiments have lead to the current model of B cell development, which includes several checkpoints for inducing tolerance and by several mechanisms:75–80 (1) deletion; elimination of autoreactive B cells (often by a cross-linking antigen); (2) anergy; autoreactive B cells are unable to respond to its antigen; (3) receptor replacement; recombination and expression of a novel VH gene; (4) receptor editing; recombination and expression of another LC. In (3 and 4), this might lead to expression of an innocuous (nonself) receptor. In the latter, the genetic information encoding the first LC is not always deleted or allelically excluded, resulting in cells expressing two LCs intracellularly although only the LC that gives rise to an innocuous BCR is found on the cell surface,81–83 although not observed in all model systems.84 By contrast to the data just discussed, B cells expressing natural autoantibodies as a transgene/knock-in are positively rather than negatively selected based on their autoreactivity,85,86 thus, do not appear to undergo tolerance induction.
A Pre-BCR Checkpoint
In addition to tolerance checkpoints based on the expression of autoreactive BCRs, a pre-BCR-mediated checkpoint has been described active already at the pre-B cell stage. The pre-BCR, as mentioned earlier, is assembled from HCs and surrogate LCs. A pre-BCR-dependent checkpoint was first observed when analyzing a patient with a mutation in one of the surrogate LC components (λ5).87 Expression of the mutant λ5 appears to affect pre-BCR surface expression, which may explain the lack of B cells in this patient.88 Nevertheless, the antibody HCs expressed by pre-B cells from this patient were unusual in that the CDR3s contained a high proportion of basic amino acids, i.e., typical of those found in anti-DNA antibodies. The presence of such pre-B cells could be related to a process where a functional pre-BCR negatively selects pro-B cells expressing such HC-CDR3s, a mechanism that evidently has failed in this patient.
In agreement with the data from humans, pre-BCR-mediated negative selection also takes place at the pro- to pre-B cell transition in mice.89 Mouse pro-B cells also express HC-CDR3s previously associated with anti-DNA antibodies and these cells undergo pre-BCR-dependent negative selection. Although the exact mechanism of this selection is unknown, the human study would suggest that the ‘anti-DNA antibody’ HCs induce apoptosis of the cells.87
By contrast to the patient expressing a mutated surrogate LC, in mice lacking surrogate LC B cells develop, albeit reduced in numbers.90 More detailed analysis of these mice uncovered a subpopulation of autoreactive mature B cells that express typical anti-DNA HC-CDR3s,89 i.e., similar to those that would normally be counter-selected by the pre-BCR. The presence of basic amino acids in the HC-CDR3s could, in most cases, be explained by the use of the D-gene segment in RF3, hence a feature also shared by anti-DNA antibodies. In addition, mice lacking surrogate LC present with higher levels of serum autoantibodies, e.g., ANA and anti-DNA antibodies, than control mice. The development of autoreactive B cells in these mice suggest that these cells either by-pass the selection normally taking place at the BCR checkpoints or, alternatively, that these cannot replace the pre-BCR checkpoint. Nevertheless, despite the development of autoreactive B cells and serum ANAs, as far as we know, these mice do not come down with autoimmune disease, although not analyzed in any detail. This could be taken to indicate that these autoantibodies are not pathogenic, the levels are too low to give rise to symptoms or that the antigen is not present.
Genetics and the Development of Pathogenic Anti-DNA Antibodies
The data discussed so far, would suggest that highly autoreactive B cells, especially those reactive to dsDNA, do not develop under normal circumstances and those recognizing ssDNA, although found in peripheral lymphoid organs, do not turn into plasma cells. However, in mouse strains such as NZB/W and MRL/lpr, there is spontaneous development of B cells that secrete relatively high levels of pathogenic autoantibodies. These autoantibodies are usually of high affinity showing evidence of SHM and are clonally related, suggesting that only a small number of autoreactive B cell clones are expanded. So how can we explain the presence of autoreactive B cells and serum autoantibodies in these mouse models and, by inference, in SLE patients? Most would agree that disease development is a multifactorial process, including genetic components, environmental factors in addition to a gender bias (9:1 women versus men of fertile age) linked to female hormones.
The influence of genetic components on B cell tolerance mechanisms in systemic autoimmunity have been directly demonstrated by crossing the above-mentioned TG mice onto genetic backgrounds in which autoimmune disease develop. It was shown that the 3H9 antibody on the MRL/lpr background91 as well as the D42 antibody on the NZB/W background92 result in high affinity anti-dsDNA autoantibodies encoded by the TG V-genes. Therefore the fourth postulate holds true for the genetically predisposed strains of mice. Furthermore, in both models, extreme clonal expansion of autoreactive B cells was observed and high affinity anti-dsDNA reactivity developed by SHM.
SLE and Genome Wide Association Studies
The development of SLE and, in particular, ANAs show a strong heritability also in humans, as twin studies have clearly indicated.93 However, the pattern of inheritance is clearly complex and an unknown number of susceptibility loci confer risk to develop SLE and ANAs. Whereas in the 1990s certain MHC, complement and Fcγ-receptor alleles have been identified as SLE susceptibility loci, most recently genome wide association studies (GWAS) identified more than 20 additional susceptibility loci, with high significance and, in most cases, reproduced in several independent studies,94 although the individual risk (odds ratio) was rather low, ranging between 1.2 to 2 for most of the susceptibility alleles. What does this reveal about the genetics of SLE? It certainly shows the complexity of the genetic basis of this systemic autoimmune disease. Most likely, in each patient an individual combination of, probably additive, susceptibility loci result in the development of autoimmune disease, together with the influence of environmental and hormonal triggers. Thus, the individual contribution of a single susceptibility allele to the development of the disease is very low.
The molecular pathways in which these susceptibility genes are involved give a clear picture about the mechanisms that can induce SLE as the susceptibility loci fall into three pathways that are apparently altered: (1) Immune complex (antibody-antigen complexes) uptake including phagocytosis of apoptotic cells; (2) Signaling alterations (mostly in B cells) and (3) Alterations in TLRs and the type I interferon pathway. Strong evidence for the importance of these three pathways in the development of SLE is also provided from mouse genetics. Here, we will focus on three examples of the involvement of these pathways, based on mouse genetics that clearly affirm the data from the GWA studies on SLE patients.
The milk fat globule epidermal growth factor (EGF) 8 (MFG-E8) is a protein that binds to apoptotic cells by recognizing phosphatidylserine and that enhances the engulfment of apoptotic cells by macrophages. Mouse mutants lacking MFG-E8 develop anti-DNA autoantibodies and glomerular disease.95 As it has been shown in these mice that apoptotic cells were not efficiently engulfed in germinal centers, the hypothesis is that B cells in the germinal center with high affinity for nuclear antigens, arising by the process of SHM, would receive survival signals, differentiate into memory B cells as well as plasma cells producing ANAs.
As an example for signaling alteration in B cells, a recently described mouse mutant carrying a single gain-of-function point mutation in the phospholipase Cγ2 gene (a signaling molecule downstream of the pre-BCR and BCR) leads to increased calcium responses in B cells after BCR stimulation and importantly, to SLE-like disease including ANAs and glomerulonephritis.96
Lastly, alterations in TLRs and the signaling components of type I interferon have recently provided strong genetic evidence for the involvement of this pathway in the development and the pathogenesis of SLE. The mere elevation of the copy number of TLR7, which recognizes single-stranded RNA, in TG mice on a normal genetic background was sufficient to induce autoimmunity towards nuclear antigens and autoimmune disease that closely resembles that of SLE.97
Concluding Remarks
Taken together, it is clear that having autoantibodies in the system is not always indicative of autoimmune disease but rather, low levels of autoantibodies appear to be the normal state in healthy individuals and may even be protective. Whereas B cells expressing natural autoantibodies seem to undergo positive selection, those that express highly autoreactive antibodies undergo negative selection, if they arise at all. Thus, in order to generate plasma cells secreting pathogenic autoantibodies a multifactorial process is required, which includes combinations of genetic components.
Acknowledgements
We acknowledge the financial support of the Swedish Cancer Foundation, Swedish Research Council (521-2009-3907), Torsten and Ragnar Söderbergs Stiftelser, Reumatikerförbundet, the Sahlgrenska Academy, the ALF fund (I.L.M.) and the Deutsche Forschungsgemeinschaft (SFB 423, T.H.W.).
Abbreviations
- ANA
anti-nuclear antibodies
- BCR
B cell receptor
- CDR
complementarity determining region
- CSR
class switch recombination
- D
diversity
- EDD
electron dense deposit
- FO
follicular
- FWR
frame work region
- GMB
glomerular basement membrane
- GP
Goodpasture's disease
- GWAS
genome wide association studies
- HC
heavy chain
- HEL
hen egg lysozyme
- Ig
immunoglobulin
- J
joining
- LC
light chain
- LPS
lipopolysaccharide
- MFG-E8
milk fat globule epidermal growth factor 8
- MZ
marginal zone
- RAG
recombination-activating-genes
- RF
reading frame
- SHM
somatic hyper mutation
- SLE
systemic lupus erythematosus
- TdT
terminal-nucleotidyl-transferase
- TG
transgenic
- V
variable
References
- 1.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 2.Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity. 2000;13:37–45. doi: 10.1016/s1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 3.Alt FW, Yancopoulos GD, Blackwell TK, Wood C, Thomas E, Boss M, et al. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 5.Burrows PD, Cooper MD. B cell development and differentiation. Curr Opin Immunol. 1997;9:239–244. doi: 10.1016/s0952-7915(97)80142-2. [DOI] [PubMed] [Google Scholar]
- 6.Conley ME, Burrows PD. Plugging the leaky pre-B cell receptor. J Immunol. 184:1127–1129. doi: 10.4049/jimmunol.0990113. [DOI] [PubMed] [Google Scholar]
- 7.Martensson IL, Almqvist N, Grimsholm O, Bernardi AI. The pre-B cell receptor checkpoint. FEBS Lett. 584:2572–2579. doi: 10.1016/j.febslet.2010.04.057. [DOI] [PubMed] [Google Scholar]
- 8.von Boehmer H, Melchers F. Checkpoints in lymphocyte development and autoimmune disease. Nat Immunol. 11:14–20. doi: 10.1038/ni.1794. [DOI] [PubMed] [Google Scholar]
- 9.Vettermann C, Herrmann K, Jack HM. Powered by pairing: the surrogate light chain amplifies immunoglobulin heavy chain signaling and pre-selects the antibody repertoire. Semin Immunol. 2006;18:44–55. doi: 10.1016/j.smim.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Duber S, Hafner M, Krey M, Lienenklaus S, Roy B, Hobeika E, et al. Induction of B-cell development in adult mice reveals the ability of bone marrow to produce B-1a cells. Blood. 2009;114:4960–4967. doi: 10.1182/blood-2009-04-218156. [DOI] [PubMed] [Google Scholar]
- 11.Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a B-1 B cell-specified progenitor. Nat Immunol. 2006;7:293–301. doi: 10.1038/ni1301. [DOI] [PubMed] [Google Scholar]
- 12.Tung JW, Mrazek MD, Yang Y, Herzenberg LA. Phenotypically distinct B cell development pathways map to the three B cell lineages in the mouse. Proc Natl Acad Sci USA. 2006;103:6293–6298. doi: 10.1073/pnas.0511305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou ZH, Notkins AL. Polyreactive antigen-binding B (PAB-) cells are widely distributed and the PAB population consists of both B-1+ and B-1− phenotypes. Clin Exp Immunol. 2004;137:88–100. doi: 10.1111/j.1365-2249.2004.02511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Notkins AL. Polyreactivity of antibody molecules. Trends Immunol. 2004;25:174–179. doi: 10.1016/j.it.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 16.Tsuiji M, Yurasov S, Velinzon K, Thomas S, Nussenzweig MC, Wardemann H. A checkpoint for autoreactivity in human IgM+ memory B cell development. J Exp Med. 2006;203:393–400. doi: 10.1084/jem.20052033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tiller T, Tsuiji M, Yurasov S, Velinzon K, Nussenzweig MC, Wardemann H. Autoreactivity in human IgG+ memory B cells. Immunity. 2007;26:205–213. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avrameas S, Ternynck T, Tsonis IA, Lymberi P. Naturally occurring B-cell autoreactivity: a critical overview. J Autoimmun. 2007;29:213–218. doi: 10.1016/j.jaut.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Elkon K, Casali P. Nature and functions of autoantibodies. Nat Clin Pract Rheumatol. 2008;4:491–498. doi: 10.1038/ncprheum0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou ZH, Tzioufas AG, Notkins AL. Properties and function of polyreactive antibodies and polyreactive antigen-binding B cells. J Autoimmun. 2007;29:219–228. doi: 10.1016/j.jaut.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avrameas S. Natural autoantibodies: from ‘horror autotoxicus’ to ‘gnothi seauton’. Immunol Today. 1991;12:154–159. doi: 10.1016/S0167-5699(05)80045-3. [DOI] [PubMed] [Google Scholar]
- 22.Haas KM, Poe JC, Steeber DA, Tedder TF. B-1a and B-1b cells exhibit distinct developmental requirements and have unique functional roles in innate and adaptive immunity to S. pneumoniae. Immunity. 2005;23:7–18. doi: 10.1016/j.immuni.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 23.Martin F, Oliver AM, Kearney JF. Marginal zone and B1 B cells unite in the early response against T-independent blood-borne particulate antigens. Immunity. 2001;14:617–629. doi: 10.1016/s1074-7613(01)00129-7. [DOI] [PubMed] [Google Scholar]
- 24.Binder CJ. Natural IgM antibodies against oxidation-specific epitopes. J Clin Immunol. 2010;30:56–60. doi: 10.1007/s10875-010-9396-3. [DOI] [PubMed] [Google Scholar]
- 25.Binder CJ, Hörkkö S, Dewan A, Chang MK, Kieu EP, Goodyear CS, et al. Pneumococcal vaccination decreases atherosclerotic lesion formation: molecular mimicry between Streptococcus pneumoniae and oxidized LDL. Nat Med. 2003;9:736–743. doi: 10.1038/nm876. [DOI] [PubMed] [Google Scholar]
- 26.Lutz HU, Binder CJ, Kaveri S. Naturally occurring auto-antibodies in homeostasis and disease. Trends Immunol. 2009;30:43–51. doi: 10.1016/j.it.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Witebsky E, Rose NR, Terplan K, Paine JR, Egan RW. Chronic thyroiditis and autoimmunization. J Am Med Assoc. 1957;164:1439–1447. doi: 10.1001/jama.1957.02980130015004. [DOI] [PubMed] [Google Scholar]
- 28.Bona CA. Postulates defining pathogenic autoantibodies and T cells. Autoimmunity. 1991;10:169–172. doi: 10.3109/08916939109001886. [DOI] [PubMed] [Google Scholar]
- 29.Ceppellini R, Polli E, Celada F. A DNA-reacting factor in serum of a patient with lupus erythematosus diffusus. Proc Soc Exp Biol Med. 1957;96:572–574. doi: 10.3181/00379727-96-23544. [DOI] [PubMed] [Google Scholar]
- 30.Friou GJ. Clinical application of a test for lupus globulin-nucleohistone interaction using fluorescent antibody. Yale J Biol Med. 1958;31:40–47. [PMC free article] [PubMed] [Google Scholar]
- 31.Miescher P, Strassle R. New serological methods for the detection of the L.E. factor. Vox Sang. 1957;2:283–287. doi: 10.1111/j.1423-0410.1957.tb03704.x. [DOI] [PubMed] [Google Scholar]
- 32.Robbins WC, Holman HR, Deicher H, Kunkel HG. Complement fixation with cell nuclei and DNA in lupus erythematosus. Proc Soc Exp Biol Med. 1957;96:575–579. doi: 10.3181/00379727-96-23545. [DOI] [PubMed] [Google Scholar]
- 33.Desai DD, Krishnan MR, Swindle JT, Marion TN. Antigen-specific induction of antibodies against native mammalian DNA in nonautoimmune mice. J Immunol. 1993;151:1614–1626. [PubMed] [Google Scholar]
- 34.Moens U, Seternes OM, Hey AW, Silsand Y, Traavik T, Johansen B, et al. In vivo expression of a single viral DNA-binding protein generates systemic lupus erythematosus-related autoimmunity to double-stranded DNA and histones. Proc Natl Acad Sci USA. 1995;92:12393–12397. doi: 10.1073/pnas.92.26.12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mevorach D, Zhou JL, Song X, Elkon KB. Systemic exposure to irradiated apoptotic cells induces autoantibody production. J Exp Med. 1998;188:387–392. doi: 10.1084/jem.188.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koffler D, Schur PH, Kunkel HG. Immunological studies concerning the nephritis of systemic lupus erythematosus. J Exp Med. 1967;126:607–624. doi: 10.1084/jem.126.4.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winfield JB, Faiferman I, Koffler D. Avidity of anti-DNA antibodies in serum and IgG glomerular eluates from patients with systemic lupus erythematosus. Association of high avidity antinative DNA antibody with glomerulonephritis. J Clin Invest. 1977;59:90–96. doi: 10.1172/JCI108626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ehrenstein MR, Katz DR, Griffiths MH, Papadaki L, Winkler TH, Kalden JR, et al. Human IgG anti-DNA antibodies deposit in kidneys and induce proteinuria in SCID mice. Kidney Int. 1995;48:705–711. doi: 10.1038/ki.1995.341. [DOI] [PubMed] [Google Scholar]
- 39.Gilkeson GS, Bernstein K, Pippen AM, Clarke SH, Marion T, Pisetsky DS, et al. The influence of variable-region somatic mutations on the specificity and pathogenicity of murine monoclonal anti-DNA antibodies. Clin Immunol Immunopathol. 1995;76:59–67. doi: 10.1006/clin.1995.1088. [DOI] [PubMed] [Google Scholar]
- 40.Madaio MP, Carlson J, Cataldo J, Ucci A, Migliorini P, Pankewycz O. Murine monoclonal anti-DNA antibodies bind directly to glomerular antigens and form immune deposits. J Immunol. 1987;138:2883–2889. [PubMed] [Google Scholar]
- 41.Vlahakos D, Foster MH, Ucci AA, Barrett KJ, Datta SK, Madaio MP. Murine monoclonal anti-DNA antibodies penetrate cells, bind to nuclei and induce glomerular proliferation and proteinuria in vivo. J Am Soc Nephrol. 1992;2:1345–1354. doi: 10.1681/ASN.V281345. [DOI] [PubMed] [Google Scholar]
- 42.Vlahakos DV, Foster MH, Adams S, Katz M, Ucci AA, Barrett KJ, et al. Anti-DNA antibodies form immune deposits at distinct glomerular and vascular sites. Kidney Int. 1992;41:1690–1700. doi: 10.1038/ki.1992.242. [DOI] [PubMed] [Google Scholar]
- 43.Briggs WA, Johnson JP, Teichman S, Yeager HC, Wilson CB. Antiglomerular basement membrane antibody-mediated glomerulonephritis and Goodpasture's syndrome. Medicine (Baltimore) 1979;58:348–361. doi: 10.1097/00005792-197909000-00002. [DOI] [PubMed] [Google Scholar]
- 44.Lerner RA, Glassock RJ, Dixon FJ. The role of antiglomerular basement membrane antibody in the pathogenesis of human glomerulonephritis. J Exp Med. 1967;126:989–1004. doi: 10.1084/jem.126.6.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui Z, Zhao MH, Segelmark M, Hellmark T. Natural autoantibodies to myeloperoxidase, proteinase 3 and the glomerular basement membrane are present in normal individuals. Kidney Int. 78:590–597. doi: 10.1038/ki.2010.198. [DOI] [PubMed] [Google Scholar]
- 46.Burlingame RW, Cervera R. Anti-chromatin (antinucleosome) autoantibodies. Autoimmun Rev. 2002;1:321–328. doi: 10.1016/s1568-9972(02)00083-6. [DOI] [PubMed] [Google Scholar]
- 47.Deocharan B, Qing X, Lichauco J, Putterman C. Alpha-actinin is a cross-reactive renal target for pathogenic anti-DNA antibodies. J Immunol. 2002;168:3072–3078. doi: 10.4049/jimmunol.168.6.3072. [DOI] [PubMed] [Google Scholar]
- 48.Mostoslavsky G, Fischel R, Yachimovich N, Yarkoni Y, Rosenmann E, Monestier M, et al. Lupus anti-DNA autoantibodies cross-react with a glomerular structural protein: a case for tissue injury by molecular mimicry. Eur J Immunol. 2001;31:1221–1227. doi: 10.1002/1521-4141(200104)31:4<1221::aid-immu1221>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 49.Faaber P, Rijke TP, van de Putte LB, Capel PJ, Berden JH. Cross-reactivity of human and murine anti-DNA antibodies with heparan sulfate. The major glycosaminoglycan in glomerular basement membranes. J Clin Invest. 1986;77:1824–1830. doi: 10.1172/JCI112508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lefkowith JB, Gilkeson GS. Nephritogenic autoantibodies in lupus: current concepts and continuing controversies. Arthritis Rheum. 1996;39:894–903. doi: 10.1002/art.1780390605. [DOI] [PubMed] [Google Scholar]
- 51.Mjelle JE, Rekvig OP, Fenton KA. Nucleosomes possess a high affinity for glomerular laminin and collagen IV and bind nephritogenic antibodies in murine lupus-like nephritis. Ann Rheum Dis. 2007;66:1661–1668. doi: 10.1136/ard.2007.070482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Bavel CC, Fenton KA, Rekvig OP, van der Vlag J, Berden JH. Glomerular targets of nephritogenic autoantibodies in systemic lupus erythematosus. Arthritis Rheum. 2008;58:1892–1899. doi: 10.1002/art.23626. [DOI] [PubMed] [Google Scholar]
- 53.Termaat RM, Brinkman K, van Gompel F, van den Heuvel LP, Veerkamp JH, Smeenk RJ, et al. Cross-reactivity of monoclonal anti-DNA antibodies with heparan sulfate is mediated via bound DNA/histone complexes. J Autoimmun. 1990;3:531–545. doi: 10.1016/s0896-8411(05)80019-8. [DOI] [PubMed] [Google Scholar]
- 54.Ben-Bassat M, Rosenfeld J, Joshua H, Hazaz B, Gura V. Lupus nephritis. Electron-dense and immunofluorescent deposits and their correlation with proteinuria and renal function. Am J Clin Pathol. 1979;72:186–193. doi: 10.1093/ajcp/72.2.186. [DOI] [PubMed] [Google Scholar]
- 55.Comerford FR, Cohen AS, Desai RG. The evolution of the glomerular lesion in NZB mice. A light and electron microscopic study. Lab Invest. 1968;19:643–651. [PubMed] [Google Scholar]
- 56.Dillard MG, Tillman RL, Sampson CC. Lupus Nephritis. Correlations between the clinical course and presence of electron-dense deposits. Lab Invest. 1975;32:261–269. [PubMed] [Google Scholar]
- 57.Kalaaji M, Mortensen E, Jorgensen L, Olsen R, Rekvig OP. Nephritogenic lupus antibodies recognize glomerular basement membrane-associated chromatin fragments released from apoptotic intraglomerular cells. Am J Pathol. 2006;168:1779–1792. doi: 10.2353/ajpath.2006.051329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dieker JW, van der Vlag J, Berden JH. Triggers for antichromatin autoantibody production in SLE. Lupus. 2002;11:856–864. doi: 10.1191/0961203302lu307rr. [DOI] [PubMed] [Google Scholar]
- 59.Krishnan MR, Jou NT, Marion TN. Correlation between the amino acid position of arginine in VH-CDR3 and specificity for native DNA among autoimmune antibodies. J Immunol. 1996;157:2430–2439. [PubMed] [Google Scholar]
- 60.Shlomchik MJ, Aucoin AH, Pisetsky DS, Weigert MG. Structure and function of anti-DNA autoantibodies derived from a single autoimmune mouse. Proc Natl Acad Sci USA. 1987;84:9150–9154. doi: 10.1073/pnas.84.24.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tillman DM, Jou NT, Hill RJ, Marion TN. Both IgM and IgG anti-DNA antibodies are the products of clonally selective B cell stimulation in (NZB × NZW) F1 mice. J Exp Med. 1992;176:761–779. doi: 10.1084/jem.176.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winkler TH, Fehr H, Kalden JR. Analysis of immunoglobulin variable region genes from human IgG anti-DNA hybridomas. Eur J Immunol. 1992;22:1719–1728. doi: 10.1002/eji.1830220709. [DOI] [PubMed] [Google Scholar]
- 63.Radic MZ, Mackle J, Erikson J, Mol C, Anderson WF, Weigert M. Residues that mediate DNA binding of autoimmune antibodies. J Immunol. 1993;150:4966–4977. [PubMed] [Google Scholar]
- 64.Radic MZ, Weigert M. Genetic and structural evidence for antigen selection of anti-DNA antibodies. Annu Rev Immunol. 1994;12:487–520. doi: 10.1146/annurev.iy.12.040194.002415. [DOI] [PubMed] [Google Scholar]
- 65.Mol CD, Muir AK, Lee JS, Anderson WF. Structure of an immunoglobulin Fab fragment specific for poly(dG).poly(dC) J Biol Chem. 1994;269:3605–3614. [PubMed] [Google Scholar]
- 66.Chukwuocha RU, Zhu M, Cho CS, Visvanathan S, Hwang KK, Rahman A, et al. Molecular and genetic characterizations of five pathogenic and two non-pathogenic monoclonal antiphospholipid antibodies. Mol Immunol. 2002;39:299–311. doi: 10.1016/s0161-5890(02)00115-3. [DOI] [PubMed] [Google Scholar]
- 67.Giles I, Lambrianides N, Pattni N, Faulkes D, Latchman D, Chen P, et al. Arginine residues are important in determining the binding of human monoclonal antiphospholipid antibodies to clinically relevant antigens. J Immunol. 2006;177:1729–1736. doi: 10.4049/jimmunol.177.3.1729. [DOI] [PubMed] [Google Scholar]
- 68.Hohmann A, Cairns E, Brisco M, Bell DA, Diamond B. Immunoglobulin gene sequence analysis of anti-cardiolipin and anti-cardiolipin idiotype (H3) human monoclonal antibodies. Autoimmunity. 1995;22:49–58. doi: 10.3109/08916939508995299. [DOI] [PubMed] [Google Scholar]
- 69.Guo W, Smith D, Aviszus K, Detanico T, Heiser RA, Wysocki LJ. Somatic hypermutation as a generator of antinuclear antibodies in a murine model of systemic autoimmunity. J Exp Med. 207:2225–2237. doi: 10.1084/jem.20092712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wellmann U, Letz M, Herrmann M, Angermuller S, Kalden JR, Winkler TH. The evolution of human anti-double-stranded DNA autoantibodies. Proc Natl Acad Sci USA. 2005;102:9258–9263. doi: 10.1073/pnas.0500132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shlomchik M, Mascelli M, Shan H, Radic MZ, Pisetsky D, Marshak-Rothstein A, et al. Anti-DNA antibodies from autoimmune mice arise by clonal expansion and somatic mutation. J Exp Med. 1990;171:265–292. doi: 10.1084/jem.171.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Erikson J, Radic MZ, Camper SA, Hardy RR, Carmack C, Weigert M. Expression of anti-DNA immunoglobulin transgenes in non-autoimmune mice. Nature. 1991;349:331–334. doi: 10.1038/349331a0. [DOI] [PubMed] [Google Scholar]
- 73.Chen C, Nagy Z, Radic MZ, Hardy RR, Huszar D, Camper SA, et al. The site and stage of anti-DNA B-cell deletion. Nature. 1995;373:252–255. doi: 10.1038/373252a0. [DOI] [PubMed] [Google Scholar]
- 74.Pewzner-Jung Y, Friedmann D, Sonoda E, Jung S, Rajewsky K, Eilat D. B cell deletion, anergy and receptor editing in “knock in” mice targeted with a germline-encoded or somatically mutated anti-DNA heavy chain. J Immunol. 1998;161:4634–4645. [PubMed] [Google Scholar]
- 75.Gay D, Saunders T, Camper S, Weigert M. Receptor editing: an approach by autoreactive B cells to escape tolerance. J Exp Med. 1993;177:999–1008. doi: 10.1084/jem.177.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goodnow CC, Crosbie J, Adelstein S, Lavoie TB, Smith-Gill SJ, Brink RA, et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 1988;334:676–682. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 77.Hartley SB, Cooke MP, Fulcher DA, Harris AW, Cory S, Basten A, et al. Elimination of self-reactive B lymphocytes proceeds in two stages: arrested development and cell death. Cell. 1993;72:325–335. doi: 10.1016/0092-8674(93)90111-3. [DOI] [PubMed] [Google Scholar]
- 78.Nemazee DA, Burki K. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature. 1989;337:562–566. doi: 10.1038/337562a0. [DOI] [PubMed] [Google Scholar]
- 79.Tiegs SL, Russell DM, Nemazee D. Receptor editing in self-reactive bone marrow B cells. J Exp Med. 1993;177:1009–1020. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li H, Jiang Y, Prak EL, Radic M, Weigert M. Editors and editing of anti-DNA receptors. Immunity. 2001;15:947–957. doi: 10.1016/s1074-7613(01)00251-5. [DOI] [PubMed] [Google Scholar]
- 81.Liu S, Velez MG, Humann J, Rowland S, Conrad FJ, Halverson R, et al. Receptor editing can lead to allelic inclusion and development of B cells that retain antibodies reacting with high avidity autoantigens. J Immunol. 2005;175:5067–5076. doi: 10.4049/jimmunol.175.8.5067. [DOI] [PubMed] [Google Scholar]
- 82.Velez MG, Kane M, Liu S, Gauld SB, Cambier JC, Torres RM, et al. Ig allotypic inclusion does not prevent B cell development or response. J Immunol. 2007;179:1049–1057. doi: 10.4049/jimmunol.179.2.1049. [DOI] [PubMed] [Google Scholar]
- 83.Witsch EJ, Cao H, Fukuyama H, Weigert M. Light chain editing generates polyreactive antibodies in chronic graft-versus-host reaction. J Exp Med. 2006;203:1761–1772. doi: 10.1084/jem.20060075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Makdasi E, Fischel R, Kat I, Eilat D. Autoreactive anti-DNA transgenic B cells in lupus-prone New Zealand black/New Zealand white mice show near perfect L chain allelic exclusion. J Immunol. 2009;182:6143–6148. doi: 10.4049/jimmunol.0803610. [DOI] [PubMed] [Google Scholar]
- 85.Hayakawa K, Asano M, Shinton SA, Gui M, Allman D, Stewart CL, et al. Positive selection of natural autoreactive B cells. Science. 1999;285:113–116. doi: 10.1126/science.285.5424.113. [DOI] [PubMed] [Google Scholar]
- 86.Tian Q, Beardall M, Xu Y, Li J, Parker DC, Casanova N, et al. B cells expressing a natural polyreactive autoantibody have a distinct phenotype and are over-represented in immunoglobulin heavy chain transgenic mice. J Immunol. 2006;177:2412–2422. doi: 10.4049/jimmunol.177.4.2412. [DOI] [PubMed] [Google Scholar]
- 87.Minegishi Y, Conley ME. Negative selection at the pre-BCR checkpoint elicited by human mu heavy chains with unusual CDR3 regions. Immunity. 2001;14:631–641. doi: 10.1016/s1074-7613(01)00131-5. [DOI] [PubMed] [Google Scholar]
- 88.Minegishi Y, Coustan-Smith E, Wang YH, Cooper MD, Campana D, Conley ME. Mutations in the human lambda5/14.1 gene result in B cell deficiency and agammaglobulinemia. J Exp Med. 1998;187:71–77. doi: 10.1084/jem.187.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Keenan RA, De Riva A, Corleis B, Hepburn L, Licence S, Winkler TH, et al. Censoring of autoreactive B cell development by the pre-B cell receptor. Science. 2008;321:696–699. doi: 10.1126/science.1157533. [DOI] [PubMed] [Google Scholar]
- 90.Shimizu T, Mundt C, Licence S, Melchers F, Martensson IL. VpreB1/VpreB2/lambda 5 triple-deficient mice show impaired B cell development but functional allelic exclusion of the IgH locus. J Immunol. 2002;168:6286–6293. doi: 10.4049/jimmunol.168.12.6286. [DOI] [PubMed] [Google Scholar]
- 91.Brard F, Shannon M, Prak EL, Litwin S, Weigert M. Somatic mutation and light chain rearrangement generate autoimmunity in anti-single-stranded DNA transgenic MRL/lpr mice. J Exp Med. 1999;190:691–704. doi: 10.1084/jem.190.5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Friedmann D, Yachimovich N, Mostoslavsky G, Pewzner-Jung Y, Ben-Yehuda A, Rajewsky K, et al. Production of high affinity autoantibodies in autoimmune New Zealand Black/New Zealand white F1 mice targeted with an anti-DNA heavy chain. J Immunol. 1999;162:4406–4416. [PubMed] [Google Scholar]
- 93.Reichlin M, Harley JB, Lockshin MD. Serologic studies of monozygotic twins with systemic lupus erythematosus. Arthritis Rheum. 1992;35:457–464. doi: 10.1002/art.1780350416. [DOI] [PubMed] [Google Scholar]
- 94.Harley ITW, Kaufman KM, Langefeld CD, Harley JB, Kelly JA. Genetic susceptibility to SLE: new insights from fine mapping and genome-wide association studies. Nat Rev Genet. 2009;10:285–290. doi: 10.1038/nrg2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hanayama R, Tanaka M, Miyasaka K, Aozasa K, Koike M, Uchiyama Y, et al. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science. 2004;304:1147–1150. doi: 10.1126/science.1094359. [DOI] [PubMed] [Google Scholar]
- 96.Yu P, Constien R, Dear N, Katan M, Hanke P, Bunney TD, et al. Autoimmunity and inflammation due to a gain-of-function mutation in phospholipase C gamma 2 that specifically increases external Ca2+ entry. Immunity. 2005;22:451–465. doi: 10.1016/j.immuni.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 97.Deane JA, Pisitkun P, Barrett RS, Feigenbaum L, Town T, Ward JM, et al. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity. 2007;27:801–810. doi: 10.1016/j.immuni.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]