Abstract
We explore the pentapeptide overlapping between human immunodeficiency virus (HIV) proteins and the human proteome. Our intent was to define viral peptides to be used in vaccines effective against different HIV strains, vaccines that are able to overcome the difficulties posed by the tendency of HIV to mutate, and that are also exempt from harmful collateral cross-reactions, as well as being repeatedly administrable to the global population. Analysis of HIV-1 envelope glycoprotein 160 (Env gp160) sequences revealed a set of 15 pentapeptides highly conserved among a number of retroviral sequences, and absent in the human proteome, thus representing unique molecular retroviral signatures. Use of these short viral peptide modules may represent the first concrete step toward the goal of a universal, safe and effective anti-HIV vaccine.
Key words: HIV versus human peptide overlap, unique viral pentapeptides, conserved viral pentapeptides, peptide-based anti-HIV vaccine, universal anti-HIV vaccine
Introduction
Both ongoing, strenuous efforts and discouraging results have characterized the more than two-decade long search for an HIV vaccine.1–5 The main obstacles to effective anti-HIV immunotherapies are: (1) the tendency of human retroviruses to rapidly mutate, resulting in a high amino acid sequence variability6,7 and (2) the concern of inducing collateral autoimmune phenomena through responses cross-reactive with the host proteome. Cardiolipin polyspecific autoreactivity by two broadly neutralizing HIV-1 monoclonal antibodies is just such an example of potential cross-reactivity.8 In point of fact, using sequence-to-sequence peptide matching to analyze the peptide commonality between HIV and human proteins, we found that HIV pentapeptides are widely, repeatedly and intensively represented in human proteins, with only a relatively limited number of viral pentameric fragments (about 10%) being unique to the retroviruses.9–11 The extensive peptide identity pattern between HIV-1 and humans equates to a high risk of cross-reactivity in the course of immune anti-HIV-1 responses, and possibly explains the link between HIV-1 infection and AIDS.11
With the ambitious aim of developing a safe, effective and universal anti-HIV-1 vaccine, the present study explores the HIV-1 Env gp160 primary sequence and describes a viral pentapeptide set highly conserved among HIV sequences from different strains, and not represented in the human proteome. We propose use of such unique viral peptide signatures for designing global anti-HIV-1 vaccines that are both efficacious and exempt from harmful collateral cross-reactions.
Results
In this study we analyzed four HIV-1 Env gp160 primary sequences. Env gp160 seems to contribute to T-cell depletion during HIV-1 infection and allows rapid transcytosis of the virus through CD4 negative cells such as the simple epithelial monolayers of the intestinal, rectal and endocervical epithelial barriers.12 Also, it seems involved in cell-to-cell spreading of HIV-1.13 Therefore, finding vaccines able to specifically target Env gp160 might inhibit HIV-1 infection and the related immunodeficiency. Moreover, HIV-1 Env gp160 was chosen as an experimental model for this study because of its high level of sequence variability.14
The four HIV-1 Env gp160 primary sequences were derived, respectively, from (1) a major HIV-1 lineage isolated in France (X01762, group M, subtype B, isolate BH10); (2 and 3) two minor variants found in Yaounde, the capital city of Cameroon (AJ291719 and AJ291720);15,16 and (4) an infectious molecular clone derived from a Spanish HIV-1 isolate (AJ006287, subtype B).17 In this way, we tried to compare common and rare HIV-1 sequences as well as HIV-1 sequences from different geographical areas.
Pentapeptides were used as scanning probes since modules of five to six amino acids represent minimal determinants involved in B- and T-cell immune recognition.18 As a matter of fact, already in 1939 Landsteiner and van der Scheer demonstrated that “antibodies may be formed which are specific for peptide chains consisting of five amino acid residues”.19 Also, Landsteiner and van der Scheer reported that antisera against the pentapeptide Gly-Gly-Gly-Gly-Leu distinguished between Gly-Gly-Gly-Gly-Leu and Gly-Gly-Leu-Gly-Gly, thus indicating that anti-pentapeptide antibodies show a high degree of specificity. This foundational paper has been followed by a number of reports, all congruent in defining pentapeptides as the minimum chain length for immune recognition.20–31
Definition of unique HIV-1 Env gp160 pentapeptides for non cross-reactive vaccine formulations.
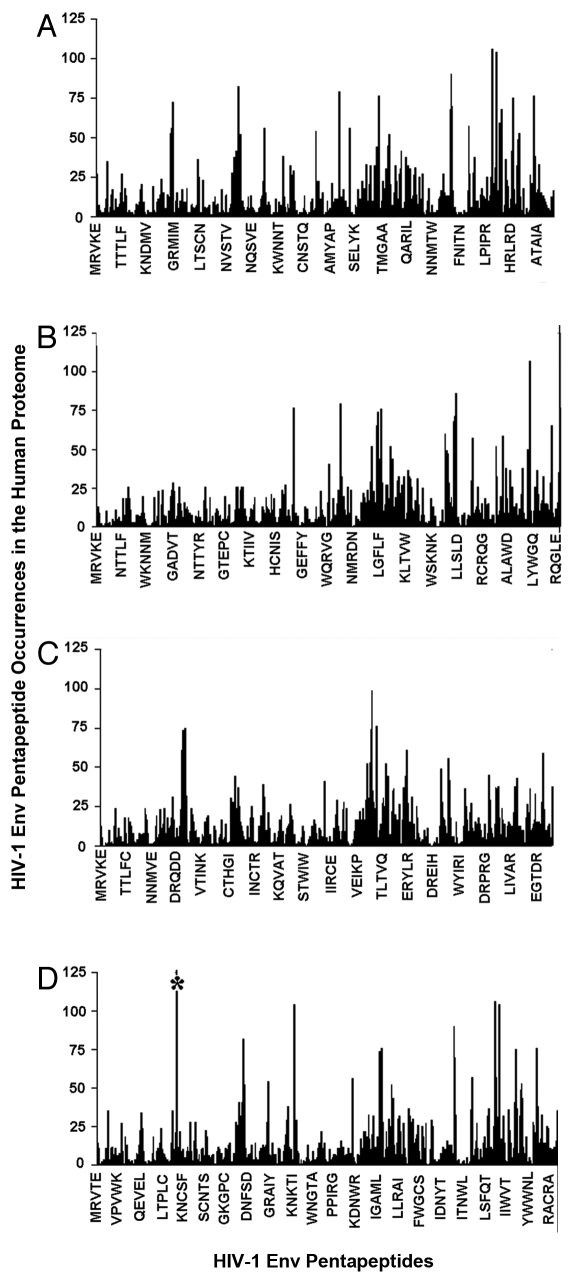
As advocated by Kanduc,32–37 only vaccines based on unique antigenic peptides might guarantee no/low cross-reactivity and the highest specificity. Hence, as a first step in this study, we searched for pentapeptides unique to the viral proteins. Figure 1 illustrates the pentapeptide identity profile of the four HIV-1 Env gp160 sequences versus the human proteome, with the x-axis reporting viral pentapeptides sequentially overlapping by four residues, and the y-axis indicating the numbers of matches of each viral pentapeptide to the human proteome. Figure 1 clearly documents that, firstly, the pentapeptide identity profile to the human proteome has a wave pattern in the four viral proteins, with high similarity sequence areas alternating with those of low similarity. Secondly, almost all of the pentapeptide blocks forming the HIV-1 Env gp160 sequences are also repeatedly present in human proteins; thus, only a limited number of pentamers are unique to the viral proteins. A high degree of peptide matching persists even when hexapeptide motifs were used as probes for sequence identity scanning (data not shown).
Figure 1.
Pentapeptide identity profile of four Env gp160 sequences to the human proteome. (A–D) refer to Env gp160 UniProtKB/Swiss-Prot accession: (A) P03375; (B) Q90DZ7; (C) Q8UMG1; and (D) O93024. (D) the ast erisk indicates the viral SSSGG pentapeptide, which is represented more than 200 times in the human proteome.
Quantification of the pentapeptide identity platform between the HIV-1 Env gp160 sequences and the human proteome is reported in Table 1. Around 90% of the pentapeptides forming the HIV-1 Env gp160 sequences also occur in the human proteome. Moreover, the shared HIV-1 Env gp160 pentapeptides are dispersed throughout the human proteome. Indeed, the mean number of times each shared 5-mer from the Env gp160 sequences occurs in the whole human proteome is ∼11 (i.e., the number of multiple occurrences divided the number of shared viral pentapeptides). According to Figure 1 and Table 1, a vaccine formulation using an entire HIV-1 Env gp160 as an antigen would produce a risk of cross-reactivity with human proteins amounting to thousands of hits. Only about 10% of viral pentapeptides do not have a match in the human proteome; these represent, therefore, molecular signatures of the retroviral antigens. By being unique to the HIV-1 Env gp160 sequences, these zero similarity pentapeptides constitute peptide sets usable in pharmaceutical vaccine formulations theoretically devoid of any cross-reactivity potential.
Table 1.
Pentapeptide overlapping between four HIV-1 Env gp160 proteins and the human proteome
| Env gp1601 | HIV-12 | Env gp160 pentapeptides: | |||
| Total | Shared with human proteins | Occurrences in human proteins3 | Unique to the viral protein | ||
| P03375 | X01762 | 852 | 768 | 8907 | 84 |
| Q90DZ7 | AJ291719 | 851 | 768 | 8424 | 83 |
| Q8UMG1 | AJ291720 | 845 | 761 | 8229 | 84 |
| O93024 | AJ006287 | 858 | 775 | 8940 | 83 |
Env gp160 given as UniProtKB/Swiss-Prot accession number.
HIV-1 given as GenBank accession number.
Number of total Env gp160 pentapeptide occurrences in the human proteome, including multiple occurrences.
Definition of conserved HIV-1 Env gp160 pentapeptides for worldwide effective vaccine formulations.
HIV-1 is extraordinarily variable,38 and this variability represents a major obstacle to AIDS vaccine development. To solve the diversity problem, country- and isolate-specific vaccines can be considered, but even these solutions appear of doubtful efficacy in light of the extreme polymorphism shown by HIV-1.39 Moreover, they would add a further economic burden to the population to be vaccinated.
With the aim of identifying consensus sequences to be used in globally-effective vaccines, we analyzed HIV-1 Env gp160 unique pentapeptides for conserved sequences. That is, the four HIV-1 Env gp160 sequences were aligned and the common unique viral 5-mers were singled out.
As visualized in Figure 2, alignment of the four HIV-1 Env gp160 sequences reveals a high level of conservation on the whole. Indeed, only 15 pentapeptides are both unique to the virus and possess a conserved sequence. These pentapeptides are: GMLMI, WVTVY, TLFCA, LFCAS, LKPCV, KPCVK, IPIHY, PIHYC, HYCAP, YCAPA, SFNCG, NCGGE, GEFFY, VWGIK and HIPRR.
Figure 2.
Sequence alignment of the four Env gp160 sequences under study. First residue of pentapeptides unique to the viral sequences and not found in human proteins is given red. Pentapeptides unique to the viral sequences and conserved among the four Env gp160 sequences are underlined. Env gp160 sequences refer to UniProtKB/Swiss-Prot accession: (A) P03375; (B) Q90DZ7; (C) Q8UMG1; and (D) O93024.
These 15 unique viral pentapeptides, absent in the human proteome and common to the four Env gp160, were used to scan the PIR database of retroviral proteomes. The purpose was to ascertain whether these pentapeptides were also present in Env gp160s from other HIV strain/group/subtype isolates. An example of the data is reported in Table 2, which describes the HIVs hosting the pentapeptide KPCVK. The HIV lists relative to the remaining 14 pentapeptides are reported in Supplemental Table 3.
Table 2.
HIVs hosting the pentapeptide KPCVK
| Type | Group | Subtype | Isolate | Taxonomy | ID |
| 1 | M | J | SE9173 | 388904 | |
| 1 | M | J | SE9280 | 388905 | |
| 1 | M | F1 | VI850 | 388813 | |
| 1 | M | K | 97ZR-EQTB11 | 388907 | |
| 1 | M | F2 | MP255 | 388815 | |
| 1 | M | F2 | MP257 | 388823 | |
| 1 | N | - | YBF106 | 388819 | |
| 2 | B | - | EHO | 388821 | |
| 2 | B | - | UC1 | 388822 | |
| 1 | M | C | ETH2220 | 388796 | |
| 2 | A | - | KR | 73484 | |
| 1 | M | B | LW123 | 82834 | |
| 1 | M | B | YU-2 | 362651 | |
| 2 | A | - | ST/24.1C#2 | 31681 | |
| 1 | M | B | WMJ1 | 31678 | |
| 1 | M | B | KB-1/ETR | 36375 | |
| 2 | A | - | CAM2 | 11715 | |
| 1 | M | B | OYI | 11699 | |
| 2 | A | - | ST | 11721 | |
| 1 | M | B | JRCSF | 11688 | |
| 1 | M | B | MFA | 11704 | |
| 1 | M | B | PIRSF162 | 11691 | |
| 1 | M | B | PIRSF33 | 11690 | |
| 1 | M | D | NDK | 11695 | |
| 2 | A | - | BEN | 11714 | |
| 2 | A | - | Ghana-1 | 11717 | |
| 2 | A | - | D194 | 11713 | |
| 2 | B | - | D205 | 11716 | |
| 1 | M | U | Z3 | 11680 | |
| 1 | M | B | NY5 | 11698 | |
| 1 | M | B | JH32 | 11694 | |
| 1 | M | B | BRVA | 11693 | |
| 1 | M | D | Z2/CDC-Z34 | 11683 | |
| 2 | A | - | SBLISY | 11718 | |
| 2 | A | - | NIH-Z | 11719 | |
| 1 | M | D | Z84 | 11681 | |
| 1 | M | A | Z321 | 11692 | |
| 1 | M | B | WMJ22 | 11705 | |
| 1 | M | B | CDC-451 | 11687 | |
| 1 | M | B | SC | 11702 | |
| 1 | M | B | MN | 11696 | |
| 1 | M | B | HXB3 | 11707 | |
| 1 | M | A | MAL | 11697 | |
| 1 | M | B | BH8 | 11684 | |
| 1 | M | D | ELI | 11689 | |
| 1 | M | D | Z6 | 11708 | |
| 1 | M | B | RF/HAT3 | 11701 | |
| 1 | M | B | HXB2 | 11706 | |
| 2 | A | - | ROD | 11720 | |
| 1 | M | B | ARV2/PIRSF2 | 11685 | |
| 1 | M | B | BRU/LAI | 11686 | |
| 1 | M | B | BH10 | 11678 | |
| 1 | N | - | YBF30 | 388818 | |
| 1 | M | F1 | 93BR020 | 388814 | |
| 1 | M | H | 90CF056 | 388826 | |
| 1 | M | G | 92NG083 | 388825 | |
| 1 | M | C | 92BR025 | 388812 |
The data in Table 2 and Supplemental Table 3 are striking. Indeed, Table 2 documents that antibodies against a small peptide module formed by only five amino acid residues, i.e., the pentapeptide KPCVK, would have the potential to target 57 HIV isolates from different strains (HIV-1 and HIV-2), groups (M, O and N as well as A and B) and numerous isolates of diverse geographical origins. Also, and most importantly, antibodies specific for such a small peptide module would have no cross-reactivity, i.e., they would not induce autoimmune reactions since this sequence is not represented in the human proteome.
Discussion
The search for an HIV-1 vaccine remains a challenge for science and medicine. A number of anti-HIV-1 vaccines, such as AIDSVAX, AIDSVAX B/B, AIDSVAX B/E and HIV gp120, did not pass the final test.40 Recently, and representing a substantial investment of money and effort, the STEP and Phambili trials were launched. The immunotherapeutic approach of the STEP and Phambili trials was based on a complex replicationdefective adenovirus type 5 (Ad5) MRK gag/pol/nef HIV vaccine, and hopes for its success were high.41 However, the trials failed and were halted at the first interim analysis due to an absence of an even minimally positive effect. Indeed, increased susceptibility to HIV-1 was found in men receiving the vaccine.42 The question—“Is an HIV vaccine possible?”43—thus appears to be legitimate.
The present work proposes a possible answer to this question. Using only four HIV sequences, a major isolate and three minor variants, we identified a minimal determinant—the pentapeptide KPCVK—present in 57 different HIV sequences. That is, antibodies designed to hit the pentapeptide KPCVK would target a large range of retroviral sequences, thus opening the way to a global approach to the prevention and cure of HIV infection. Hence, it is logical to postulate that applying this rationale and using the data illustrated in Supplemental Table 3, a universal anti-HIV weapon may be developed using only a small number of peptides. Likewise, this methodological approach might also be applied to other viral proteins. Of special interest, it has to be noted that many of the 15 pentapeptides absent in the human proteome and shared by numerous HIVs are also present in simian immunodeficiency viruses (SIVs). For example, the pentapeptide KPCVK is found in SIV-wrc Pbt-05GM-X02 (Tax ID: 498715), SIV-mnd 1 (Tax ID: 358183), SIV African green monkey vervet (isolate AGM3) (Tax ID: 11730), SIV 17E-Fr (Tax ID: 160753), SIVmac (Tax ID: 11711). Clearly, utilization of pentapeptides shared by SIVs and HIVs would imply a fast transfer of results from animal models to clinical human trials in vaccine testing and evaluation, with a paramount gain in time and economical investment. In any case, and most importantly, the eventual vaccine preparations would be safe. Being based on peptide sequences not present in the human proteome, these vaccines will probably be free of cross-reactivity,32–37 therefore nullifying the risk of autoimmune collateral events, even following repeated administration. In conclusion, global and safe protection from HIV infection appears to be at hand.
Methods
UniProtKB/Swiss-Prot accession of the analyzed Env gp160 sequences are: P03375, from HIV-1 sequence X01762, BH10 group M subtype B, 11678; Q90DZ7, from AJ291719, a minor variant from Yaounde, the capital city of Cameroon;15,16 Q8UMG1 from AJ291720, a minor variant from Yaounde;15,16 O93024 from HIV-1 AJ006287, subtype B, an infectious molecular clone derived from a Spanish HIV-1 isolate.17 The sequences were downloaded from www.uniprot.org.44 Each viral sequence was dissected into consecutive pentapeptides offset by one amino acid residue. Pentapeptides were then utilized as probes to scan the human and retroviral proteomes for exact matches using the Protein Information Resource perfect match program (pir.georgetown.edu/pirwww/search/peptide.shtml).45 Sequence alignment was performed using the T-COFFEE Software (www.tcoffee.org).46
Authors' Contributions
G.L. and A.S. performed the computational analyses. D.K. proposed the original idea, interpreted the data, developed the research project and wrote the manuscript. All authors discussed the results, and revised and commented on the manuscript.
Supplementary Material
References
- 1.Boasso A, Shearer GM, Clerici M. The hunt for an HIV vaccine: Time to rethink recent failures. Lancet. 2008;371:1897–1898. doi: 10.1016/S0140-6736(08)60812-0. [DOI] [PubMed] [Google Scholar]
- 2.Ledford H. HIV vaccine developers battle on, despite high-profile failures. Nat Biotechnol. 2008;26:591–592. doi: 10.1038/nbt0608-591. [DOI] [PubMed] [Google Scholar]
- 3.Bradac J, Dieffenbach CW. HIV vaccine development: Lessons from the past, informing the future. IDrugs. 2009;12:435–439. [PubMed] [Google Scholar]
- 4.Thomas C. Roadblocks in HIV research: Five questions. Nat Med. 2009;15:855–859. doi: 10.1038/nm0809-855. [DOI] [PubMed] [Google Scholar]
- 5.Wijesundara DK, Jackson RJ, Ramshaw IA, Ranasinghe C. Human immunodeficiency virus-1 vaccine design: where do we go now? Immunol Cell Biol. 2010;89:367–374. doi: 10.1038/icb.2010.118. [DOI] [PubMed] [Google Scholar]
- 6.Stingl G. Anti-retroviral therapy in HIV-1-infected persons: Concepts and strategies. Curr Probl Dermatol. 1989;18:250–268. doi: 10.1159/000416863. [DOI] [PubMed] [Google Scholar]
- 7.Trabaud MA, Cotte L, Labernardière JL, Lebel-Binay S, Icard V, Tardy JC, et al. Variants with different mutation patterns persist in the quasispecies of enfuvirtide-resistant HIV-1 population during and after treatment in vivo. J Acquir Immune Defic Syndr. 2007;46:134–144. doi: 10.1097/QAI.0b013e3181354710. [DOI] [PubMed] [Google Scholar]
- 8.Haynes BF, Fleming J, St. Clair EW, Katinger H, Stiegler G, Kunert R, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 9.Kanduc D, Stufano A, Lucchese G, Kusalik A. Massive peptide sharing between viral and human proteomes. Peptides. 2008;29:1755–1766. doi: 10.1016/j.peptides.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanduc D. Pentapeptide overlapping between human immunodeficiency viruses and Homo sapiens proteomes is higher than 90% Retrovirology. 2009;6:44. [Google Scholar]
- 11.Lucchese G, Stufano A, Calabro' M, Kanduc D. Charting the peptide crossreactome between HIV-1 and the human proteome. Front Biosci. 2011;E3:1385–1400. doi: 10.2741/e341. [DOI] [PubMed] [Google Scholar]
- 12.Crise B, Buonocore L, Rose JK. CD4 is retained in the endoplasmic reticulum by the human immunodeficiency virus type 1 glycoprotein precursor. J Virol. 1990;64:5585–5593. doi: 10.1128/jvi.64.11.5585-5593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alfsen A, Iniguez P, Bouguyon E, Bomsel M. Secretory IgA specific for a conserved epitope on gp41 envelope glycoprotein inhibits epithelial transcytosis of HIV-1. J Immunol. 2001;166:6257–6265. doi: 10.4049/jimmunol.166.10.6257. [DOI] [PubMed] [Google Scholar]
- 14.Vanden Haesevelde M, Decourt JL, De Leys RJ, Vanderborght B, van der Groen G, van Heuverswijn H, et al. Genomic cloning and complete sequence analysis of a highly divergent African human immunodeficiency virus isolate. J Virol. 1994;68:1586–1596. doi: 10.1128/jvi.68.3.1586-1596.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montavon C, Vergne L, Bourgeois A, Mpoudi-Ngole E, Malonga-Mouellet G, Butel C, et al. Identification of a new circulating recombinant form of HIV type 1, CRF11-cpx, involving subtypes A, G, J and CRF01-AE, in Central Africa. AIDS Res Hum Retroviruses. 2002;18:231–236. doi: 10.1089/08892220252781301. [DOI] [PubMed] [Google Scholar]
- 16.Vergne L, Bourgeois A, Mpoudi-Ngole E, Mougnutou R, Mbuagbaw J, Liegeois F, et al. Biological and genetic characteristics of HIV infections in Cameroon reveals dual group M and O infections and a correlation between SI-inducing phenotype of the predominant CRF02_AG variant and disease stage. Virology. 2003;310:254–266. doi: 10.1016/s0042-6822(03)00167-3. [DOI] [PubMed] [Google Scholar]
- 17.Olivares I, Casado Herrero C, Iglesias-Ussel MD, Dietrich U, Lopez Galindez C. Complete sequence of an infectious molecular clone derived from a Spanish HIV type I isolate. AIDS Res Hum Retroviruses. 1998;14:1649–1651. doi: 10.1089/aid.1998.14.1649. [DOI] [PubMed] [Google Scholar]
- 18.Lucchese G, Stufano A, Trost B, Kusalik A, Kanduc D. Peptidology: short amino acid modules in cell biology and immunology. Amino Acids. 2007;33:703–707. doi: 10.1007/s00726-006-0458-z. [DOI] [PubMed] [Google Scholar]
- 19.Landsteiner K, van der Scheer J. On the serological specificity of peptides. III. J Exp Med. 1939;69:705–719. doi: 10.1084/jem.69.5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith SC, McAdam WJ, Kemp BE, Morgan FJ, Cotton RG. A monoclonal antibody to the phosphorylated form of phenylalanine hydroxylase: Definition of the phosphopeptide epitope. Biochem J. 1987;244:625–631. doi: 10.1042/bj2440625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woods JP, Spinola SM, Strobel SM, Cannon JG. Conserved lipoprotein H.8 of pathogenic Neisseria consists entirely of pentapeptide repeats. Mol Microbiol. 1989;3:43–48. doi: 10.1111/j.1365-2958.1989.tb00102.x. [DOI] [PubMed] [Google Scholar]
- 22.Reddehase MJ, Rothbard JB, Koszinowski UH. A pentapeptide as minimal antigenic determinant for MHC class I-restricted T lymphocytes. Nature. 1989;337:651–653. doi: 10.1038/337651a0. [DOI] [PubMed] [Google Scholar]
- 23.Okada Y, Nakayama S, Iguchi S, Kikuchi Y, Irie M, Sawada J, et al. Amino acids and peptides. XXXIII. Synthesis of N-terminal epitope peptides of mammalian metallothioneins (MTs) Chem Pharm Bull. 1992;40:1029–1032. doi: 10.1248/cpb.40.1029. [DOI] [PubMed] [Google Scholar]
- 24.Sheets RL, Pandey R, Klement V, Grant CK, Roy-Burman P. Biologically selected recombinants between feline leukemia virus (FeLV) subgroup A and an endogenous FeLV element. Virology. 1992;190:849–855. doi: 10.1016/0042-6822(92)90924-e. [DOI] [PubMed] [Google Scholar]
- 25.Papadouli I, Sakarellos C, Tzartos SJ. High-resolution epitope mapping and fine antigenic characterization of the main immunogenic region of the acetylcholine receptor: Improving the binding activity of synthetic analogues of the region. Eur J Biochem. 1993;211:227–234. doi: 10.1111/j.1432-1033.1993.tb19890.x. [DOI] [PubMed] [Google Scholar]
- 26.Endo M, Nunomura W, Takakuwa Y, Hatakeyama M, Higashi T. A novel epitope (pentapeptide) in the human hemoglobin beta chain. Hemoglobin. 1998;22:321–331. doi: 10.3109/03630269809071527. [DOI] [PubMed] [Google Scholar]
- 27.Tiwari R, Geliebter J, Lucchese A, Mittelman A, Kanduc D. Computational peptide dissection of Melan-a/MART-1 oncoprotein antigenicity. Peptides. 2004;25:1865–1871. doi: 10.1016/j.peptides.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Kanduc D, Lucchese A, Mittelman A. Non-redundant peptidomes from DAPs: Towards “the vaccine”? Autoimmun Rev. 2007;6:290–294. doi: 10.1016/j.autrev.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Stufano A, Capone G, Pesetti B, Polimeno L, Kanduc D. Clustering of rare peptide segments in the HCV immunome. Self/Nonself. 2010;1:154–162. doi: 10.4161/self.1.2.11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stufano A, Kanduc D. Proteome-based epitopic peptide scanning along PSA. Exp Mol Pathol. 2009;86:36–40. doi: 10.1016/j.yexmp.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Lucchese G, Stufano A, Kanduc D. Proposing lowsimilarity peptide vaccines against Mycobacterium tuberculosis. J Biomed Biotechnol. 2010:832341. doi: 10.1155/2010/832341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanduc D. Immunogenicity in peptide-immunotherapy. From self/nonself to similar/dissimilar sequences. Adv Exp Med Biol. 2008;640:198–207. doi: 10.1007/978-0-387-09789-3_15. [DOI] [PubMed] [Google Scholar]
- 33.Kanduc D. Correlating low-similarity peptide sequences and allergenic epitopes. Curr Pharm Des. 2008;14:289–295. doi: 10.2174/138161208783413257. [DOI] [PubMed] [Google Scholar]
- 34.Kanduc D. Epitopic peptides with low similarity to the host proteome: Towards biological therapies without side effects. Expert Opin Biol Ther. 2009;9:45–53. doi: 10.1517/14712590802614041. [DOI] [PubMed] [Google Scholar]
- 35.Kanduc D. “Self-nonself” peptides in the design of vaccines. Curr Pharm Des. 2009;15:3283–3289. doi: 10.2174/138161209789105135. [DOI] [PubMed] [Google Scholar]
- 36.Kanduc D. Protein information content resides in rare peptide segments. Peptides. 2010;31:983–988. doi: 10.1016/j.peptides.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Kanduc D. The self/nonself issue: A confrontation between proteomes. SelfNonself. 2010;1:255–258. doi: 10.4161/self.1.3.11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McBurney SP, Ross TM. Viral sequence diversity: challenges for AIDS vaccine designs. Expert Rev Vaccines. 2008;7:1405–1417. doi: 10.1586/14760584.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura KJ, Gach JS, Jones L, Semrau K, Walter J, Bibollet-Ruche F, et al. 4E10-resistant HIV-1 isolated from four subjects with rare membrane-proximal external region polymorphisms. PLoS One. 2010;5:9786. doi: 10.1371/journal.pone.0009786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adis International Ltd., author HIV gp120 vaccine—VaxGen: AIDSVAX, AIDSVAX B/B, AIDSVAX B/E, HIV gp120 vaccine—Genentech, HIV gp120 vaccine AIDSVAX—VaxGen, HIV vaccine AIDSVAX—VaxGen. Drugs R D. 2003;4:249–253. doi: 10.2165/00126839-200304040-00007. [DOI] [PubMed] [Google Scholar]
- 41.Casimiro DR, Chen L, Fu TM, Evans RK, Caulfield MJ, Davies ME, et al. Comparative immunogenicity in rhesus monkeys of DNA plasmid, recombinant vaccinia virus and replication-defective adenovirus vectors expressing a human immunodeficiency virus type 1 gag gene. J Virol. 2003;77:6305–6313. doi: 10.1128/JVI.77.11.6305-6313.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gray G, Buchbinder S, Duerr A. Overview of STEP and Phambili trial results: two phase IIb test-of-concept studies investigating the efficacy of MRK adenovirus type 5 gag/pol/nef subtype B HIV vaccine. Curr Opin HIV AIDS. 2010;5:357–361. doi: 10.1097/COH.0b013e32833d2d2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson NA, Watkins DI. Is an HIV vaccine possible? Braz J Infect Dis. 2009;13:304–310. doi: 10.1590/s1413-86702009000400013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.UniProt consortium: The universal protein resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:142–148. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barker WC, Garavelli JS, Huang H, McGarvey PB, Orcutt BC, Srinivasarao GY, et al. The protein information resource (PIR) Nucleic Acids Res. 2000;28:41–44. doi: 10.1093/nar/28.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poirot O, O'Toole E, Notredame C. Tcoffee@igs: A web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003;31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.