Abstract
In human membrane proteins, intrinsically disordered regions, the regions that lack a well-defined three-dimensional structure under physiological conditions, preferentially occur in the cytoplasmic tails. Many of these proteins represent cell receptors that function by recognizing their cognate ligand outside the cell and translating this binding information into an intracellular activation signal. Based on location of recognition and signaling (effector) domains, functionally diverse and unrelated cell receptors can be classified into two main families: those in which binding and signaling domains are located on the same protein chain, the so-called single-chain receptors (SRs), and those in which these domains are intriguingly located on separate subunits, the so-called multichain receptors (MRs). Recognition domains of both SRs and MRs are known to be well ordered. In contrast, while cytoplasmic signaling domains of SRs are well-structured as well, those of MRs are intrinsically disordered. Despite important role of receptor signaling in health and disease, extensive comparative structural analysis of receptor signaling domains has not been carried out as of yet. In this study, using a variety of prediction algorithms, we show that protein disorder is a characteristic and distinctive feature of receptors with recognition and signaling functions distributed between separate protein chains. We also reveal that disorder distribution patterns are rather similar within SR subclasses suggesting potential functional explanations. Why did nature select protein disorder to provide intracellular signaling for MRs? Is there any correlation between disorder profiles of signaling domains and receptor function? These and other questions are addressed in this article.
Key words: intrinsically disordered proteins, immune signaling, protein disorder, single-chain receptors, multichain immune recognition receptors, MIRR, T cell receptor, B cell receptor, RTK, receptor tyrosine kinases
Introduction
In order to receive and respond to signals from their environment, cells maintain a diversity of receptors on their surface that respond specifically to individual stimuli. Cell surface receptors are integral membrane proteins and, as such, consist of three basic domains: extracellular ligand-recognition/binding domains, transmembrane domains and cytoplasmic signaling (or effector) domains. These receptors mediate transmembrane signal transduction, a complex fundamental process by which extracellular binding information is translated into intracellular signaling sequences and further into physiological cell response. This process plays an important role in health and disease and is central to therapeutic control of multiple diseases.1,2 It is therefore fundamentally and clinically important to structurally understand how signal transduction occurs.
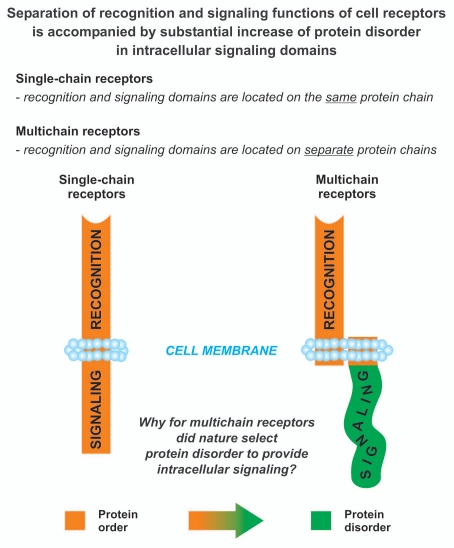
Intrinsically disordered proteins (IDPs) and regions (IDRs), i.e., entire proteins and localized protein regions that lack a well-defined ordered structure under physiological conditions,3 are critically involved in receptor signaling.4,5 Intrinsic disorder serves as the native and functional state for many signaling proteins4 with phosphorylation, one of the critical and obligatory events in cell signaling, occurring predominantly within IDRs.5 Importantly, long IDRs preferentially reside on the cytoplasmic side of many human transmembrane proteins including cell membrane receptors.6–8 Based on location of binding and signaling (effector) domains, functionally diverse and unrelated cell receptors can be structurally classified into two main families: those in which binding and signaling domains are located on the same protein chain, the so-called single-chain receptors (SRs), and those in which binding and signaling domains are intriguingly located on separate subunits, the so-called multichain receptors (MRs).9–13 Because of the fact that most multichain receptors are immune receptors, they are all commonly referred to as multichain immune recognition receptors (MIRRs).2,9,10 Nevertheless, members of this family are not necessarily immune-related (an example is the major collagen receptor on platelets, glycoprotein VI, GPVI).
Recently, using a variety of biophysical techniques, we found that the cytoplasmic domains of MIRR signaling subunits represent a novel class of IDPs.14–16 Our classification of these domains as a novel class of IDPs17,18 is based on the following logics: (a) they all contain an immunoreceptor tyrosine-based activation motif (ITAM), tyrosines of which are phosphorylated upon receptor triggering (structural criteria),9 (b) they all function through similar molecular mechanisms (mechanistic criteria),9,11,12,19 (c) they all represent functionally important regions of more than 25 receptors that belong to one family of receptors (functional criteria),9,10,12,13 (d) they all are characterized by unusual biophysical phenomenon for IDPs—ability to homodimerize,14–16,18,20 and what is especially important, they all homodimerize without disorder-to-order structural transition14–16,18 (distinctive criteria). Protein disorder of MIRR cytoplasmic signaling domains is in strong contrast to protein order of SR cytoplasmic domains. Recently, we have highlighted these observations and suggested signaling-related functional connections between protein order, disorder and oligomericity.17,21 However, extensive comparative structural analysis of receptor signaling domains has not been carried out as of yet.
In this study, using a variety of prediction algorithms, we analyze signaling (effector) domains of functionally diverse single- and multichain receptors that are expressed on various cells (Table 1). We show that in contrast to disordered cytoplasmic signaling domains of MIRRs, those of SRs are ordered. This clearly demonstrates that separation of recognition and signaling functions of cell receptors is accompanied by substantial increase of protein disorder in intracellular signaling domains. We also reveal that order-disorder distribution patterns are rather similar within SR subclasses suggesting potential functional explanations. The biological significance of these findings is discussed.
Table 1.
Cytoplasmic signaling (effector) domains of various cell receptors studied in this work
| Receptor/signaling subunita | Functional roleb | Swiss-Prot accession numberc | alpha-MoRF predictiond |
| Single-chain receptors | |||
| Receptor tyrosine kinases | |||
| ErbB-1 | Receptor for EGF but also for other members of the EGF family, as TGFalpha, amphiregulin, betacellulin, heparin-binding EGF-like growth factor, GP30 and vaccinia virus growth factor. Control of cell growth and differentiation. |
P00533 (669–210) |
1–18 385–402 414–431 |
| ErbB-2 | Essential component of a neuregulin-receptor complex, although neuregulins do not interact with it alone. GP30 is a potential ligand for this receptor. Binds to the MT-CO2 promoter and activates its transcription. |
P04626 (676–1255) |
370–387 409–426 500–517 |
| ErbB-3 | Binds and is activated by neuregulins and NTAK. |
P21860 (665–1342) |
1–18 555–572 595–612 |
| ErbB-4 | Specifically binds and is activated by neuregulins, heparin-binding EGF-like growth factor, betacellulin and NTAK. Interaction with these factors induces cell differentiation. Not activated by EGF, TGF-A and amphiregulin. |
Q15303 (676–1308) |
351–368 424–441 612–629 |
| PDGFR-A | Receptor that binds both PDGFA and PDGFB and has a tyrosine-protein kinase activity. |
P16234 (550–1089) |
523–540 |
| PDGFR-B | Receptor that binds specifically to PDGFB and PDGFD and has a tyrosine-protein kinase activity. Phosphorylates Tyr residues at the C-terminus of PTPN11 creating a binding site for the SH2 domain of GRB2. |
P09619 (557–1106) |
451–468 |
| FGFR-1 | Receptor for basic fibroblast growth factor. Receptor for FGF23 in the presence of KL. |
P11362 (398–822) |
10–27 |
| FGFR-2 | Receptor for acidic and basic fibroblast growth factors. |
P21802 (399–821) |
- |
| FGFR-3 | Receptor for acidic and basic fibroblast growth factors. Preferentially binds FGF1. |
P22607 (397–806) |
15–32 |
| FGFR-4 | Receptor for acidic fibroblast growth factor. Does not bind to basic fibroblast growth factor. Binds FGF19. |
P22455 (391–802) |
16–33 |
| VEGFR-1 | Receptor for VEGF, VEGFB and PGF. The VEGF-kinase ligand/receptor signaling system plays a key role in vascular development and regulation of vascular permeability. |
P17948 (781–1338) |
522–539 |
| VEGFR-2 | Receptor for VEGF or VEGFC. In case of HIV-1 infection, the interaction with extracellular viral Tat protein seems to enhance angiogenesis in Kaposi's sarcoma lesions. |
P35968 (790–1356) |
167–184 485–502 525–542 |
| VEGFR-3 | Receptor for VEGFC. |
P35916 (798–1298) |
151–168 183–200 455–472 |
| HGFR | Receptor for hepatocyte growth factor and scatter factor. Functions in cell proliferation, scattering, morphogenesis and survival. |
P08581 (956–1390) |
10–27 |
| Trk-A | Required for high-affinity binding to NGF, neurotrophin-3 and neurotrophin-4/5 but not BDNF. Known substrates for the TRK receptors are SHC1, PI3-kinase, and PLCgamma-1. Has a crucial role in the development and function of the nociceptive reception system as well as establishment of thermal regulation via sweating. Activates ERK1 by either SHC1- or PLC-gamma-1-dependent signaling pathway. |
P04629 (440–796) |
- |
| Trk-B | Receptor for BDNF, neurotrophin-3 and neurotrophin-4/5 but not NGF. Involved in the development and/or maintenance of the nervous system. Known substrates for the TRK receptors are SHC1, PI-3 kinase and PLCgamma-1. |
Q16620 (455–822) |
- |
| Trk-C | Receptor for NT-3. Known substrates for the Trk receptors are SHC1, PI-3 kinase and PLCG1. The different isoforms do not have identical signaling properties. |
Q16288 (454–839) |
- |
| LTK | The exact function of this protein is not known. |
P29376 (450–864) |
1–18 372–389 398–415 |
| Tie-1 | The exact function of this protein is not known. |
P35590 (785–1138) |
- |
| Tie-2 | Receptor for angiopoietin 1. It may constitute the earliest mammalian endothelial cell lineage marker. Probably regulates endothelial cell proliferation, differentiation and guides the proper patterning of endothelial cells during blood vessel formation. |
Q02763 (771–1124) |
- |
| ROR1 | Functional role is not yet clear. |
Q01973 (428–937) |
1–18 27–44 315–332 376–393 |
| ROR2 | May be involved in the early formation of the chondrocytes. It seems to be required for cartilage and growth plate development. Phosphorylates YWHAB, leading to induction of osteogenesis and bone formation. |
Q01974 (425–943) |
1–18 30–47 366–383 401–418 502–519 |
| DDR1 | Interacts (via PPxY motif) with WWC1 (via WW domains) in a collagen-regulated manner. Forms a tripartite complex with WWC1 and PRKCZ, but predominantly in the absence of collagen. May be involved in cell-cell interactions and recognition. |
Q08345 (444–913) |
1–18 59–76 92–109 126–143 |
| DDR2 | Receptor for fibrillar collagen mediates fibroblast migration and proliferation. Contributes to cutaneous wound healing. |
Q16832 (422–855) |
1–18 47–64 |
| RYK | Potential growth factor receptor protein tyrosine kinase. |
P34925 (253–604) |
1–18 |
| MuSK | Key mediator of agrin's action and is involved in NMJ organization. |
O15146 (517–869) |
11–28 |
| INSR | Binds insulin. Mediates the metabolic functions of insulin. Binding to insulin stimulates association of the receptor with downstream mediators including IRS1 and PI3K. Can activate PI3K either directly by binding to the p85 regulatory subunit, or indirectly via IRS1. |
P06213 (980–1382) |
362–379 |
| C-Ret receptor | Probable receptor with tyrosine-protein kinase activity. Important for development. |
P07949 (658–1114) |
356–373 |
| EPHA-1 | Receptor for members of the ephrin-A family. Binds with a low affinity to ephrin-A1. |
P21709 (569–976) |
- |
| EPHA-2 | Receptor for members of the ephrin-A family. Binds to ephrin-A1, -A3, -A4 and -A5. Plays an important role in angiogenesis and tumor neovascularization. |
P29317 (559–976) |
- |
| EPHA-3 | Receptor for members of the ephrin-A family. Binds to ephrin-A2, -A3, -A4 and -A5. Could play a role in lymphoid function. |
P29320 (566–983) |
- |
| EPHA-4 | Receptor for members of the ephrin-A family. Binds to ephrin-A1, -A4 and -A5. May play a role in a signal transduction process involved in hindbrain pattern formation. |
P54764 (570–986) |
- |
| EPHA-5 | Receptor for members of the ephrin-A family. Binds to ephrin-A1, -A2, -A3, -A4 and -A5. |
P54756 (595–1037) |
- |
| EPHA-6 | Receptor for members of the ephrin-A family. |
Q9UF33 (571–1035) |
- |
| EPHA-7 | Receptor for members of the ephrin-A family. Binds to ephrin-A1, -A2, -A3, -A4 and -A5. |
Q15375 (578–998) |
- |
| EPHA-8 | Receptor for members of the ephrin-A family. Interacts with FYN. Interacts with ANKS1B. |
P29322 (564–1005) |
19–36 |
| EPHA-10 | Receptor for members of the ephrin-A family. Binds to ephrin-A3, -A4 and -A5. |
Q5JZY3 (587–1008) |
- |
| EPHB-1 | Receptor for members of the ephrin-B family. Binds to ephrin-B1, -B2 and -B3. May be involved in cell-cell interactions in the nervous system. |
P54762 (564–984) |
- |
| EPHB-2 | Receptor for members of the ephrin-B family. Acts as a tumor suppressor. |
P29323 (565–1055) |
474–491 |
| EPHB-3 | Receptor for members of the ephrin-B family. Binds to ephrin-B1 and -B2. |
P54753 (581–998) |
- |
| EPHB-4 | Receptor for members of the ephrin-B family. Binds to ephrin-B2. May have a role in events mediating differentiation and development. |
P54760 (561–987) |
- |
| EPHB-6 | Kinase-defective receptor for members of the ephrin-B family. Binds to ephrin-B1 and ephrin-B2. Modulates cell adhesion and migration. |
O15197 (601–1006) |
- |
| TNF receptors | |||
| TNF-R1 | Receptor for TNFSF2/TNFalpha and homotrimeric TNFSF1/lymphotoxin-alpha. Contributes to the induction of non-cytocidal TNF effects including anti-viral state and activation of the acid sphingomyelinase. |
P19438 (235–455) |
188–205 |
| TNF-R2 | Receptor with high affinity for TNFSF2/TNFalpha and approximately 5-fold lower affinity for homotrimeric TNFSF1/lymphotoxin-alpha. Mediates most of the metabolic effects of TNFalpha. |
P20333 (288–461) |
1–18 91–108 |
| TNF-R3 (LTBR) | Receptor for the heterotrimeric lymphotoxin containing LTA and LTB, and for TNFS14/LIGHT. Promotes apoptosis via TRAF3 and TRAF5. May play a role in the development of lymphoid organs. |
P36941 (249–435) |
30–47 158–175 |
| TNF-R4 | Receptor for TNFSF4/OX40L/GP34. Interacts with TRAF2, TRAF3 and TRAF5. |
P43489 (236–277) |
1–18 25–42 |
| TNF-R5 | Receptor for TNFSF5/CD40LG. Interacts with TRAF1, TRAF2, TRAF3, TRAF5 and TRAF6 |
P25942 (216–277) |
5–22 |
| TNF-R6 (FAS receptor) | Receptor for TNFSF6/FASLG. FAS-mediated apoptosis may have a role in the induction of peripheral tolerance, in the antigen-stimulated suicide of mature T cells, or both. |
P25445 (191–335) |
1–18 |
| TNF-R7 | Receptor for CD70/CD27L. Interacts with SIVA1 and TRAF2. May play a role in survival of activated T cells. May play a role in apoptosis. |
P26842 (213–260) |
1–18 |
| TNF-R8 | Receptor for TNFSF8/CD30L. Interacts with TRAF1, TRAF2, TRAF3 and TRAF5. May play a role in the regulation of cellular growth and transformation of activated lymphoblasts. Regulates gene expression. |
P28908 (408–595) |
6–23 104–121 |
| TNF-R9 | Receptor for TNFSF14/4-1BBL. Interacts with TRAF1, TRAF2 and TRAF3. Interacts with LRR-repeat protein 1/LRR-1. Possibly active during T cell activation. |
Q07011 (214–255) |
6–23 |
| TNF-R10A | Receptor for the cytotoxic ligand TNFSF10/TRAIL. Can interact with TRADD and RIPK1. Interacts with ARAP1. Promotes the activation of NFkappaB. |
O00220 (263–468) |
- |
| TNF-R10B | Receptor for the cytotoxic ligand TNFSF10/TRAIL. Can interact with TRADD and RIPK1. Promotes the activation of NFkappaB. |
O14763 (232–440) |
- |
| TNF-R10D | Receptor for the cytotoxic ligand TRAIL. It is not capable of inducing apoptosis but protects against TRAIL-mediated apoptosis. Reports are contradictory with regards to its ability to induce the NFkappaB pathway. |
Q9UBN6 (233–386) |
29–46 94–111 127–144 |
| TNF-R11A (RANK) | Receptor for TNFSF11/RANKL/TRANCE/OPGL. Interacts with TRAF1, TRAF2, TRAF3, TRAF5 and TRAF6. Interacts (via cytoplasmic domain) with GAB2. Essential for RANKL-mediated osteoclastogenesis. |
Q9Y6Q6 (234–616) |
256–273 301–318 341–358 |
| TNF-R12A | Receptor for TNFSF12/TWEAK. Associates with TRAF1 and TRAF2, and probably also with TRAF3. Weak inducer of apoptosis in some cell types. Promotes angiogenesis and the proliferation of endothelial cells. |
Q9NP84 (102–129) |
- |
| TNF-R13B | Receptor for TNFSF13/APRIL and TNFSF13B/TALL1/BAFF/BLYS that binds both ligands with similar high affinity. Binds TRAF2, TRAF5 and TRAF6. Binds the NH2-terminal domain of CAMLG with its C-terminus. Involved in the stimulation of B- and T cell function and the regulation of humoral immunity. |
O14836 (187–293) |
1–18 44–61 |
| TNF-R13C | B-cell receptor specific for TNFSF13B/TALL1/BAFF/BLyS. Promotes the survival of mature B-cells and the B-cell response. |
Q96RJ3 (100–184) |
1–18 68–85 |
| TNF-R14 | Receptor for BTLA. Receptor for TNFSF14/LIGHT and homotrimeric TNFSF1/lymphotoxin-alpha. Involved in lymphocyte activation. Interacts with TRAF2, TRAF3 and TRAF5. Interacts with HHV-1 and HHV-2 envelope glycoprotein D. Functions as an entry receptor for these viruses. Enhances the entry of several wild-type HSV strains of both serotypes into CHO cells and mediates HSV entry into activated human T cells. |
Q92956 (224–283) |
- |
| TNF-R16 (NGF receptor) | Low affinity receptor which can bind to NGF, BDNF, NT-3 and NT-4. Interacts with p75NTR-associated cell death executor. Interacts with TRAF2, TRAF4, TRAF6, PTPN13 and RANBP9. Interacts with LINGO1. Can mediate cell survival as well as cell death of neural cells. |
P08138 (273–427) |
1–18 |
| TNF-R17 | Receptor for TNFSF13B/BLyS/BAFF and TNFSF13/APRIL. Associates with TRAF1, TRAF2, TRAF3, TRAF5 and TRAF6. Promotes B-cell survival and plays a role in the regulation of humoral immunity. Activates NFkappaB and JNK. |
Q02223 (78–184) |
62–79 |
| TNF-R18 | Receptor for TNFSF18. Binds to TRAF1, TRAF2 and TRAF3, but not TRAF5 and TRAF6. Binds through its C-terminus to SIVA1/SIVA. Seems to be involved in interactions between activated T-lymphocytes and endothelial cells and in the regulation of TCR-mediated cell death. Mediates NFkappaB activation via the TRAF2/NIK pathway. |
Q9Y5U5 (184–241) |
3–20 41–58 |
| TNF-R19 | Associates with TRAF1, TRAF2, TRAF3 and TRAF5. Interacts with LINGO1. Can mediate activation of JNK and NFkappaB. May promote caspase-independent cell death. |
Q9NS68 (192–423) |
172–189 |
| TNF-R21 | Associates with TRADD. Interacts with N-APP. May activate NFkappaB and promote apoptosis. May activate JNK and be involved in T-cell differentiation. Required for both normal cell body death and axonal pruning. |
O75509 (371–655) |
162–179 |
| TNF-R25 | Receptor for TNFSF12/APO3L/TWEAK. Interacts directly with the adapter TRADD. Interacts with BAG4. Mediates activation of NFkappaB and induces apoptosis. May play a role in regulating lymphocyte homeostasis. |
Q93038 (221–417) |
- |
| TNF-R27 (EDA-A2 receptor) | Receptor for EDA isoform A2, but not for EDA isoform A1. Associates with TRAF1, TRAF3 and TRAF6. Mediates the activation of the NFkappaB and JNK pathways. |
Q9HAV5 (160–297) |
11–28 50–67 82–99 |
| Type I Cytokine Receptors | |||
| EpoR | Receptor for erythropoietin. Mediates erythropoietin-induced erythroblast proliferation and differentiation. Triggers the JAK2/STAT5 signaling cascade. In some cell types, can also activate STAT1 and STAT3. May also activate the LYN tyrosine kinase. |
P19235 (274–508) |
90–107 165–182 218–235 |
| G-CSF-R | Receptor for granulocyte colony-stimulating factor (CSF3), essential for granulocytic maturation. Plays a crucial role in the proliferation, differentiation and survival of cells along the neutrophilic lineage. May function in some adhesion or recognition events at the cell surface. |
Q99062 (651–836) |
20–37 52–69 |
| GM-CSF/IL-3/IL-5 receptor common beta-chain | High affinity receptor for IL-3, IL-5 and granulocyte-macrophage colony-stimulating factor. The beta subunit is common to the IL-3, IL-5 and GM-CSF receptors. |
P32927 (461–897) |
48–65 122–139 166–183 252–269 355–372 |
| Growth hormone receptor | Receptor for pituitary gland growth hormone involved in regulating postnatal body growth. On ligand binding, couples to the JAK2/STAT5 pathway. The soluble form (GHBP) acts as a reservoir of growth hormone in plasma and may be a modulator/inhibitor of GH signaling. |
P10912 (289–638) |
93–110 131–148 189–206 |
| Prolactin receptor | Receptor for the anterior PRL. Interacts with SMARCA1. Interacts with GH1. Interacts with CSH. Isoform 4 is unable to transduce prolactin signaling. Isoform 6 is unable to transduce prolactin signaling. |
P16471 (259–622) |
67–84 111–128 138–155 180–197 239–256 |
| Type II Cytokine Receptors | |||
| IFNgamma receptor alpha-chain | Receptor for IFNgamma. Two receptors bind one IFNgamma dimer. |
P15260 (267–489) |
135–152 |
| IFNgamma receptor beta-chain | Part of the receptor for IFNgamma. Required for signal transduction. This accessory factor is an integral part of the IFNgamma signal transduction pathway and is likely to interact with GAF, JAK1 and/or JAK2. |
P38484 (269–337) |
40–57 |
| IFNalpha/beta receptor alpha-chain | Associates with IFNAR2 to form the type I IFN receptor. Receptor for IFNsalpha and beta. Interacts with IFNAR2. Binding to type I IFNs triggers tyrosine phosphorylation of a number of proteins including JAKs, TYK2, STAT proteins and IFNRalpha- and beta-subunits themselves. |
P17181 (458–557) |
61–78 |
| IFNalpha/beta receptor beta-chain | Associates with IFNAR1 to form the type I IFN receptor. Receptor for IFNalpha and beta. Involved in IFNmediated STAT1, STAT2 and STAT3 activation. Isoform 1 and isoform 2 are directly involved in signal transduction due to their association with the TYR kinase, JAK1. Isoform 3 is a potent inhibitor of type I IFN receptor activity. |
P48551 (265–515) |
157–174 234–251 |
| TGFbeta receptors | |||
| TGFbeta type I receptor | On ligand binding, forms a receptor complex consisting of two type II and two type I transmembrane serine/threonine kinases. Interacts with CD109 and RBPMS. Interacts with SMAD2 when phosphorylated on several residues in the GS region. Interacts with TRAF6. Receptor for TGFbeta. |
P36897 (148–503) |
- |
| TGFbeta type II receptor | On ligand binding, forms a receptor complex consisting of two type II and two type I transmembrane serine/threonine kinases. Binds to DAXX. Interacts with TCTEX1D4. Receptor for TGFbeta. |
P37173 (188–567) |
363–380 |
| Other single chain receptors | |||
| Beta-2 AR | Binds SLC9A3R1 and GPRASP1. Interacts with ARRB1 and ARRB2. Interacts with SRC, USP20 and USP33. Mediates the catecholamine-induced activation of adenylate cyclase through the action of G proteins. Binds epinephrine with an approximately 30-fold greater affinity than it does norepinephrine. |
P07550 (59–71) (130–150) (221–274) (330–413) |
- - 18–35 34–51 63–80 |
| Multichain receptors | |||
| T cell receptor | |||
| Zeta | Contains 3 ITAM domains. Plays a role in assembly and expression of the TCR complex as well as signal transduction upon antigen triggering. Interacts with SLA and SLA2. Interacts with DOCK2 and TRAT1. Interacts with HIV-1and HIV-2 Nef protein; this interaction induces downregulation of cell surface TCR/CD3 complexes. |
P20963 (52–164) |
18–35 58–75 |
| CD3epsilon | Contains 1 ITAM domain. Mediates signal transduction. |
P07766 (153–207) |
- |
| CD3delta | Contains 1 ITAM domain. Mediates signal transduction. |
P04234 (127–171) |
15–32 |
| CD3gamma | Contains 1 ITAM domain. Mediates signal transduction. |
P09693 (138–182) |
22–39 |
| B cell receptor | |||
| Igalpha | Contains 1 ITAM domain. Mediates signal transduction. Required for BCR surface expression and for efficient differentiation of pro- and pre-B-cells. Stimulates SYK autophosphorylation and activation. Binds to BLNK. Represses BCR signaling during development of immature B cells. |
P11912 (166–226) |
24–41 |
| Igbeta | Contains 1 ITAM domain. Mediates signal transduction. Enhances phosphorylation of Ig alpha. |
P40259 (181–229) |
- |
| Fc receptors | |||
| Fcepsilon RI beta chain | Signaling subunit of Fcepsilon type I receptor (Fcepsilon RI). Contains 1 ITAM domain (in: 201–244). Mediates signal transduction and initiation of the allergic response. |
Q01362 (1–59) (118–130) (201–244) |
42–59 - - |
| FcRgamma chain | Signaling subunit of Fc epsilon RI and other Fc receptors. Contains 1 ITAM domain. Mediates signal transduction. Regulates several aspects of the immune response. |
P30273 (45–86) |
- |
| Other multichain receptors | |||
| DAP-10 | Signaling subunit of NKG2D and other multichain receptors. Contains 1 tyrosine-phosphorylated YINM motif. Mediates signal transduction. |
Q9UBK5 (70–93) |
- |
| DAP-12 | Signaling subunit of TREM-1 and other multichain receptors. ITAM domain. Mediates signal transduction. |
O43914 (62–113) |
- |
For multichain receptors including MIRRs.
As provided in the Swiss-Prot database [Bairoch A, et al. The Universal Protein Resource (UniProt). Nucleic Acids Res 2005; 33:154–9].
Numbers in parentheses are amino acid residues of cytoplasmic domain.
Amino acid residues. Predicted using the algorithms described in Mohan A, et al. Analysis of molecular recognition features (MoRFs). J Mol Biol 2006; 362:1043–59.
Abbreviations: MIRR, multichain immune recognition receptor; alpha-MoRF, alpha-Helix-forming Molecular Recognition Feature; EGF, epidermal growth factor; TGF, transforming growth factor; GP, glycoprotein; NTAK, neural- and thymus-derived activator for ErbB kinases; PDGFR, platelet-derived growth factor receptor; PTPN11, the SH2-containing tyrosine phosphatase Shp2; GRB2, growth factor receptor-bound protein 2; FGFR, fibroblast growth factor receptor; KL, Klotho; VEGFR, vascular endothelial growth factor receptor; MuSK, muscle specific kinase; PGF, prostaglandin F; HGFR, hepatocyte growth factor receptor; TRK, tyrosine kinase receptor; NGF, nerve growth factor; BDNF, brain-derived neurotrophic factor; SHC, Src homologous and collagen protein; PI, phosphatidylinositol; PLC, phospholipase C; ERK, extracellular signal-regulated kinase; NT, neurotrophin; LTK, leukocyte tyrosine kinase; ROR, receptor tyrosine kinase-like orphan receptor; YWHAB, the beta polypeptide of tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein; DDR, discoidin domain receptor; WWC, WW domain-containing protein; PRKCZ, protein kinase Czeta; MuSK; muscle-specific kinase; NMJ, neuromuscular junction; INSR, insulin receptor; IRS, insulin receptor substrate; PI3K, phosphatidylinositol 3′-kinase; EPH, ephrin; TNF, tumor necrosis factor; LTBR, lymphotoxin-b receptor; TRAF, tumor necrosis factor receptor-associated factor; LRR, leucine-rich repeat; RANK, receptor activator of NFκB; HSV, herpes simplex virus; HHV, human herpes virus; TCR, T cell receptor; EDA, ectodermal dysplasin; EpoR, erythropoietin receptor; G-CSF-R, granulocyte colony-stimulating factor receptor; IL, interleukin; GMCSF, granulocyte-macrophage colony stimulating factor; GHBP, growth hormone binding protein; PRL, pituitary hormone prolactin; CsH, cyclosporin H; IFN, interferon; TGF, transforming growth factor; AR, adrenergic receptor; ITAM, immunoreceptor tyrosine-based activation motif; HIV, human immunodeficiency virus; Ig, immunoglobulin; BCR, B cell receptor; DAP-12, DNAX-activating protein of molecular mass 12 kilodaltons; TREM-1, triggering receptor expressed on myeloid cells 1.
Results
Cytoplasmic signaling (effector) domains of vast majority of single-chain receptors are mostly ordered, whereas those of multichain receptors are intrinsically disordered.
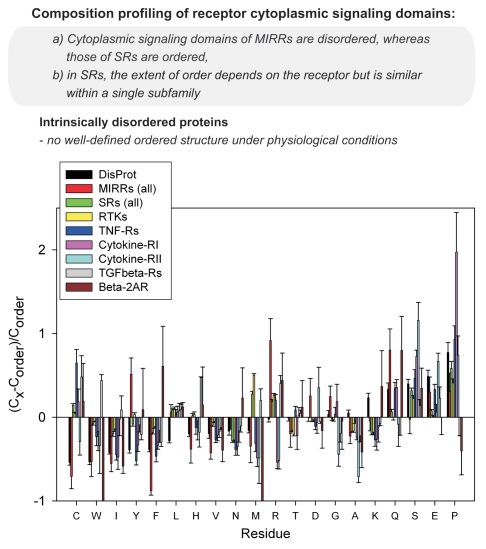
Since the amino acid sequences of ordered and intrinsically disordered proteins are noticeably different,3,22–25 at the first stage, we analyzed the amino acid compositions of the cytoplasmic signaling domains of MIRRs and SRs. The results of this analysis are shown in Figure 1 as the relative composition profiles calculated for various datasets as described by Vacic and colleagues.23 Here, the fractional difference in composition between a given set of proteins and a set of completely ordered proteins was calculated for each amino acid residue. The fractional difference was evaluated as (CX - Corder)/Corder, where CX is the content of a given amino acid in a given set of proteins, and Corder is the corresponding content in the set of fully ordered proteins from PDB. The fractional difference in composition between typical IDPs from the DisProt Database26 and a set of ordered proteins for comparison is also shown (Fig. 1). Amino acid residues are arranged by the order of their enrichment in typical IDPs, from less abundant to the left, to more abundant to the right.
Figure 1.
Relative amino acid compositions for cytoplasmic signaling (effector) domains of single- and multichain receptors. The bar for a given amino acid represents the fractional difference in composition between a given set of analyzed sequences and a set of ordered proteins. The fractional difference is calculated as (CX - Corder)/Corder, where CX is the composition of a given amino acid in a given set of proteins and Corder is the corresponding composition in a set of ordered proteins, and plotted for each amino acid. Black bars correspond to a set of the experimentally characterized disordered proteins in the DisProt database. Positive and negative values indicate residues in a given set that have more and less order, respectively. Confidence intervals were estimated using per-protein bootstrapping with 1,000 iterations. Abbreviations: DisProt, disordered proteins; MIRR, multichain immune recognition receptor; SR, single-chain receptor; RTK, receptor tyrosine kinase; TNF-R, tumor necrosis factor receptor; cytokine-R, cytokine receptor; TGFbeta-R, transforming growth factor-beta receptor; beta-2AR, beta 2-adrenergic receptor.
The usefulness of this analysis is determined by the fact that the propensity of a given protein to be intrinsically disordered is determined by a set of specific features of its amino acid sequence and composition.3,22–25 For example, IDPs are significantly depleted in bulky hydrophobic (I, L and V) and aromatic amino acid residues (W, Y and F), which would normally form the hydrophobic core of a folded globular protein. IDPs also possess a low content of C and N residues. These less abundant residues, namely I, L, V, W, F, Y, C and N, are proposed to be called order-promoting amino acids. On the other hand, IDPs and IDRs are shown to be substantially enriched in disorder-promoting amino acids: E, K, R, G, Q, S, P and A.
Application of this tool to the analysis of the cytoplasmic signaling domains of MIRRs and SRs revealed a number of interesting features. SRs, if considered all together, possess the amino acid composition biases typical for ordered proteins. This is illustrated by the fact that the corresponding bars (shown by green) are generally scattered around the zero line (Fig. 1). However, if we consider amino acid compositions of SRs in different subfamilies, something interesting is obvious: receptor tyrosine kinases (RTKs, yellow bars) and transforming growth factor-beta receptors (TGF-Rs, gray bars) are the most ordered since their bars show the smallest deviations from the zero line (Fig. 1). On the other hand, tumor necrosis factor receptors (TNF-Rs, dark blue bars) have some features of disordered proteins, being depleted in major order-promoting residues and showing enrichment in some disorder-promoting residues. This tendency is even more obvious for both (I and II) types of cytokine receptors. For MIRRs, our data demonstrate that the cytoplasmic domains of MIRR signaling subunits have many amino acid composition features typical for IDPs, being systematically depleted in order-promoting residues and systematically enriched in the majority of disorder-promoting residues (Fig. 1).
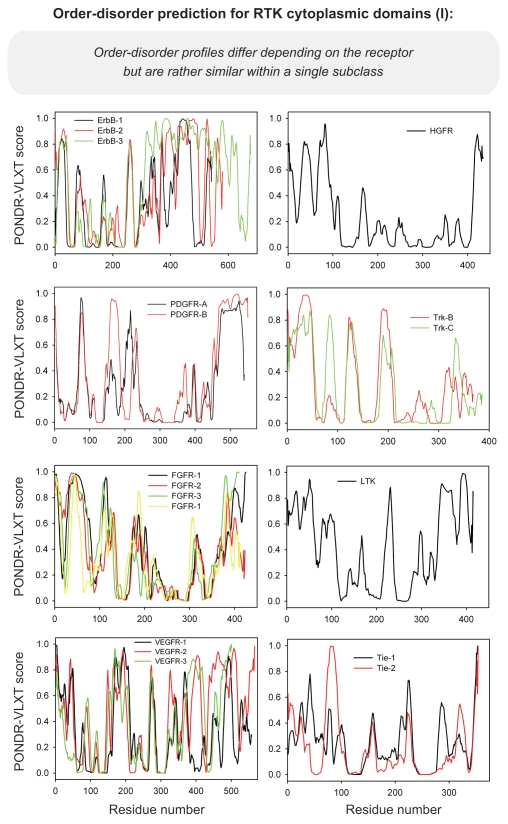
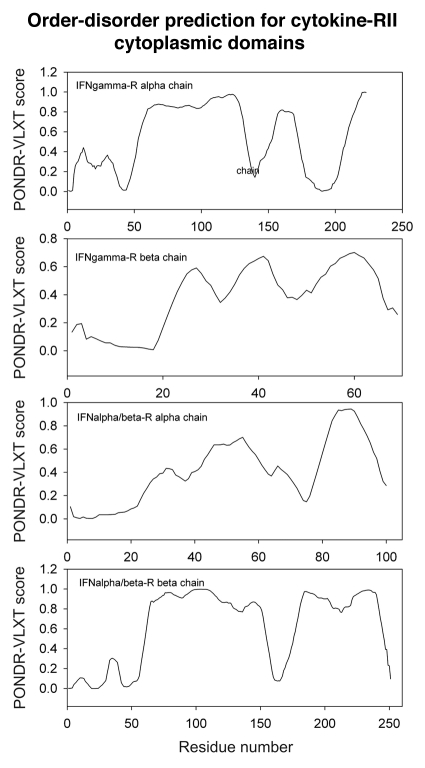
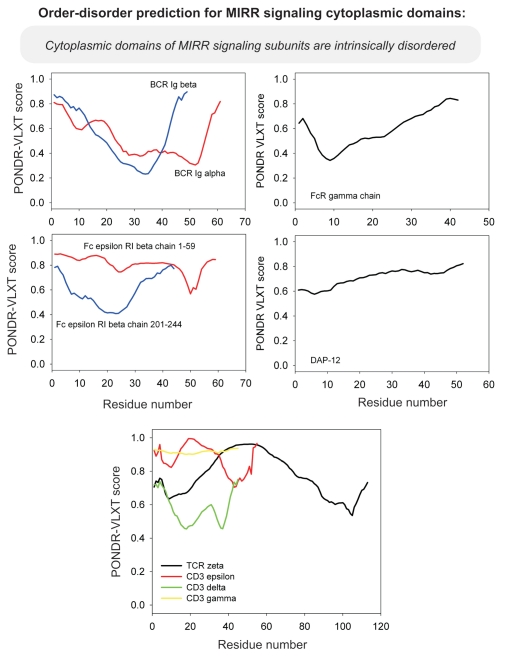
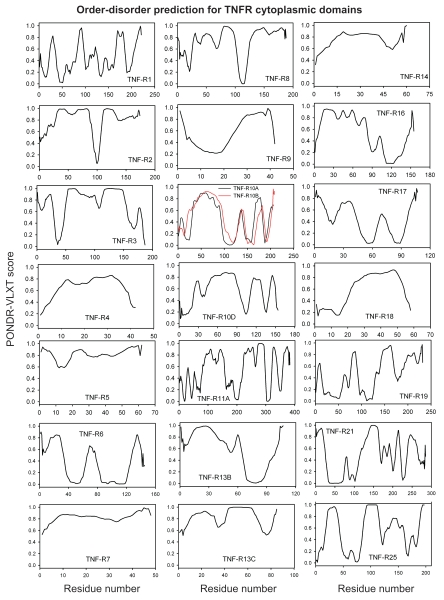
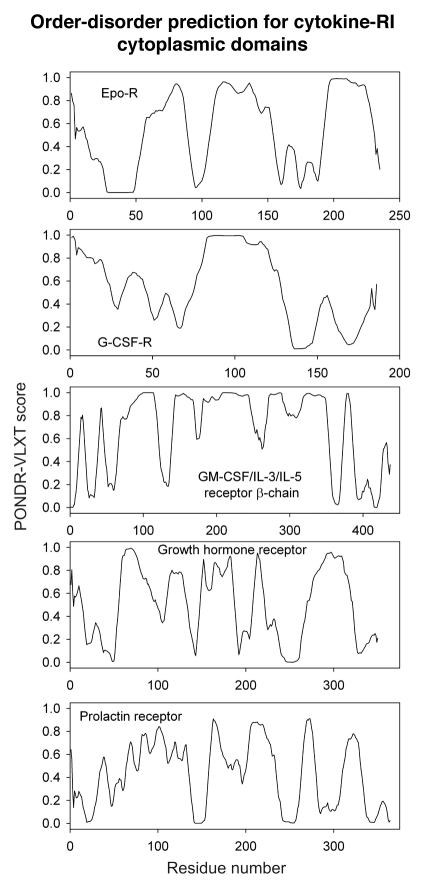
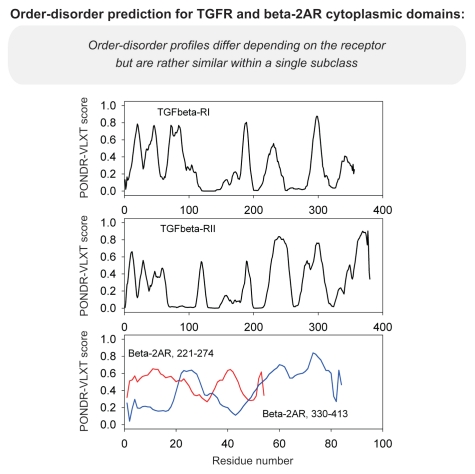
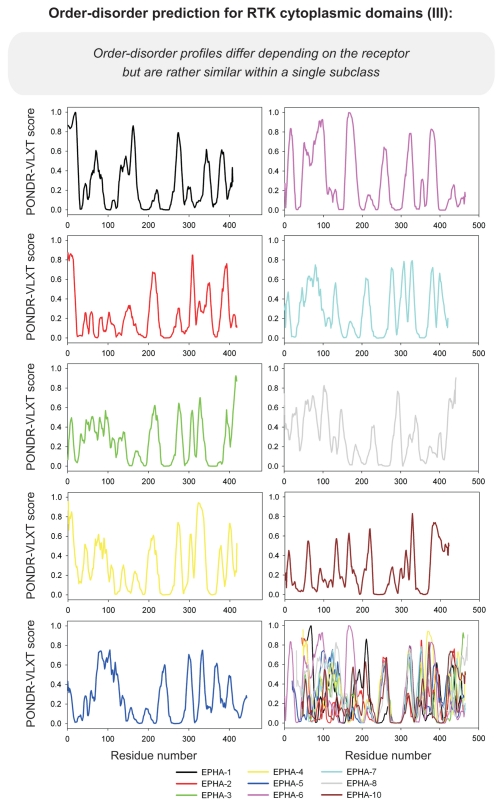
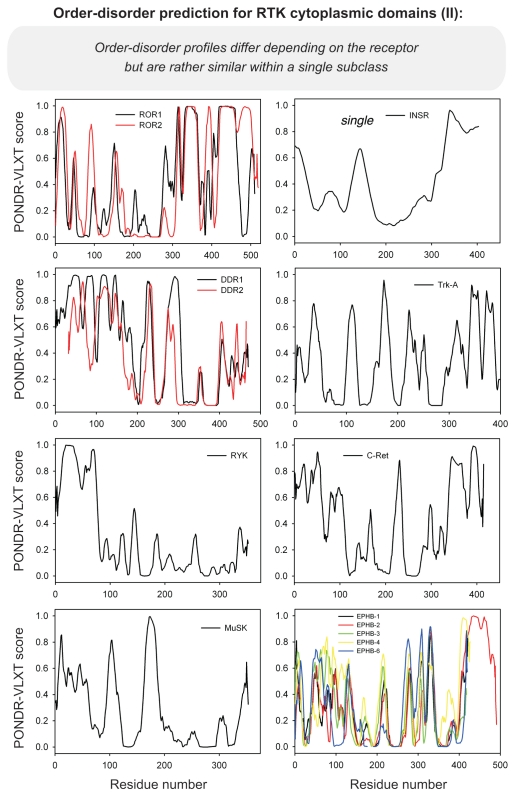
Differential occurrence of protein intrinsic disorder in the cytoplasmic signaling (effector) domains of cell receptors is further supported by the results of the per-residue disorder predictions in various SRs and MIRRs by the PONDR® VLXT algorithm.27 Figures 2 through 8 clearly show that all SRs have some disordered regions. On the other hand, Figure 9 reemphasizes the important fact that MIRRs are highly disordered and contain very little of predicted order. In fact, the MIRR disorder distribution curves are mostly located in the upper halves of PONDR plots (Fig. 9). As far as different cytoplasmic domains of SRs are concerned, the amount of disorder varies both between the subfamilies and subclasses and within the subfamilies and subclasses (Figs. 2–8). In agreement with composition profiling data discussed above, some SRs such as TNF-Rs (Fig. 5), cytokine receptors (Figs. 7 and 8) and beta2-adrenergic receptor, beta-2AR (Fig. 6), have more disorder whereas other SRs such as RTKs (Figs. 2–4) and TGF-Rs (Fig. 6), have less. In fact, disorder profiles for several TNF-Rs (e.g., see receptors R2, R3, R4, R5, R7, R8, R9, R10, R13, R14 and R18; Fig. 5) are characteristic for proteins with high intrinsic disorder propensity. Data also show that typically disorder profiles significantly differ for different SR subfamilies and subclasses but show less variation within a subfamily or a subclass. Within several subfamilies and subclasses, SRs are characterized by rather similar disorder profiles. This is clearly seen from the plots containing PONDR® VLXT curves for several proteins of one subfamily/subclass. In fact, for such proteins one can find a conserved feature and use it as a starting point for alignment. This is done in Figures 2–5 for several subclasses just by shifting sequences relative each other, without introduction of any gaps. The conservation of intrinsic disorder profiles within several SR subfamilies and subclasses is very important since it suggests that this disorder distribution within a given protein sequence might be conserved for functional reasons.
Figure 2.
PONDR VL-XT prediction analysis of cytoplasmic domains of receptor tyrosine kinases. HGFR, hepatocyte growth factor receptor; PDGFR, platelet-derived growth factor receptor; TRK, tyrosine kinase receptor; FGFR, fibroblast growth factor receptor; LTK, leukocyte tyrosine kinase; VEGFR, vascular endothelial growth factor receptor.
Figure 8.
PONDR VL-XT prediction analysis of cytoplasmic domains of type II cytokine receptors. IFN, interferon.
Figure 9.
PONDR VL-XT prediction analysis of cytoplasmic domains of multichain receptor signaling subunits. MIRR, multichain immune recognition receptor; TCR, T cell receptor; DAP-12, DNAX-activating protein of molecular mass 12 kilodaltons.
Figure 5.
PONDR VL-XT prediction analysis of cytoplasmic domains of tumor necrosis factor receptors.
Figure 7.
PONDR VL-XT prediction analysis of cytoplasmic domains of type I cytokine receptors. EpoR, erythropoietin receptor; G-CSF-R, granulocyte colony-stimulating factor receptor; GMCSF, granulocyte-macrophage colony stimulating factor; IL, interleukin.
Figure 6.
PONDR VL-XT prediction analysis of cytoplasmic domains of transforming growth factor receptors and beta 2-adrenergic receptor.
Figure 4.
PONDR VL-XT prediction analysis of cytoplasmic domains of receptor tyrosine kinases (continuation). EPH, ephrin.
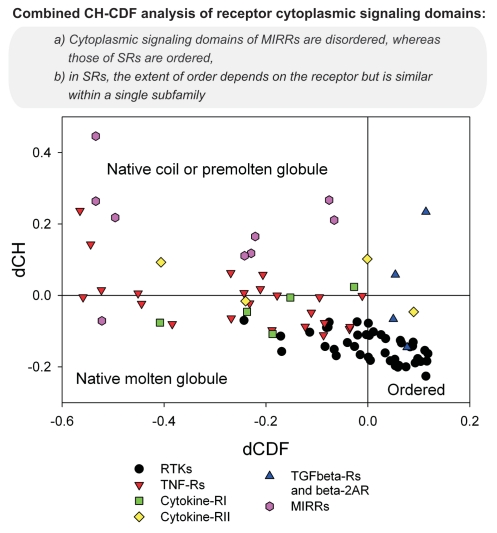
Next, we analyzed the overall disorderedness of various signaling domains of MIRRs and SRs by a CH-CDF plot (Fig. 10). This is a global disorder analysis method that combines the outputs of charge-hydropathy (CH),3,28 and cumulative distribution function (CDF) plot tools.28,29 CH-plot is a binary disorder predictor that puts a given protein onto a 2D graph by taking the mean Kay-Doolittle hydrophobicity of a protein as an X-coordinate and its mean absolute net charges as a Y-coordinate. In the corresponding CH-plot, fully structured proteins and fully disordered proteins can be separated by a boundary line. All proteins located above this boundary line are highly likely to be extended, while proteins located below this line are likely to be compact. The vertical distance on the CH-plot from the location of the protein to the boundary line is then used as a measure of disorder (or structure) tendency of a given protein. This distance is further referred to as CH-distance.
Figure 10.
Combined charge-hydropathy (CH) and cumulative distribution function (CDF) plot analysis of receptor cytoplasmic domains. Each spot represents a single sequence whose coordinates are calculated as a distance of this protein from the boundary in the corresponding CH-plot (Y-coordinate) and an average distance of the corresponding CDF curve from the boundary (X-coordinate). RTK, receptor tyrosine kinase; TNF-R, tumor necrosis factor receptor; cytokine-RI, type I cytokine receptor; cytokine-RII, type II cytokine receptor; TGFbeta-R, transforming growth factor-beta receptor; beta-2AR, beta 2-adrenergic receptor; MIRR, multichain immune recognition receptor.
CDF is another binary disorder predictor which describes the disorder status of an entire protein.28,29 In brief, it is a cumulative histogram of disordered residues at various disordered score. By definition, ordered proteins have many structure-promoting residues and less disorder-promoting residues. Therefore, the CDF curve of an ordered protein increases very sharply at the side of low disordered scores, and then goes flat on the side of high disordered scores. For disordered proteins, the CDF curve moves upward slightly at regions of low disordered scores, then jumps up at the regions with high disordered scores. Hence, on the CDF plot, structured proteins tend to stay on the upper left half, while disordered proteins locate at the lower right half of the plot. By comparing the locations of CDFs for a group of fully disordered and fully structured proteins, a boundary line between these two groups of proteins can be identified. Proteins whose CDF is above the boundary line are predicted to be structured, while proteins with lower CDF than the boundary are predicted to be disordered. The distance from CDF boundary is also a kind of measurement of disorder (structure) status of the protein and is referred as CDF distance. To evaluate the CDF distance in this study, we applied the PONDR® VLXT-based CDF boundary since the disorder status of the studied domains was evaluated by PONDR® VLXT.
Combination of both CH- and CDF-distances represents a new analytical technique, the CH-CDF plot analysis, that provides very useful information on the general status of the protein at the entire sequence level.30 Results of the CH-CDF analysis of various cytoplasmic signaling domains are shown in Figure 10. CH-CDF plot can be split into four quadrants: Q1 (upper right), Q2 (lower right), Q3 (lower left) and Q4 (upper left). Proteins in Q1 are structured by CDF, but disordered by CH; proteins in Q2 are predicted to be structured by both CDF and CH; proteins in Q3 are disordered by CDF but structured by CH; and Q4 is for proteins predicted to be disordered by both methods. Therefore, quadrants Q3 and Q4 include proteins disordered as a whole, with Q3 containing native molten globules, and with Q4 containing native coils or native pre-molten globules.30 The order-disorder status of SRs in CH-CDF plot roughly matches the results of the per-residue disorder predictions (Fig. 10). In fact, TGF-Rs are located in quadrants Q1 and Q2. The majority of RTKs are located in quadrant Q2, with some RTKs being found in Q3 (mostly grouped close to the boundary between Q2 and Q3). The majority of TNF-Rs are in Q3, with some in Q4. Interestingly, although type I cytokine receptors are found mostly in Q3, type II cytokine receptors are spread over Q2, Q3 and Q4. Finally, all cytoplasmic domains of MIRR signaling subunits are noticeably more disordered than the majority of SR cytoplasmic domains and represent mostly native coils or native pre-molten globules (Fig. 10).
Previous studies revealed a tight correlation between the protein intrinsic disorder and protein binding.31–33 A special type of intrinsic disorder-based interaction-prone sequence motifs known as molecular recognition features (MoRFs) was shown to play an important role in protein-protein interactions.34,35 MoRF is a short structure-prone segment of a long disordered region which usually undergoes a disorder-to-order transition upon binding to a partner. MoRFs are frequently found in proteins involved in recognition, signaling and regulation. Based on the structure adopted upon binding, at least three basic types of MoRFs are found: α-MoRFs, β-MoRFs and ι-MoRFs, which form α-helices, β-strands and irregular secondary structure when bound, respectively.36 Complex-MoRFs have also been reported.37 In this study, over 40% of eukaryotic proteins were estimated to contain at least one α-helical MoRF.34,36
Therefore, to elucidate the role of intrinsic disorder in protein-protein interactions of SR and MIRR signaling domains, their potential binding sites were evaluated by the MoRF-II predictor.34,35 Results of this analysis show that more than half of RTKs have predicted MoRFs, with some RTKs possessing more than one recognition motif (Table 1). On the other hand, 43% RTKs, being mostly ordered, do not have any MoRFs. Similarly, half of signaling domains from TGF-Rs and beta-2AR are MoRF-less, with another half of these receptors typically possessing just one MoRF per chain. On the contrary, 82% TNF-Rs have MoRFs, with the vast majority of these receptors possessing more than one MoRF (Table 1). Although all cytokine receptors analyzed are shown to have MoRFs, the number of recognition features per chain is clearly subclass-dependent, with the majority of the type II cytokine receptors possessing one MoRF per chain and with all type I receptors having more than one MoRF per chain. Surprisingly, the cytoplasmic domains of MIRR signaling subunits in general were less enriched in MoRFs. This could be due to the fact that these domains are noticeably shorter and significantly more disordered than proteins from the SR family. Intriguingly, biophysical studies of these domains revealed a previously unknown phenomenon of binding of IDPs to intrinsically disordered or well-ordered protein partners without a disorder-to-order transition.14–16,21,38
One also should keep in mind that the algorithm utilized in this study is able to predict only α-helical MoRFs. Considering four groups of MoRFs: α-MoRFs, β-MoRFs, i-MoRFs (irregular-MoRFs) and complex-MoRFs,36,37 it is likely that highly disordered signaling domains of MIRRs might contain recognition features different from α-MoRFs, e.g., i-MoRFs. MoRFs are functional binding motifs. Thus, the high abundance of MoRFs in effector domains of SRs and MIRRs suggests that these sequences might play an important role in their biological actions.
Discussion
Signaling concerns the transfer of information from one body, a source, to another, a receiver in order to stimulate activity.39 In the context of receptor signaling, the process involves transmembrane transduction of the recognition information through the membrane. Interestingly, recent evolutionary studies of glycosyltransferases revealed important aspects of recognition through homooligomerization.40 A recently proposed novel model of transmembrane signaling, the Signaling Chain HOmoOLigomerization (SCHOOL) model, suggests that formation of competent signaling homooligomers in cytoplasmic milieu is the necessary and sufficient event to trigger receptors of both structural families (SRs and MIRRs) and to induce cell activation.9,12,17,41 Within this model, receptor homooligomerization induced or tuned upon ligand binding outside the cell is translated across the membrane into protein oligomerization in cytoplasmic milieu, thus providing a general platform for receptor-mediated signaling. Thus, one can expect that further evolutionary and structural studies of receptor cytoplasmic domains will reveal important aspects of signaling through homooligomerization. This can not only further explain differential occurrence of protein intrinsic disorder in receptor signaling domains but also can significantly advance the general knowledge of transmembrane signal transduction.
Why do intrinsic disorder profiles substantially differ for different SR subfamilies and subclasses but show much less variation within the subfamily or the subclass?
It should be noted that classification of cell receptors as SRs and MIRRs is based only on their structural assembly but not their functions (Fig. 11).9,10 In the SR family, receptors can be further divided into different subfamilies, such as RTKs or TNF-Rs. Within such subfamilies there can exist subclasses that include functionally related receptors. Example is the RTK subfamily that has several different subclasses such as ErbB receptors, fibroblast growth factor receptors (FGFRs) and others (Table 1). In this study, we found fact that protein order-disorder profiles are substantially different for different SR subfamilies and subclasses. This can reflect the lack of functional relations between their members. In this context, rather similar profiles within several receptor subfamilies and subclasses can be readily explained by similarities in functions of their members.
Figure 11.
Differential occurrence of protein intrinsic disorder in the cytoplasmic signaling domains of single- and multichain cell receptors.
Is there any correlation between disorder profiles of signaling domains and receptor function? Intrinsic protein disorder is thought to confer many functional advantages that include the increased speed of interaction, specificity without excessive binding strength and the binding promiscuity, i.e., ability to bind to multiple partners.32,42 On the other hand, cytoplasmic signaling (effector) domains of SRs may need to interact with multiple partners to induce diverse signaling. Thus, one can hypothesize that differential occurrence of protein disorder between different SR subfamilies might correlate with diversity of the induced signaling pathways that result in different functional outcomes. In other words, the more diverse are the receptor-mediated signaling pathways, the more protein disorder is found in the members of this subfamily and/or subclass. In this context, the most ordered cytoplasmic signaling (effector) domains should be in those receptors that provide only a single ON/OFF signal. Further structural and functional studies will be required to confirm this hypothesis.
Why for MIRRs, i.e., the receptors with extracellular recognition and intracellular signaling functions provided by the separate protein chains, did nature select protein disorder of signaling domains?
In MIRRs, signaling is achieved through receptor-associated subunits that contain in their cytoplasmic domains one or more copies of the ITAM regions with two appropriately spaced tyrosines (YxxL/Ix6–8YxxL/I; where x denotes non-conserved residues; Table 2),43 or the YxxM motif, found in the DAP10 cytoplasmic domain.44 Upon receptor triggering, tyrosine residues of the ITAM/YxxM regions are phosphorylated in an early and obligatory event in the signaling cascade. Primary sequences of MIRR ITAMs are shown in Tables 1 and 2. Intriguingly, the common motif, ITAM, provides diverse activation signals in the context of different signaling subunits of one receptor (e.g., T cell receptor, TCR, that contains 4 different ITAM-containing signaling subunits) or one subunit of different receptors (e.g., various MIRRs that signal through the ITAM-containing FcRγ signaling chain). This suggests a potential need for interaction of the cytoplasmic domains of MIRR signaling subunits with multiple binding partners to induce different downstream sequences and, as a result, different functional outcomes. This can be one of the reasons why to select intrinsic protein disorder for MIRR-mediated signaling.
Table 2.
Primary sequences of MIRR ITAMs
| Signaling subunit | MIRR | ITAM sequencea,b |
| TCRζ | TCR, Fc receptors, NK receptors |
YNE LNL GRR EEY DVL YNE LQK DKM AEA YSE I YQG LST ATK DTY DAL |
| CD3ε | TCR | YEP IRK GQR DLY SGL |
| CD3δ | TCR | YQP LRD RDD AQY SHL |
| CD3γ | TCR | YQP LKD RED DQY SHL |
| Igα | BCR | YEG LNL DDC SMY EDI |
| Igβ | BCR | YEG LDI DQT ATY EDI |
| FcRγ | Fc receptors, ILT/LIR receptors, GPVI, myeloid CLRs, NK receptors | YTG LST RNQ ETY ETL |
| DAP-12 | TREM receptors, MAIR-II, SIRPβ1, myeloid CLRs, NK receptors | YQE LQG QRS DVY SDL |
ITAM tyrosine residues are underlined.
Three ITAMs of TCRζ are shown.
In the context of the SCHOOL,9,12 protein disorder of the ITAM-containing signaling domains also provides a molecular basis to explain high specificity, selectivity and sensitivity of immune cells in recognition and discrimination of different antigens/ligands and how this recognition/discrimination results in different functional outcomes. Recent studies discovered that all ITAM-containing cytoplasmic domains of MIRR signaling subunits form homooligomers.14,16 Homooligomerization of these intrinsically disordered domains is a highly dynamic process that is characterized by micromolar affinity and a rapid association and dissociation kinetics.14,16 This is typical for the proteins that associate and dissociate in response to changes in their environment, such as the majority of signal transduction mediators. Further, the SCHOOL model assumes that the diversity of the immune cell response is partly provided by the combinatorial nature of MIRR-mediated signaling. Signal diversification may be achieved through different patterns of MIRR signaling subunit oligomerization9,11 in combination with distinct activation signals provided by different MIRR signaling modules45–56 and/or different ITAMs located on the same signaling module (e.g., TCRζ chain that contains 3 ITAMs).57 Thus, according to the model, the diversity of cell functional outcomes in response to different ligands is related to intracellular protein disorder of MIRR signaling subunits and is higher with the more different signaling subunits the MIRR complex has. Thus, TCR-mediated signaling and cell activation has the highest combinatorial potential as compared to other MIRRs, explaining a high variability of distinct TCR-triggered intracellular signaling pathways and therefore distinct T cell functional responses depending on the nature of the stimulus.9,12
In conclusion, one can expect that further multidisciplinary studies will clarify the raised questions of great interest and practical utility. This can substantially improve our understanding of receptor-mediated ligand recognition and signal transduction through protein order, disorder and homooligomerization.
Materials and Methods
Sequences and data sets.
The data set of cell receptor-associated signaling sequences was extracted from SWISS-PROT (http://www.expasy.org/sprot/). The corresponding SWISS-PROT accession numbers and amino acid residues of the analyzed cytoplasmic domains are indicated in Table 1.
Compositional profiling.
To gain insight into the relationships between sequence and disorder, the amino acid compositions in different data sets were compared using an approach recently developed for IDPs.22 The fractional difference in composition between a given set of the analyzed cytoplasmic sequences, intrinsically disordered proteins,27 and a set of ordered proteins27 was calculated for each amino acid residue. The fractional difference is calculated as (CX - Corder)/Corder, where CX is the composition of a given amino acid in a given protein set and Corder is the corresponding composition in a set of ordered proteins and plotted for each amino acid. Positive and negative values indicate residues in a given set that have more and less order, respectively. Confidence intervals were estimated using per-protein bootstrapping with 1,000 iterations.
Prediction of intrinsic disorder.
Prediction of intrinsic disorder in cytoplasmic signaling domains of cell receptors was performed using PONDR VLXT,27 CDF,29 and charge-hydropathy plots.3
PONDR (predictor of natural disordered regions) is a set of neural network predictors of disordered regions on the basis of local amino acid composition, flexibility, hydropathy, coordination number and other factors. These predictors classify each residue within a sequence as either ordered or disordered. PONDR VL-XT combines three neural networks, one for internal sequences and two for either terminus of the sequence. The output of the XT predictor provides predictions up to 14 amino acids from their respective ends. A simple average is taken for the overlapping predictions, and a sliding window of nine amino acids is used to smooth the prediction values along the length of the sequence. Unsmoothed prediction values from the XT predictors are used for the first and last four sequence positions.
CDF (cumulative distribution function) analysis summarizes the per-residue disorder predictions by plotting PONDR VL-XT scores against their cumulative frequency, which allows ordered and disordered proteins to be distinguished on the basis of the distribution of prediction scores.28 At any given point on the CDF curve, the ordinate gives the proportion of residues with a PONDR score less than or equal to the abscissa. The optimal boundary that provided the most accurate order-disorder classification was shown to represent seven points located in the 12th through 18th bins.28 Thus, for CDF analysis, order-disorder classification is based on whether a CDF curve is above or below a majority of boundary points.
CH (charge-hydropathy)-plot analysis is based on the fact that naturally folded and IDPs occupy non-overlapping regions in the CH-plots, with natively unfolded proteins being specifically localized within a particular region of CH phase space, and was performed as previously described in reference 3 and 28.
Statistical analysis.
An analysis of variability in the percentage of proteins with predicted disorder was performed by bootstrap re-sampling as described previously in reference 58. Briefly, for each data set, proteins were sampled randomly with replacement. The number of randomly sampled proteins for each data set was equal to the number of proteins in the data set. The fraction of proteins with disordered regions of a given length was determined for each sample. The data sets were sampled 1,000 times, and these values were used to calculate the standard error of the fractions for each data set. The 95% confidence intervals were calculated from the standard errors and are shown as error bars in corresponding Figures. Non-overlapping confidence intervals indicated that the fractions were significantly different.
α-MoRF predictions.
Indicator of α-helix forming molecular recognition fragments (α-MoRFs),34 is based on observations that predictions of order in otherwise highly disordered proteins correspond to protein regions that mediate interaction with other proteins or DNA.59,60 This predictor focuses on short binding regions within long IDRs that are likely to form helical structure upon binding. Analyses of α-MoRFs in the cytoplasmic signaling domains of cell receptors was performed as described in reference 36.
Figure 3.
PONDR VL-XT prediction analysis of cytoplasmic domains of receptor tyrosine kinases (continuation). ROR, receptor tyrosine kinase-like orphan receptor; INSR, insulin receptor; TRK, tyrosine kinase receptor; MuSK, muscle specific kinase; DDR, discoidin domain receptor; EPH, ephrin.
Abbreviations
- MIRR
multichain immune recognition receptor
- ITAM
immunoreceptor tyrosine-based activation motif
- TCR
T cell receptor
- BCR
B cell receptor
- DAP-12
DNAX adapter proteins of 12 kD
- CLR
C-type lectin receptor
- GPVI
glycoprotein VI
- ILT
Ig-like transcript
- LIR
leukocyte Ig-like receptor
- MAIR-II
myeloid-associated Ig-like receptor
- NITR
novel immune-type receptor
- NK
natural killer cells
- SIRP
signal regulatory protein
- TREM receptors
triggering receptors expressed on myeloid cells
References
- 1.Rudd CE. Disabled receptor signaling and new primary immunodeficiency disorders. N Engl J Med. 2006;354:1874–1877. doi: 10.1056/NEJMp068062. [DOI] [PubMed] [Google Scholar]
- 2.Sigalov AB, editor. Multichain immune recognition receptor signaling: From spatiotemporal organization to human disease. New York: Springer-Verlag; 2008. p. 357. [PubMed] [Google Scholar]
- 3.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 4.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 5.Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minezaki Y, Homma K, Nishikawa K. Intrinsically disordered regions of human plasma membrane proteins preferentially occur in the cytoplasmic segment. J Mol Biol. 2007;368:902–913. doi: 10.1016/j.jmb.2007.02.033. [DOI] [PubMed] [Google Scholar]
- 7.De Biasio A, Guarnaccia C, Popovic M, Uversky VN, Pintar A, Pongor S. Prevalence of intrinsic disorder in the intracellular region of human single-pass type I proteins: The case of the notch ligand Delta-4. J Proteome Res. 2008;7:2496–2506. doi: 10.1021/pr800063u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kukhtina NB, Arefieva TI, Krasnikova TL. Intracellular signal cascade in CD4+ T-lymphocyte migration stimulated by interferon-gamma-inducible protein-10. Biochemistry (Mosc) 2005;70:652–656. doi: 10.1007/s10541-005-0165-5. [DOI] [PubMed] [Google Scholar]
- 9.Sigalov AB. The SCHOOL of nature. I. Transmembrane signaling. Self/Nonself—Immune Recognition and Signaling. 2010;1:4–39. doi: 10.4161/self.1.1.10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keegan AD, Paul WE. Multichain immune recognition receptors: Similarities in structure and signaling pathways. Immunol Today. 1992;13:63–68. doi: 10.1016/0167-5699(92)90136-U. [DOI] [PubMed] [Google Scholar]
- 11.Sigalov A. Multi-chain immune recognition receptors: Spatial organization and signal transduction. Semin Immunol. 2005;17:51–64. doi: 10.1016/j.smim.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Sigalov AB. Multichain immune recognition receptor signaling: different players, same game? Trends Immunol. 2004;25:583–589. doi: 10.1016/j.it.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Sigalov AB. Protein intrinsic disorder and oligomericity in cell signaling. Mol Biosyst. 2010;6:451–461. doi: 10.1039/b916030m. [DOI] [PubMed] [Google Scholar]
- 14.Sigalov A, Aivazian D, Stern L. Homooligomerization of the cytoplasmic domain of the T cell receptor zeta chain and of other proteins containing the immunoreceptor tyrosine-based activation motif. Biochemistry. 2004;43:2049–2061. doi: 10.1021/bi035900h. [DOI] [PubMed] [Google Scholar]
- 15.Sigalov AB, Aivazian DA, Uversky VN, Stern LJ. Lipid-binding activity of intrinsically unstructured cytoplasmic domains of multichain immune recognition receptor signaling subunits. Biochemistry. 2006;45:15731–15739. doi: 10.1021/bi061108f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sigalov AB, Zhuravleva AV, Orekhov VY. Binding of intrinsically disordered proteins is not necessarily accompanied by a structural transition to a folded form. Biochimie. 2007;89:419–421. doi: 10.1016/j.biochi.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sigalov AB. The SCHOOL of nature. II. Protein order, disorder and oligomericity in transmembrane signaling. Self/Nonself—Immune Recognition and Signaling. 2010;1:89–102. doi: 10.4161/self.1.2.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sigalov AB. Unusual biophysics of immune signaling-related intrinsically disordered proteins. Self/Nonself—Immune Recognition and Signaling. 2010;1:271–281. doi: 10.4161/self.1.4.13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ortega E. How do multichain immune recognition receptors signal? A structural hypothesis. Mol Immunol. 1995;32:941–945. doi: 10.1016/0161-5890(95)00070-u. [DOI] [PubMed] [Google Scholar]
- 20.Berko D, Carmi Y, Cafri G, Ben-Zaken S, Sheikhet HM, Tzehoval E, et al. Membrane-anchored beta 2-microglobulin stabilizes a highly receptive state of MHC class I molecules. J Immunol. 2005;174:2116–2123. doi: 10.4049/jimmunol.174.4.2116. [DOI] [PubMed] [Google Scholar]
- 21.Sigalov AB. Protein intrinsic disorder and oligomericity in cell signaling. Mol Biosyst. 2010;6:451–461. doi: 10.1039/b916030m. [DOI] [PubMed] [Google Scholar]
- 22.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 23.Vacic V, Uversky VN, Dunker AK, Lonardi S. Composition Profiler: A tool for discovery and visualization of amino acid composition differences. BMC Bioinformatics. 2007;8:211. doi: 10.1186/1471-2105-8-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campen A, Williams RM, Brown CJ, Meng J, Uversky VN, Dunker AK. TOP-IDP-scale: a new amino acid scale measuring propensity for intrinsic disorder. Protein Pept Lett. 2008;15:956–963. doi: 10.2174/092986608785849164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, et al. DisProt: The database of disordered proteins. Nucleic Acids Res. 2007;35:786–793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 28.Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005;44:1989–2000. doi: 10.1021/bi047993o. [DOI] [PubMed] [Google Scholar]
- 29.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- 30.Mohan A, Sullivan W, Jr, Radivojac P, Dunker AK, Uversky VN. Intrinsic disorder in pathogenic and non-pathogenic microbes: discovering and analyzing the unfoldomes of early-branching eukaryotes. Mol Biosyst. 2008;4:328–340. doi: 10.1039/b719168e. [DOI] [PubMed] [Google Scholar]
- 31.Kim PM, Sboner A, Xia Y, Gerstein M. The role of disorder in interaction networks: A structural analysis. Mol Syst Biol. 2008;4:179. doi: 10.1038/msb.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fong JH, Panchenko AR. Intrinsic disorder and protein multibinding in domain, terminal and linker regions. Mol Biosyst. 2010;6:1821–1828. doi: 10.1039/c005144f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fong JH, Shoemaker BA, Garbuzynskiy SO, Lobanov MY, Galzitskaya OV, Panchenko AR. Intrinsic disorder in protein interactions: insights from a comprehensive structural analysis. PLoS Comput Biol. 2009;5:1000316. doi: 10.1371/journal.pcbi.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 35.Cheng Y, Oldfield CJ, Meng J, Romero P, Uversky VN, Dunker AK. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry. 2007;46:13468–13477. doi: 10.1021/bi7012273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, et al. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 37.Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, et al. Characterization of molecular recognition features, MoRFs and their binding partners. J Proteome Res. 2007;6:2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sigalov AB, Kim WM, Saline M, Stern LJ. The intrinsically disordered cytoplasmic domain of the T Cell receptor zeta chain binds to the Nef protein of Simian immunodeficiency virus without a disorder-to-order transition. Biochemistry. 2008;47:12942–12944. doi: 10.1021/bi801602p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams RJ. Signalling: Basics and evolution. Acta Biochim Pol. 2004;51:281–298. [PubMed] [Google Scholar]
- 40.Hashimoto K, Madej T, Bryant SH, Panchenko AR. Functional states of homooligomers: Insights from the evolution of glycosyltransferases. J Mol Biol. 2010;399:196–206. doi: 10.1016/j.jmb.2010.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sigalov AB. Signaling chain homooligomerization (SCHOOL) model. Adv Exp Med Biol. 2008;640:121–163. doi: 10.1007/978-0-387-09789-3_12. [DOI] [PubMed] [Google Scholar]
- 42.Hegyi H, Schad E, Tompa P. Structural disorder promotes assembly of protein complexes. BMC Struct Biol. 2007;7:65. doi: 10.1186/1472-6807-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reth M. Antigen receptor tail clue. Nature. 1989;338:383–384. [PubMed] [Google Scholar]
- 44.Wu J, Cherwinski H, Spies T, Phillips JH, Lanier LL. DAP10 and DAP12 form distinct, but functionally cooperative, receptor complexes in natural killer cells. J Exp Med. 2000;192:1059–1068. doi: 10.1084/jem.192.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pitcher LA, van Oers NS. T-cell receptor signal transmission: Who gives an ITAM? Trends Immunol. 2003;24:554–560. doi: 10.1016/j.it.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 46.Pike KA, Baig E, Ratcliffe MJ. The avian B-cell receptor complex: Distinct roles of Igalpha and Igbeta in B-cell development. Immunol Rev. 2004;197:10–25. doi: 10.1111/j.0105-2896.2004.0111.x. [DOI] [PubMed] [Google Scholar]
- 47.Storch B, Meixlsperger S, Jumaa H. The Igalpha ITAM is required for efficient differentiation but not proliferation of pre-B cells. Eur J Immunol. 2007;37:252–260. doi: 10.1002/eji.200636667. [DOI] [PubMed] [Google Scholar]
- 48.Gazumyan A, Reichlin A, Nussenzweig MC. Igbeta tyrosine residues contribute to the control of B cell receptor signaling by regulating receptor internalization. J Exp Med. 2006;203:1785–1794. doi: 10.1084/jem.20060221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin S, Cicala C, Scharenberg AM, Kinet JP. The Fc(epsilon)RIbeta subunit functions as an amplifier of Fc(epsilon)RIgamma-mediated cell activation signals. Cell. 1996;85:985–995. doi: 10.1016/s0092-8674(00)81300-8. [DOI] [PubMed] [Google Scholar]
- 50.Sanchez-Mejorada G, Rosales C. Signal transduction by immunoglobulin Fc receptors. J Leukoc Biol. 1998;63:521–533. doi: 10.1002/jlb.63.5.521. [DOI] [PubMed] [Google Scholar]
- 51.Lysechko TL, Ostergaard HL. Differential Src family kinase activity requirements for CD3zeta phosphorylation/ZAP70 recruitment and CD3epsilon phosphorylation. J Immunol. 2005;174:7807–7814. doi: 10.4049/jimmunol.174.12.7807. [DOI] [PubMed] [Google Scholar]
- 52.Kuhns MS, Davis MM. Disruption of extracellular interactions impairs T cell receptor-CD3 complex stability and signaling. Immunity. 2007;26:357–369. doi: 10.1016/j.immuni.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 53.Chau LA, Bluestone JA, Madrenas J. Dissociation of intracellular signaling pathways in response to partial agonist ligands of the T cell receptor. J Exp Med. 1998;187:1699–1709. doi: 10.1084/jem.187.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jensen WA, Pleiman CM, Beaufils P, Wegener AM, Malissen B, Cambier JC. Qualitatively distinct signaling through T cell antigen receptor subunits. Eur J Immunol. 1997;27:707–716. doi: 10.1002/eji.1830270320. [DOI] [PubMed] [Google Scholar]
- 55.Pitcher LA, Mathis MA, Young JA, Deford LM, Purtic B, Wulfing C, et al. The CD3gammaepsilon/deltaepsilon signaling module provides normal T cell functions in the absence of the TCRzeta immunoreceptor tyrosine-based activation motifs. Eur J Immunol. 2005;35:3643–3654. doi: 10.1002/eji.200535136. [DOI] [PubMed] [Google Scholar]
- 56.Kesti T, Ruppelt A, Wang JH, Liss M, Wagner R, Tasken K, et al. Reciprocal regulation of SH3 and SH2 domain binding via tyrosine phosphorylation of a common site in CD3{epsilon} J Immunol. 2007;179:878–885. doi: 10.4049/jimmunol.179.2.878. [DOI] [PubMed] [Google Scholar]
- 57.Chae WJ, Lee HK, Han JH, Kim SW, Bothwell AL, Morio T, et al. Qualitatively differential regulation of T cell activation and apoptosis by T cell receptor zeta chain ITAMs and their tyrosine residues. Int Immunol. 2004;16:1225–1236. doi: 10.1093/intimm/dxh120. [DOI] [PubMed] [Google Scholar]
- 58.Cheng Y, LeGall T, Oldfield CJ, Dunker AK, Uversky VN. Abundance of intrinsic disorder in protein associated with cardiovascular disease. Biochemistry. 2006;45:10448–10460. doi: 10.1021/bi060981d. [DOI] [PubMed] [Google Scholar]
- 59.Garner E, Romero P, Dunker AK, Brown C, Obradovic Z. Predicting binding regions within disordered proteins. Genome Inform Ser Workshop Genome Inform. 1999;10:41–50. [PubMed] [Google Scholar]
- 60.Callaghan AJ, Aurikko JP, Ilag LL, Gunter Grossmann J, Chandran V, Kuhnel K, et al. Studies of the RNA degradosome-organizing domain of the Escherichia coli ribonuclease RNase E. J Mol Biol. 2004;340:965–979. doi: 10.1016/j.jmb.2004.05.046. [DOI] [PubMed] [Google Scholar]