Abstract
Due to the variety and complexity of microorganisms, the mechanisms needed for pathogen recognition are diverse. Innate immune recognition is mainly based on a series of germ-line encoded receptors that have been selected by evolution to recognize nonself molecules present in microorganisms. Innate immunity also recognizes changes in our cells caused by infection, such as the lack or induction of self molecules. Adaptative immunity somatically generates large repertories of receptors which collectively recognize any nonself antigen. These receptors are randomly generated, and the adaptative immune system has to learn how to eliminate or inactivate cells with high avidity receptors for self molecules. Given the enormous variety of microbe structures and immune receptors, the difference between self and nonself is not absolute; it depends on the threshold of activation. In genetically diverse populations, individuals who have this activation threshold too far from the average may suffer an autoimmune reaction. Accumulation of mutations in cancer cells generates neoantigens that may be also recognized as nonself molecules, but the extent of self and nonself discrimination limits immune responsiveness to them. Surprisingly, most of the molecules expressed by cancer cells recognized by the immune system are non mutated self molecules.
Key words: self, nonself, adaptative immunity, innate immunity, T cells, autoimmunity, cancer
Introduction
Complex animals live in nutrient rich and protected environments which are attractive habitats for other smaller organisms. Some internal microbes are not harmful and are even beneficial for their hosts, but others are pathogens, which directly may harm or even kill the host. The complexity of higher organisms augments the necessity to maintain their self integrity in an increasing hostile environment. Virtually all organisms, from bacteria to higher animals, possess recognition systems that allow them to discriminate between self and nonself and possess effector mechanisms to defend themselves from nonself. Complex immune systems have evolved in vertebrates, but the strategies and immune mechanisms involved in the discrimination of self and nonself are essentially the same in all vertebrates. Two different arms, the innate and adaptative immune system, have emerged at different moments in evolution, and they are conceptually different. Innate immunity is the dominant immune system found in plants, fungi, insects and primitive multicellular organisms1–3 (Table 1). The innate immune system recognition is based on a series of germ-line encoded receptors which have been selected during the evolution to specifically recognize pathogens. Its response to microbes is innate as its action does not depend upon prior exposure to particular pathogens. The innate immune system provides immediate defense against infection, but it does not confer long-lasting or protective immunity to the host. Innate discrimination between self and nonself is mainly based on receptors, which recognize nonself molecules present in pathogens, but not present in the host. These nonself antigens are key molecules in the survival and/or virulence of these pathogens, which are difficult to mutate without affecting viability or infectivity of pathogens. They are frequently conserved in whole groups or families of microbes.
Table 1.
A summary of properties distinguishing the innate and adaptative immune system (3)
| Property | Defense mechanism (innate) | Immune system (adaptative) |
| Present in: | All organisms | Vertebrates only |
| Self-nonself discrimination is: | Germline selected | Somatically selected |
| Receptors are: | Germline selected | Somatically selected |
| Antiself selection purges: | Individuals | Cells |
| Defects in antiself selection: | Does not causes autoimmunity | Causes autoimmunity |
| Unresponsive to the: | Self-of-the-species | Self-of-the-individual |
| Effector mechanisms are: | Innate | Coupled to innate effector mechanisms |
However, a central point of the co-evolution of hosts and pathogens is the fact that microbes may evolve faster than their hosts. Innate immunity is able to recognize and eliminate a vast number of pathogens; however, germ-line encoded mechanisms of defense can not compete with rapidly dividing microbes. A major goal of the evolution of vertebrates was the appearance of the adaptative immune system, thought to have arisen in the first jawed vertebrates. The adaptive immune response is not innate, but it has the ability to generate a specific immune response against any microbe we encounter and to mount stronger attacks every time the pathogen is encountered. Adaptative immunity somatically generates large repertories of receptors of T and B lymphocytes, [T Cell Receptor (TCR) and B Cell Receptor (BCR)], which may be able to virtually recognize any nonself antigen. The generation of these large repertoires of adaptative receptors for nonself created two new problems during evolution. First, this random process may yield some receptors that have high avidity for our own self molecules. Consequently, the adaptative immune system had to learn how to discriminate self from nonself to avoid an anti-self reaction. Lymphocytes bearing these high avidity autoreactive receptors have to be eliminated or regulated. However, the imperfection of this process is clearly seen in the high frequency of autoimmune diseases that exists. Second, the adaptative immune system of recognition had to develop effector mechanisms capable of eliminating pathogens. To solve this problem, the adaptive immune system was coupled to an evolutionarily older effector mechanism of the innate immune system, and uses the same system for elimination of pathogens.
Self and Nonself Paradigm
Janeway formulated the theory of “extended self and nonself” which hypothesized that microbes are distinguished from self molecules by a variety of germ-line encoded receptors that recognize molecular signatures present in the microorganisms but absent in the host.2 The recognition of microbes by the innate immunity depends on a system of receptors that recognize pathogen-associated molecular patterns (PAMPs) unique to microbes and distinct from self. The classic example of this system of recognition is the toll like receptors (TLRs), which recognize structurally specific molecules conserved among families or groups of pathogens.4 TLRs ligands are quite invariant molecules, conserved from Drosophila melanogaster to mammals, despite that microbes are constantly mutating their antigens. However, TLR ligands are generally key molecules involved in the survival and virulence of these pathogens.4 Pathogens that mutate these structures may escape from TLR recognition, but they are less virulent, less viable or both. For instance, TLR4 detects lipopolysaccharide (LPS), which is the major component of in the outer membrane of all Gram-negative bacteria. LPS contributes greatly to the structural integrity of the bacteria, increases the negative charge of the cell membrane and helps stabilize the overall membrane structure. LPS is a large molecule consisting of a lipid covalently bound to a polysaccharide. The polysaccharide part varies in different gram negative bacteria and this part of LPS may be recognized by adaptative immunity. The lipid part is highly conserved among all Gram negative bacteria and is recognized by the innate immune system. Many immune innate receptors directly engage microbial molecules of different biochemical structures including lipids, proteins and sugars. Some sugars are recognized by the receptors of the mannan-binding lectin-associated serine protease (MASP) pathway, which include soluble mannose binding receptors (ficolins and other collectins) that activate complement via the MASP-2 pathway. The most structurally variable molecules in nature are proteins and alterations in protein structure most consistently distinguish one specie from another. Proteins are usually recognized by the adaptive immunity, which is well suited to recognize such differences; but innate immunity also recognizes some proteins, such as flagellin (a ligand for TLR5), which are widely conserved across many microbial taxa.
The classic view of the innate immune system shaped through evolution to discriminate self from nonself has been expanded due to the description of additional functions. Innate immunity not only recognizes nonself antigens, but also utilizes more sophisticated mechanisms of recognition. Modification of self proteins by microbial enzymes may also lead to recognition by the innate immune system. For example, the coagulases of Staphylococci may initiate a clotting cascade. Fungal proteases can activate a proteolytic cascade that eventuates the production of spaetzle, the ligand for Toll. Along the same lines, the action of microbial toxins, such as the adenosine diphosphate (ADP)-ribosyltransferase of diphtheria toxin, may cause cell death and so produce awareness of infection.
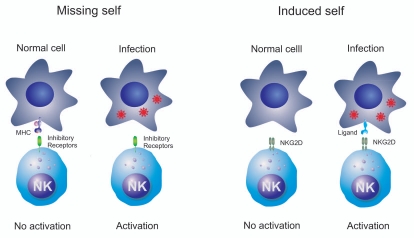
Innate receptors may also recognize self molecules not present in a healthy state, but appear associated with certain diseases. In other words, the innate immune system may also recognize changes in our cells caused by infection. The “missing self” hypothesis proposed by Karre et al. provided the first description on how Natural Killer (NK) cell function is regulated (Fig. 1).5 Target cells expressing major histocompatibility complex (MHC) class I molecules are more resistant to NK cell mediated killing than virally infected cells that have lost the expression of MHC class I molecules. Thus, NK cells utilize the inhibitory receptors to differentiate “self” from “missing self”. Inhibitory receptors are represented in humans by immunoglobulin (Ig)-like receptor-(KIRs) and lectin-like CD94/NKG2A/B heterodimer and in mouse by Ly49. Interaction of MHC class I with these inhibitory receptors prevents the activation of NK cells and the lysis of the target cell.6,7
Figure 1.
Innate immunity may also recognize changes in cells caused by infection. NK cells utilize inhibitory receptors to differentiate “self” from “missing self”. The lack of expression of MHC class I molecules (missing self) promotes the activation of NK cells and the lysis of the target cell. NK cells also express activating receptors, such as NKG2D, which may directly recognize ligands induced in response to the infection (induced self).
Innate immunity also recognizes expression of self molecules that are induced by infection. NKG2D is an activating receptor expressed by NK cells and T cells which interacts with stress-inducible ligands.7–12 In mice, NKG2D ligands are not expressed by most benign cells, but are upregulated on some virally infected cells, which thus become susceptible to NK cell killing. This finding has originated the “induced or stressed-self” hypothesis which postulates that NKG2D ligands are not expressed in normal cells, but they are upregulated in response to cellular changes caused by infection or malignant cell transformation (Fig. 1). Consequently, the missing self hypothesis and the induced self hypothesis postulated that innate immune system does not only recognize nonself molecules but also many cellular changes caused by infection, including the suppression or induction of self molecules. Additionally, the existence of other unknown mechanisms to recognize microbes different from the self and nonself discrimination can not be excluded.
The increasing evidence that the functions of the immune system exceed the self and self nonself discrimination has prompted several theories about its functions. One of the most renowned is the danger model proposed by Polly Matzinger. She postulated the potential range of stimuli that can trigger the innate arm of immunity.13,14 She claimed that the immune system does not respond to nonself antigens, but instead it responds to dangerous molecules, cellular damage and destruction regardless of a self or nonself origin. Some statements of this model are inarguable, such as the immune response against a sterile injury.15 This response is only focused on the potential danger, but it does not respond to a nonself pathogen. Nevertheless, the term danger is nebulous, the mechanisms involved in the response to danger are obscure, and molecular events responsible for the danger response remain to be defined.16 Many “danger” ligands have been proposed, including uric acid, heat shock protein 70 (HSP70) and high mobility group box 1 (HMGB1). Some of these ligands are clearly inflammatory, whereas the case has not been convincingly made for others. However, this model proposes what triggers innate immune system, since clearly the function of all receptors of the innate immune system do not fit into the self and nonself paradigm.
Discrimination of Self and Nonself is Somatically Learned in Adaptative Immunity
When the vertebrates appeared, the germ-line evolution of the recognition elements of defense systems became too slow to compete with rapidly dividing pathogens, which have the ability to mutate and escape from immune recognition. A major overhaul of the recognition machinery of the defense system became necessary.3,17,18 The solution was the appearance of the adaptative immunity in the first vertebrates. To compete with the fast evolving microbes, adaptative immunity creates a nearly unlimited variability of receptors by random recombination to recognize any foreign antigen. Adaptative immunity somatically generates large repertories of receptors, TCR and BCR, which together may be able to recognize any nonself molecule. During their development, each B and T lymphocyte generates a unique receptor by rearranging of its receptor genes. Random variability generates a repertoire with potential anti-self B and T cells. These putative anti-self T and B cells need to be sorted by a somatic learning process and eventually deleted or inactivated.19 The ability of lymphocytes to discriminate self and nonself is shaped within each individual by clonal deletion of T cells and B cells with high avidity receptors for self antigens present in primary lymphoid tissues (thymus and bone marrow, respectively). Only those lymphocytes with low or intermediate avidity to self are present in peripheral tissues. Consequently, the self and nonself discrimination by the adaptive immune system is not directed by the germ-line, but instead it is somatically selected. Tissue transplants between non-vertebrate individuals (with only an innate immune system) are accepted because the recognition of nonself is germ-line selected, whereas transplants of tissues between vertebrate individuals (with adaptative immune systems) are rejected because anti-alloantigens specificities are not purged from the random repertoire. However, in spite of the adaptative immune system having emerged a new way of discrimination between self and nonself, most of the effector mechanisms used by vertebrates to eliminate pathogens are essentially identical to those generated by the non-vertebrates. The essential difference between innate and adaptative immune system is the way that they recognize nonself microbes, rather than the way for eliminating them.
The definition of self and nonself in the adaptative immune system is arbitrary. For instance, foreign antigens presented during foetal life are considered self because adaptative immunity learns to discriminate self from nonself during their maturation in primary lymphoid organs and any antigen present during this selection process is consider as self. Deletion in the thymus of high avidity T cell clones specific for the majority of self antigens generates a truncated peripheral self reactive repertoire of T cells with mainly intermediate and low avidity TCRs. Consequently, there are only high avidity T cells for nonself antigens. The activation of intermediate avidity, self-reactive T cells in the periphery may represent a potential danger for the host; self and nonself discrimination also need to be achieved in peripheral tissues by immune regulation. The “avidity model of self and nonself discrimination” postulated that survival of T cell clones in both the thymus and the periphery is determined by the avidity of TCR for specific MHC/antigen peptides presented by antigen-presenting cells.20 This model postulates that the adaptative immune system achieves self and nonself discrimination, not by discriminating self from nonself, but rather by perceiving the avidity of T cell activation. Discrimination of self from nonself in peripheral tissues is achieved by selective downregulation of intermediate avidity T cells to both self and nonself antigens and eliminates T cells containing the potentially pathogenic self-reactive T cells. This regulation enables the immune system to control the antiself responses without damaging the effective anti-nonself immunity, which is, largely, mediated by high avidity T cells specific to microbes. Thus, this model postulates that adaptative immune system does not discriminate foreign antigens by discriminating self from nonself in the periphery, but rather by perceiving the avidity of T cell activation.21
The Imperfection of Self and Nonself Discrimination Leads to Autoimmunity
The evolution of the immune system had to solve many compromises between conflicting demands of destroying nonself but tolerating self. Due to the enormous variety of microbial structures and immune receptors, the difference between self and nonself is not absolute, but based on choices that depend on the threshold for activation. The solution is optimized for the survival of a population as a whole. In genetically diverse populations, individuals who have this threshold of activation at one or the other extreme would suffer to an anti-self reaction or fail to respond to some microbes. There is no reason to think that there is an evolutionary pressure to assure the tolerance of self antigens that are unlikely to cause problems in most of the individuals of the population. Consequently, the self and nonself discrimination process of adaptative immune system is clearly imperfect and a relatively high frequency of anti-self reaction and autoimmune disease exists.
In humans, autoimmune diseases usually arise spontaneously, are usually sustained and persistent reactions, and lead to long term tissue damage, presumably because self antigens that drive the autoimmune responses can only be removed from the organism by destroying the cells that produce them. The events that trigger autoimmunity usually precedes the clinical symptoms of the disease and the early events are poorly studied and understood. Although many animal models of autoimmune diseases have been developed, some of which depend on a single gene mutation, the relevance of these animal models to human autoimmune disease is uncertain in most cases. The mechanisms involved in the trigger of autoimmune disease are largely unknown.
Autoimmunity is caused by the breakdown in self and nonself discrimination. Autoimmunity is not caused by the lost of discrimination of self by innate immunity since non-vertebrates do not develop autoimmune disease, as evolution has only selected germ-line system of discrimination of nonself pathogens. Current dogma infers that autoimmunity is caused by a break in the mechanisms of self and nonself discrimination of the adaptative immunity and that auto-reactive T cells that have not been properly inactivated, play a leading role in this process. The strong association of nearly all (if not all) autoimmune diseases with a particular HLA allele is clear evidence towards a primary involvement of T cells in the trigger of the disease.2 However, although HLA genes are the most relevant, many different genes may be implicated in the susceptibility to an autoimmune disease. Presumably an exogenous trigger that acts on a genetically predisposed individual can provide the first step in the activation of autoreactive T cells. Microbes may be a major environmental factor involved in the development of autoimmune diseases.22 A paradoxical observation has been the strong association of certain microbial organisms with autoimmune diseases. For example, Klebsiella pneumoniae and coxsackievirus B have been correlated with ankylosing spondylitis and diabetes mellitus type 1, respectively. Nevertheless, after an exhaustive search for at least the last five decades, compelling evidence for the pathogens responsible for the autoimmune disease has not been obtained. Similarly, the inciting self antigens which clearly trigger these diseases also remain elusive.
Several mechanisms have been proposed to be involved in the pathogenesis of autoimmune diseases, such as molecular mimicry, exposure of hidden antigens, T cell and B cell dysfunction, loss of suppressor function, polyclonal B cell activation by superantigens, epitope spreading and epitope drift. However, the clearest evidence of the origin of any autoimmune disease arose in the context of rheumatic fever, which follows infection with Group A beta-haemolytic streptococci. Rheumatic fever is an inflammatory disease, which typically develops two to three weeks after a streptococcal infection and is believed to be caused by antibodies generated against streptococcus antigens, which cross-react with antigens of the heart valve.15,23 These antibodies cause damage that impairs cardiac function, but the illness is so named because its presentation is similar to rheumatism. Consequently, if there is an exogenous nonself antigen which shares structural similarities with certain self antigens (which mimics the self antigens), the immune response generated against it can also, in theory, bind to the host antigens and amplify the immune response. Infectious agents may mimic host antigens and induce cross-reactive autoimmune responses to epitopes within host proteins which, in susceptible individuals, may tip the balance toward immunological responses versus tolerance and subsequently lead to autoimmune disease. Despite clear evidence that vaccination with mimetic microbial antigens has the potential to activate autoreactive T cells, crucial evidence for triggering of autoimmunity by mimetic sequences in natural pathogens remains lacking, although they may provoke a prolonged inflammatory response when occurring a subject with a susceptible immunological background.
More surprisingly, infections may also protect from autoimmune diseases.24 An interesting inverse relationship exists between infections and autoimmune diseases. In areas where multiple infectious diseases are endemic, autoimmune diseases are quite rarely seen. In contrast, a higher incidence of most immune disorders including autoimmune and allergic diseases, inflammatory bowel diseases and some lymphocyte malignancies has been observed in western countries. These epidemiological and clinical data have supported the hygiene hypothesis which postulates that the fewer infections observed over the last three decades in developed countries is the main cause of the incessant increase in immune disorders.24 Many mechanisms to explain this protection have been proposed including antigenic competition, immune regulation and stimulation of a large variety of regulatory cells (Th2, CD25+, Tr1 and NKT). However, the hygiene hypothesis does not exclude an etiological role for specific pathogens in a given autoimmune disorder, but instead, it adds another layer of complexity to the self and nonself discrimination paradigm. It postulates that self and nonself discrimination not only depends on the infectious agent itself, but also in the complex interplay between hosts and microbes.
Recognition of Cancer Cells by the Host's Immune System
The relationship between cancer and the immune system is complex and has been the subject of much historical controversy. In 1909, Paul Ehrlich predicted that the immune system repressed the growth of carcinomas that would otherwise occur with greater frequency. In 1957, Frank Macfarlane Burnet established the “immune surveillance theory,” which postulated that the immune system recognizes and eliminates transformed cells, and described the extent to which self and nonself discrimination limits immune responsiveness to emerging tumors. Despite subsequent challenges to this hypothesis over the next several decades, recent studies in immunodeficient mice validated the cancer immune surveillance theory. Research clearly demonstrates that both innate and adaptative immunity have been implicated in the immune response to tumors.25–27
How does the innate immune system discriminate cancer cells from their normal counterparts? The immune mechanisms against spontaneous cancer remain to be fully elucidated. Much data on the specific mechanisms of immune surveillance was acquired in experimental animal models using cancers induced by carcinogens. These animal models do not accurately reflect the pathogenesis of human spontaneous cancer, in which mutations are accumulated over decades. Nevertheless, current clinical data and the analysis of immunodeficient mice clearly indicate that innate NK cells and adaptative cytotoxic T cells are the major players in the immune surveillance against cancer. The potential role of other immune cells in the immune surveillance of cancer remains controversial.
Current evidence suggests that the innate immunity represents the first line of defense against tumor development. Innate immunity does not recognize tumor antigens, but it recognizes changes in cells caused by transformation, such as the lack of expression of self molecules or the induction of stress-inducible self molecules, in a similar way that NK cells recognize some infected cells. NK cells were named “natural killers” because of the initial notion that they are able to kill transformed cells that lacked self markers of MHC class I (missing self hypothesis) (Fig. 1).5 Accumulation of genetic alterations in cancer cells may impair the expression of MHC class I molecules, favoring the evasion of cancer cells from cytotoxic T cells; however, NK cells utilize the inhibitory receptors to differentiate normal self cells from self cells that lack MHC, and eliminate those MHC-negative transformed cells. NK cells also express activating receptors, such as NKG2D, which may directly recognize stressinducible ligands in cancer cells (induced self hypothesis). The human NKG2D ligands, MICA, MICB and ULBPs are not expressed by most normal cells, but are expressed in response to cellular stress and are expressed in a high proportion of epithelial tumors and haematological malignancies.7–12 NKG2D ligand expression is induced by carcinogens and genotoxic stress, and tumor cells expressing these proteins are readily eliminated by NK and CD8 T cells.6,28,29 NKG2D-deficient mice are defective in tumor immune surveillance. Similarly, mice lacking γδT cells, which normally express NKG2D, are highly susceptible to epithelial tumors. NK cells could kill these skin carcinoma cells by a NKG2D dependent mechanism.30,31 Thus, NKG2D system is able to recognize stress-inducible self molecules expressed in cancer. Together, these data clearly suggest a key role of NKG2D in the immune surveillance of cancer and highlight the relevance of the recognition of changes in transformed cells as a central point in the innate immune response against cancer.
Adaptative immunity also plays a significant role in the elimination of established tumors and it may be an important target in immunotherapy. Experiments using immune deficient knockout mice indicate that T cells play a leading role in the immune response to tumors.25–27,32 How do T cells eliminate cancer cells? Cancer does not fit neatly into the self and nonself paradigm, because cancer is not a foreign pathogen, but rather arises from self cells. However, cancers are caused by an accumulation of genetic and epigenetic abnormalities which typically affect oncogenes, genes involved in programmed cell death and tumor suppressor genes.33 Mutations of self proteins may be viewed as comparable to nonself proteins from microbes. Given the vast number of genetic alterations associated with transformation, tumor cells may be envisioned to express many neoantigens. Paradoxically, T cells and antibodies from cancer patients largely recognize non mutated self antigens.34 T cells, especially CD8 and CD4 Tαβ cells are able to recognize tumor antigens in the context of MHC class I and class II molecules. Some of these tumor antigens are derived from mutated self proteins, including tumor antigens derived from oncogenic virus, mutated oncogenes, tumor suppressor genes or other mutated genes.35 However, one of the surprises of tumor immunology was the finding that much of the adaptive immune response to tumor cells was directed against non mutated self antigens rather than against mutated self proteins that differentiated them from normal sequence.6,28,36 Indeed, some of the tumor antigens turned out to be self proteins that were expressed in normal tissues, as well as in their tumor counterparts. These targeted self antigens included normal proteins that are aberrantly expressed or overexpressed in cancer cells, differentiation antigens and cell type-specific oncofetal antigens. Thus, recognition of cancer cells is not mainly based in self and nonself discrimination, but rather based on the threshold of T cell activation.36
Nevertheless, in spite of compelling evidence of the role of the immune system in the surveillance of cancer, a strong immune response against tumor antigens in established tumors is lacking. Many reasons are proposed for this phenomena. First, many antigenic changes created by individual mutations can be rather subtle changes to the structure and do not induce a strong immune response. Additionally, only self-reactive T cells that have TCRs with low or intermediate avidity for self antigens survive thymus maturation and regulation in the periphery. Thus, tumor antigen-specific T cells commonly exhibit lower affinity against their antigenic ligands than the affinity of T cells specific for nonself antigens of microbes. The lack of significant T cell response to established tumors is also due to the complex dynamic interaction between cancer and the immune system. There is growing evidence that immune surveillance represents only one dimension of the complex relationship between the immune system and cancer.37 The immune system is able to eliminate some tumors, but the failure of the immune system to eliminate other primary tumors promotes their capability to escape immune recognition and destruction. To explain these findings, the cancer immunoediting hypothesis has been postulated.35,38,39 Cancer immunoediting is a dynamic process composed of three phases: elimination, equilibrium and escape. Tumor cell variants which have survived the elimination phase enter the equilibrium phase. The equilibrium phase has a selection pressure on tumor cells which are genetically unstable and rapidly mutating. Tumor cell variants which have acquired resistance to elimination then enter the escape phase and the tumor cells continue to grow and expand in an uncontrolled manner and may eventually lead to terminal malignancies. Consequently, many of the genetic abnormalities accumulated in established tumors have been selected because they are less immunogenic or even impair the immune response. Likewise, established cancers present numerous mechanisms to impair the anti-cancer T cell response. However, in spite of the lack of a natural strong immune response against established cancer, if T cells with intermediate avidity for tumor antigens can be activated through immunotherapy (i.e., vaccination or other means), then these T cells can reject tumors that present these mutated self antigens.37
The self and nonself discrimination further emphasizes the relationship between tumor immunity and autoimmunity.40 A clinical corollary of this interesting relationship is the relatively common finding of vitiligo in patients with melanoma responding to immunotherapy of their tumor.41 However, fundamental differences between tumor cells and their normal counterparts provide the immune system an opportunity for discriminating tumor cells from self normal cells.
Acknowledgements
This work was supported by the Spanish grants of Fondo de Investigaciones Sanitarias (Institute Carlos III) PS09/00420 and FIS PI08/0566.
Abbreviations
- TCR
T cell receptor
- BCR
B cell receptor
- TLRs
toll like receptors
- MHC
major histocompatibility complex
- NK
natural killer
References
- 1.Murphy KM, Travers P, Walport M. Immunobiology; Seventh Edition. New York and London: Garland Science; 2007. [Google Scholar]
- 2.Janeway CA., Jr The immune system evolved to discriminate infectious nonself from noninfectious self. Immunol Today. 1992;13:11–16. doi: 10.1016/0167-5699(92)90198-G. [DOI] [PubMed] [Google Scholar]
- 3.Cohn M. The common sense of the self-nonself discrimination. Springer Semin Immunopathol. 2005;27:3–17. doi: 10.1007/s00281-005-0199-1. [DOI] [PubMed] [Google Scholar]
- 4.Roach JC, Glusman G, Rowen L, Kaur A, Purcell MK, Smith KD, et al. The evolution of vertebrate Toll-like receptors. Proc Natl Acad Sci USA. 2005;102:9577–9582. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kärre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 6.Diefenbach A, Raulet DH. Strategies for target cell recognition by natural killer cells. Immunol Rev. 2001;181:170–184. doi: 10.1034/j.1600-065x.2001.1810114.x. [DOI] [PubMed] [Google Scholar]
- 7.Cerwenka A, Lanier LL. Ligands for natural killer cell receptors: Redundancy or specificity. Immunol Rev. 2001;181:158–169. doi: 10.1034/j.1600-065x.2001.1810113.x. [DOI] [PubMed] [Google Scholar]
- 8.Gasser S, Raulet DH. Activation and self-tolerance of natural killer cells. Immunol Rev. 2006;214:130–142. doi: 10.1111/j.1600-065X.2006.00460.x. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez S, Groh V, Spies T. Immunobiology of human NKG2D and its ligands. Curr Top Microbiol Immunol. 2006;298:121–138. doi: 10.1007/3-540-27743-9_6. [DOI] [PubMed] [Google Scholar]
- 10.López-Larrea C, Suárez-Alvarez B, López-Soto A, López-Vázquez A, Gonzalez S. The NKG2D receptor: Sensing stressed cells. Trends Mol Med. 2008;14:179–189. doi: 10.1016/j.molmed.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez S, López-Soto A, Suarez-Alvarez B, López-Vazquez A, Lopez-Larrea C. NKG2D ligands: Key targets of the immune response. Trends Immunol. 2008;29:397–403. doi: 10.1016/j.it.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol Rev. 2010;235:267–285. doi: 10.1111/j.0105-2896.2010.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matzinger P. Tolerance, danger and the extended family. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 14.Matzinger P. The danger model: A renewed sense of self. Science. 2002;296:301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 15.Chen GY, Nuñez G. Sterile inflammation: Sensing and reacting to damage. Nat Rev Immunol. 2010;10:826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beutler B. Microbe sensing, positive feedback loops and the pathogenesis of inflammatory diseases. Immunol Rev. 2009;227:248–263. doi: 10.1111/j.1600-065X.2008.00733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Litman GW, Rast JP, Fugmann SD. The origins of vertebrate adaptive immunity. Nat Rev Immunol. 2010;10:543–553. doi: 10.1038/nri2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper MD, Herrin BR. How did our complex immune system evolve? Nat Rev Immunol. 2010;10:2–3. doi: 10.1038/nri2686. [DOI] [PubMed] [Google Scholar]
- 19.Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol. 2009;9:833–844. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- 20.Jiang H, Chess L. How the immune system achieves self-nonself discrimination during adaptive immunity. Adv Immunol. 2009;102:95–133. doi: 10.1016/S0065-2776(09)01202-4. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y, Zheng Z, Jiang Y, Chess L, Jiang H. The specificity of T cell regulation that enables self-nonself discrimination in the periphery. Proc Natl Acad Sci USA. 2009;106:534–539. doi: 10.1073/pnas.0811843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chervonsky AV. Influence of microbial environment on autoimmunity. Nat Immunol. 2009;11:28–35. doi: 10.1038/ni.1801. [DOI] [PubMed] [Google Scholar]
- 23.Kumar V, Abbas AK, Fausto N, Mitchell RN. Robbins Basic Pathology. 8th Ed. Saunders Elsevier; 2007. pp. 403–406. [Google Scholar]
- 24.Strachan DP. Hay fever, hygiene and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Street SE, Trapani JA, Macgregor D, Smyth MJ. Suppression of lymphoma and epithelial malignancies effected by interferon gamma. J Exp Med. 2002;196:129–134. doi: 10.1084/jem.20020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Broek E, Kagi D, Ossendorp F, Toes R, Vamvakas S, Lutz WK, et al. Decreased tumor surveillance in perforin-deficient mice. J Exp Med. 1996;184:1781–1790. doi: 10.1084/jem.184.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smyth Mj, Thia KY, Street SE, MacGregor D, Godfrey DI, Trapani JA. Perforin-mediated cytotoxicity is critical for surveillance of spontaneous lymphoma. J Exp Med. 2000;192:755–760. doi: 10.1084/jem.192.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutherland CL, Rabinovich B, Chalupny NJ, Brawand P, Miller R, Cosman D. ULBPs, human ligands of the NKG2D receptor, stimulate tumor immunity with enhancement by IL-15. Blood. 2006;108:1313–1319. doi: 10.1182/blood-2005-11-011320. [DOI] [PubMed] [Google Scholar]
- 30.Girardi M, Oppenheim DE, Steele CR, Lewis JM, Glusac E, Filler R, et al. Regulation of cutaneous malignancy by gammadelta T cells. Science. 2001;294:605–609. doi: 10.1126/science.1063916. [DOI] [PubMed] [Google Scholar]
- 31.Guerra N, Tan YX, Joncker NT, Choy A, Gallardo F, Xiong N, et al. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28:571–580. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, et al. IFNgamma and lymphocytes prevent primary tumor development and shape tumor immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 33.Kinzler KW, Vogelstein B. The genetic basis of human cancer. revised Ed. New York: McGraw-Hill, Medical Pub Division; 2002. “Introduction”. 2nd, illustrated. [Google Scholar]
- 34.Houghton AN, Guevara-Patiño JA. Immune recognition of self in immunity against cancer. J Clin Invest. 2004;114:468–471. doi: 10.1172/JCI22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: From immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 36.Sharma RA, Browning MJ. Mechanisms of the self/non-self-survey in the defense against cancer: Potential for chemoprevention? Crit Rev Oncol Hematol. 2005;56:5–22. doi: 10.1016/j.critrevonc.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 37.Wu Y, Zheng Z, Jiang Y, Chess L, Jiang H. The specificity of T cell regulation that enables self-nonself discrimination in the periphery. Proc Natl Acad Sci USA. 2009;106:534–539. doi: 10.1073/pnas.0811843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smyth MJ, Dunn GP, Schreiber RD. Cancer immunosurveillance and immunoediting: The roles of immunity in suppressing tumor development and shaping tumor immunogenicity. Adv Immunol. 2006;90:1–50. doi: 10.1016/S0065-2776(06)90001-7. [DOI] [PubMed] [Google Scholar]
- 39.Teng MW, Swann JB, Koebel CM, Schreiber RD, Smyth MJ. Immune-mediated dormancy: An equilibrium with cancer. J Leukoc Biol. 2008;84:988–993. doi: 10.1189/jlb.1107774. [DOI] [PubMed] [Google Scholar]
- 40.Engelhorn ME, Guevara-Patiño JA, Noffz G, Hooper AT, Lou O, Gold JS, et al. Autoimmunity and tumor immunity induced by immune responses to mutations in self. Nat Med. 2006;12:198–206. doi: 10.1038/nm1363. [DOI] [PubMed] [Google Scholar]
- 41.Rosenberg SA, White DE. Vitiligo in patients with melanoma: Normal tissue antigens can be targets for cancer immunotherapy. J Immunother Emphasis Tumor Immunol. 1996;19:81–84. [PubMed] [Google Scholar]