Abstract
Rap1 is a Ras-like GTPase that has been studied with respect to its role in cadherin-based cell adhesion. Rap1 exists as two separate isoforms, Rap1A and Rap1B, which are 95% identical and yet the phenotype of the isoform-specific knockout mice is different. We and others have previously identified a role for Rap1 in regulating endothelial adhesion, junctional integrity and barrier function; however, these early studies did not distinguish a relative role for each isoform. To dissect the individual contribution of each isoform in regulating the endothelial barrier, we utilized an engineered microRNA-based approach to silence Rap1A, Rap1B or both, then analyzed barrier properties of the endothelium. Electrical impedance sensing experiments show that Rap1A is the predominant isoform involved in endothelial cell junction formation. Quantification of monolayer integrity by VE-cadherin staining revealed that knockdown of Rap1A, but not Rap1B, increased the number of gaps in the confluent monolayer. This loss of monolayer integrity could be rescued by re-expression of exogenous Rap1A protein. Expression of GFP-tagged Rap1A or 1B revealed quantifiable differences in localization of each isoform, with the junctional pool of Rap1A being greater. The junctional protein AF-6 also co-immunoprecipitates more strongly with expressed GFP-Rap1A. Our results show that Rap1A is the more critical isoform in the context of endothelial barrier function, indicating that some cellular processes differentially utilize Rap1A and 1B isoforms. Studying how Rap1 isoforms differentially regulate EC junctions may thus reveal new targets for developing therapeutic strategies during pathological situations where endothelial barrier disruption leads to disease.
Key words: Rap1A, Rap1B, GTPase, endothelial cell, cell-cell adhesion, cell-cell junctions
Introduction
Endothelial cells (ECs) lining blood vessels and the cell-cell junctions which link them act as the primary barrier of the vasculature. Strict regulation of the passage of fluids, immune cells and macromolecules across this barrier is required during normal physiological processes such as immune surveillance, antigen recognition and acute inflammation. Misregulation of the vascular barrier occurs in the context of many pathological situations, including acute and chronic inflammatory diseases, ischemia-reperfusion injury and atherosclerosis. Adjacent endothelial cells are physically connected by transmembrane proteins present at both adherens and tight junctions. Vascular endothelial cadherin (VE-cadherin) is one of the most studied transmembrane EC junctional proteins with respect to regulating permeability of ECs, but others such as occludin, claudins, nectin and the JAM family are also highly relevant. At the cytoplasmic face of cell junctions, a collection of proteins including ZO-1/ZO-2/ZO-3 and α-catenin bind to and potentially link the transmembrane proteins to the actin cytoskeleton.1–4 The association of junctional proteins with F-actin is required for the dynamic opening and resealing of EC junctions during permeability fluctuations. Because of the relationship between the cytoskeleton and junctional proteins, cellular signaling pathways that control cytoskeletal dynamics have been the focus of many studies. In ECs, the association of junctions with cortical actin bundles correlates with stable, mature junctions.5,6 Recent studies have also shown that cell-cell junctions can associate with stress fibers that insert from neighboring cells, this type of interaction is promoted by certain inflammatory signals and is thought to contribute to tension-driven opening of cell-cell junctions.7
Of the signaling molecules recruited to EC junctions, Rho family GTPases are known to be key regulators of cytoskeletal dynamics; they participate in signaling pathways that affect cell migration, adhesion, actin cytoskeleton remodeling,8 and cell-cell junctions.9 RhoA is activated downstream of several EC adhesion molecules,10–12 and the resulting actomyosin contractility and formation of stress fibers is thought to be a primary mechanism for weakening EC junctions (reviewed in ref. 13). On the other hand, active Rac1 GTPase has been shown to be both barrier protective and barrier disruptive, depending on the cell type and stimulus.14–16 In general, the presence of cortical actin bundles in the cell periphery promotes junctional stability and function.17 However, under conditions of increased permeability and junctional instability, induction of cytoplasmic stress fibers occurs. Insertion of these stress fibers into junctional sites7 and the resulting generation of tension are proposed to be major mechanisms of junctional breakdown and induction of permeability.9
Although Rho family GTPases have received the majority of attention with regard to regulation of the vascular barrier, we have had a long-standing interest in a different GTPase, Rap1, which is a member of the Ras superfamily of small GTPases. Rap1 has previously been shown by our lab and others, to be involved in regulating the assembly and permeability of EC junctions.5,18–23 Rap1 activation and subsequent EC junctional strengthening have also been implicated as mechanisms for inhibiting monocyte transendothelial migration.23 The junctional strengthening effect of Rap1 activation has been correlated with rearrangement of the actin cytoskeleton into prominent perijunctional ring structures and loss of actin stress fibers.6,18,20,24 As mentioned above, switching between these actin phenotypes (perijunctional vs. stress fibers), may provide a mechanism for regulating junctional dynamics.
Several guanine nucleotide exchange factors (GEFs) for Rap1 have been identified including Epac1, PDZGEF-1/2 and C3G; some have been specifically implicated in Rap1 activation during EC junctional regulation.20–22,24 GTPase activating proteins (GAPs) which inactivate Rap1 include Spa-1 and Rap1GAP.25,26 There are only a few known effectors for Rap1. Some of these effectors, including afadin/AF6,27 KRIT-1/CCM1,28 and MAGI-1,21 are localized to EC junctions. Of these, afadin/AF6 has recently been implicated together with Rap1 in promoting various VEGF and S-1-P mediated angiogenic responses such as migration, tube formation and PI3K signaling.29
Importantly, what is often referred to in the literature as “Rap1” is actually two separate isoforms, Rap1A30,31 and Rap1B.32,33 The isoforms share 95% amino acid sequence identity, are both post-translationally modified by geranylgeranylation, and are ubiquitously expressed in many tissues. In fact, until relatively recently, little distinction has been explicitly made between them. For example, two commonly employed means to experimentally target Rap1 GTPase are the drug 8-pCPT-O'-Me-cAMP which activates Rap GTPases via the GEF Epac and ectopic expression of Rap1GAP which inhibits Rap GTPase activity. Neither of these approaches specifically targets Rap1A or 1B isoforms, or even distinguishes between Rap1 and Rap2.34,35
Knockout mice specific for each isoform exist, although the double knockout is lethal.36,37 Surviving Rap1A-null mice develop normally, with no gross abnormalities.37,38 However, upon backcrossing for 6 generations into a C57BL/6J mouse strain, the Mendelian inheritance ratio of Rap1A−/− pups from heterozygous crosses was found to be very slightly reduced (from 25% to ∼15%),37 indicating that there is some strain dependence with respect to severity of phenotype. Rap1A-null T and B cells exhibit defects in adhesion in vitro, but this did not translate into hematopoietic or cell homing deficiencies in vivo.37 More recently, using a hind-limb ischemia model, it has been demonstrated that Rap1A-null mice have reduced neovascularization responses,39 but overall the phenotype can be considered mild. By contrast, Rap1B-null mice have a more severe phenotype, exhibiting up to 85% late embryonic and perinatal lethality, due to complications arising from embryonic hemorrhage.40 Surviving Rap1B-null mice are smaller, and exhibit a bleeding defect due to decreased integrin-mediated cell adhesion and reduced platelet aggregation. Further analysis of Rap1B−/− mice also revealed defective in vivo angiogenesis in response to VEGF and FGF2.36 Thus, the close homology of Rap1A and Rap1B isoforms at the amino acid level is at odds with the observed phenotypic differences of the mouse knockouts.
Recently, more attention has been paid to identifying Rap1 isoform-specific functions, although the results have often been conflicting. For example, it has been reported that EC proliferation, EC permeability, and/or cell migration require Rap1A only,29,41 Rap1B,36 or both Rap1A and 1B.39,42 The results have varied depending on cell type (epithelial vs. endothelial), subtype (microvascular endothelial vs. HUVEC) or method of knockdown (genetic knockout vs. various siRNA methods). In addition, not all of these studies looked at both isoforms, focusing instead on one or the other. Thus, determining the relative importance of Rap1A vs. 1B with respect to EC junctional regulation remains a key question. Given our interest in Rap1 biology and EC barrier function, we set out to establish whether Rap1A and 1B isoforms differentially regulate junctional barrier properties.
Results
As discussed in the Introduction, isoform-specific Rap1 null animals exhibit unique phenotypes despite the striking similarity between Rap1A and Rap1B at the amino acid level. Amino acid alignment reveals there are only 9 differences in a protein containing a total of 184 residues, representing an identity of 95% (Fig. 1). Even more intriguing is the fact that of these mismatches, 6 are clustered within the last 13 amino acids of the protein, in a region known as the C-terminal hypervariable region.43,44 Previous studies have shown that the localization and membrane targeting of various GTPases is, in part, dictated by the properties of this domain (reviewed in ref. 45). To explore this, the first property we examined was cellular localization of Rap1A versus Rap1B. Adenoviruses encoding expression vectors for each isoform, GFP-tagged at the N-terminus, were used to infect confluent HUVEC grown on coverslips. GFP-fluorescence imaging of a wide area of monolayer reveals that GFP-Rap1A is found localized to EC junctions throughout the field of view. GFP-Rap1B exhibits less junctional staining and a more cytoplasmic, perinuclear staining pattern (Fig. 2A). Costaining these monolayers with a VE-cadherin antibody confirms that GFP-Rap1A is present at cell-cell junctions; Rap1A colocalizes strongly with this adherens junction marker (Fig. 2B). By contrast, GFP-Rap1B is found mainly concentrated in a perinuclear distribution and displays weaker colocalization with VE-cadherin. The enlarged color merge images serve to highlight these differences in localization (Fig. 2B and lower panels). For a more quantitative analysis of the localization differences between isoforms, we performed image analysis to determine the relative junctional intensity of Rap1A compared with Rap1B. As quantified in Figure 2C, the pool of Rap1A at cell-cell junctions is significantly greater than the junctional pool of Rap1B.
Figure 1.
Rap1A and Rap1B isoforms differ in only 9 amino acid residues. Protein sequence alignment of human Rap1A and Rap1B; amino acids that are different are highlighted in red and indicated by asterisk. Alignment is based on NCBI RefSeq: Rap1A (NM_001010935.1) and Rap1B (NM_001010942.1).
Figure 2.
Rap1A and Rap1B exhibit differences in subcellular localization; Rap1A is more prominent at cell junctions, co-localizing with VE-cadherin and co-immunoprecipitating with AF6. (A) GFP-tagged Rap1A or Rap1B were exogenously expressed in HUVEC and localization was visualized by fluorescence microscopy. Rap1A isoform is concentrated at cell-cell junctions; Rap1B has both perinuclear and weaker junctional fluorescence. Scale bar = 100 µm. (B) Localization of GFP-Rap1A (left column) or GFP-Rap1B (right column) and co-staining for VE-cadherin. Boxed areas in each VE-cadherin image have been enlarged in the merged parts, identifying Rap1 isoform localization in green and VE-cadherin localization in red. Rap1A more strongly colocalizes with VE-cadherin in areas of cell-cell contact, as indicated by yellow/orange coloring in the merged part (left). Scale bar = 50 µm. (C) Quantification of GFP-Rap1A vs. Rap1B junctional pixel intensity relative to total cellular GFP fluorescence. VE-cadherin was used as a marker for cell-cell junctions. Images were analyzed using ImageJ as described in Methods. The junctional proportion of Rap1A is significantly higher than Rap1B. Graph represents average junctional intensity of >10 cells per field (n = 9 fields) from 3 independent experiments *p < 0.01. (D) GFP-tagged proteins (GFP-CA-Rap1A, CA-1B or GFP alone) were expressed in HUVEC , followed by immunoprecipitation with anti-GFP antibodies. Lysates and IP samples were blotted with indicated antibodies. The junctional protein AF-6 more strongly co-immunoprecipitates with GFP-Rap1A. Equivalent expression level of GFP-Rap1A and 1B, was confirmed; endogenous Rap and AF-6 protein present in total cell lysates indicate equal loading.
We next looked at the interaction between Rap1 isoforms and the junctional scaffold protein AF-6, which is a known Rap1 effector.26 We observed that GFP-tagged constitutively active (CA) Rap1A co-immunoprecipitates more afadin/AF6 than does GFP-CA-Rap1B (Fig. 2D). Longer exposure of the western blot reveals a weak band of AF-6 coimmunoprecipitating with GFP-Rap1B (data not shown). Confirmation that AF-6 is localized to EC junctions under these conditions is shown as supplemental data. Also apparent from these images (Sup. Fig. 1) is that expression of CA-Rap1A, but not CA-Rap1B, promotes a more linear and less jagged junctional morphology, and that CA-Rap1A is more localized to cell-cell contacts. Taken together, these data are consistent with AF-6 being a cell junction-enriched Rap1 effector protein26,27,46 and provides corroborating evidence of our observation that Rap1A is enriched at cell-cell junctions, as this isoform more efficiently co-immunoprecipitates with AF-6.
Studying the unique functions of Rap1A and Rap1B in primary ECs requires a method to efficiently knockdown expression of each isoform in a specific and effective manner. To this end, we utilized an adenovirally-encoded “engineered” microRNA-based approach. We generated adenoviruses for knockdown of Rap1A (utilizing 2 different targeting sequences), Rap1B or a non-targeting negative control shRNA sequence. For knockdown of total Rap1, both Rap1A and 1B shRNA virus was added to cells. In our hands, adenoviral infection efficiency of HUVEC approaches 100% as determined by the co-cistronic expression of a GFP marker (data not shown). Lack of commercially available isoform-specific antibodies makes it difficult to analyze knockdown by traditional western blot methods. However, it has previously been shown that in ECs, Rap1B accounts for >90% of total cellular Rap1.36 With this in mind, the western blot in Figure 3A confirms that knockdown of each isoform does occur. The remaining Rap1 signal in the 1A knockdown lane likely represents the more abundant Rap1B isoform, confirming previous observations that Rap1B is the major isoform in ECs.36 In order to conclusively determine specificity of knockdown, we additionally analyzed mRNA levels using RT-PCR with isoform-specific primers. As shown in Figure 3B, each shRNA efficiently reduced mRNA levels of the appropriate isoform. Furthermore, we characterized two separate Rap1A shRNA viruses (Rap1A#1 and #2), that targeted unique sites within the Rap1A sequence, and both were found to successfully knockdown Rap1A mRNA levels. Importantly, treatment with Rap1A shRNA had no effect on Rap1B mRNA levels and vice versa, indicating isoform specificity of the knockdown virus.
Figure 3.
Characterization of isoform-specific knockdown approach. Knockdown was achieved by using adenovirus-delivered shRNAs, designed to specifically target only one isoform. Negative control shRNA (Neg) uses a sequence identified as non-targeting for any known mammalian gene. (A) western blot of HUVEC lysates 72 hr after induction of shRNA expression. Blotting with a pan-Rap1 antibody reveals knockdown of both isoforms, with combined knockdown (1A + 1B), being most effective at reducing total Rap1 (A + B) protein levels. B-actin serves as a loading control; GFP is co-cistronically expressed with the shRNA sequence and thus serves as a marker for equivalent delivery and expression. (B) Reverse transcriptase-PCR analysis of mRNA levels using isoform-specific PCR primers. Two different Rap1A shRNA targeting sequences were tested. Human B2-microglobulin serves as loading control.
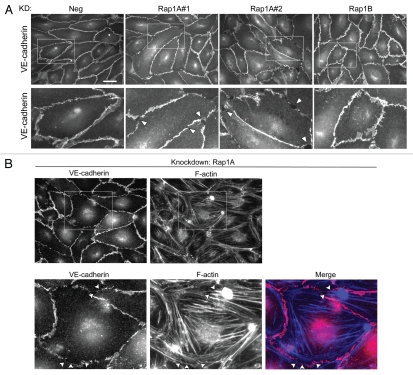
With these tools now at our disposal, we could begin to study the relative contribution of Rap1A versus 1B, with respect to EC junctional regulation. Continuous localization of EC junctional proteins along cell-cell contacts is an important indicator of junctional integrity (reviewed in ref. 47–49). Thus, HUVECs treated with the adenovirus for knockdown of Rap1A, Rap1B or negative control, were plated at confluent density and analyzed post-fixation by immunofluorescent staining of VE-cadherin (Fig. 4A). Knockdown of Rap1A with two separate shRNA sequences disrupted the junctional localization of VE-cadherin compared with control, as indicated by discontinuous staining with a jagged appearance. The Rap1B knockdown cell monolayer lacked this obvious disruption in VE-cadherin. Zoomed in areas (lower row) show that the Rap1A knockdown monolayers have major discontinuities (arrowheads) compared with negative control, while the Rap1B knockdown has more subtle differences in VE-cadherin localization that were more difficult to characterize. In addition to VE-cadherin, we also analyzed the localization of other junctional proteins (Sup. Fig. 2A–C) such as β-catenin, CD31 (PECAM-1) and ZO-1. Similarly, knockdown of Rap1A, but not Rap1B, disrupts the junctional localization of these proteins. Importantly, knockdown did not reduce the overall expression level of VE-cadherin or β-catenin in these cell monolayers (Sup. Fig. 2E).
Figure 4.
VE-cadherin and F-actin localization is disrupted following knockdown of Rap1A. (A) HUVEC s infected with the indicated shRNA viruses were grown for 3 days and replated onto Matrigel-coated coverslips. Cells were fixed 24 hrs after replating on coverslips. (Top row) Adherens junctions were visualized by anti-VE-cadherin immunofluorescent staining. Negative control shRNA-treated and Rap1B knockdown cell monolayers are visually intact; knockdown of Rap1A using two independent shRNAs causes disruption of the endothelial cell monolayer. Scale bar = 30 µm. (Bottom row) Boxed areas from top row images are enlarged to show the gaps and discontinuities in VE-cadherin staining in the Rap1A#1 and #2 knockdown monolayers (several gaps highlighted by arrowheads). (B) HUVEC were treated with Rap1A shRNA knockdown virus as in (A), followed by co-staining for VE-cadherin (red) and F-actin (blue). Boxed areas are enlarged in bottom row parts. In addition to gaps in the VE-cadherin staining pattern, the F-actin cytoskeleton reorganizes to form prominent stress fibers after knockdown of Rap1A. Arrowheads indicate gaps where F-actin fibers are inserted.
We also looked at changes in organization of the F-actin cytoskeleton, and observed that loss of Rap1A correlated with an increase in the amount of stress fibers and gaps, along with decreased perijunctional actin (Sup. Fig. 2D). The increased appearance of stress fibers in Rap1 knockdown cells was explored in more detail by co-immunofluorescence imaging with VE-cadherin (Fig. 4B). High magnification images of Rap1A knockdown cells shows that the discontinuous VE-cadherin staining at cell gaps is also where F-actin stress fibers appear to insert (arrowheads). Thus, the localized disruption of VE-cadherin at gaps may be the result of stress fiber-induced tension, as described previously in reference 7.
As a more quantitative gauge of EC junctional integrity, we determined the number of cells with gaps or discontinuities in VE-cadherin staining within a given monolayer following knockdown of each Rap1 isoform singly, and combined (“both”). To simplify the quantification, knockdown cells were cocultured with uninfected cells at a ratio (∼1:4) to ensure that a given knockdown cell would be relatively isolated from others. This allowed for a more accurate scoring of individual cells with gaps. An example image showing the criteria used to make this assessment is shown in Figure 5A, where gaps in VE-cadherin staining around a knockdown cell (GFP-positive) are indicated by arrows. Percent of knockdown cells with gaps combined from 3 independent experiments (500 cells for each condition) is quantified in Figure 5B. The graph shows that loss of Rap1A induced a significantly greater percentage of cells that exhibit junctional gaps or discontinuities (54.7% ± 6.1) compared with control (13.2% ± 8.9) or Rap1B knockdown (30.8% ± 5.9). While knockdown of Rap1B had a modest effect on junctional disruption, importantly, the combined knockdown of both isoforms together was not additive.
Figure 5.
Quantification of monolayer gaps following knockdown of either Rap1A or Rap1B singly or both together. HUVEC were infected with shRNA virus for 3 days, trypsinized and replated in co-culture with uninfected HUVEC at a ratio of ∼1:4 (infected:uninfected). (A) Representative image of scoring criteria for quantification. Image is of GFP-positive cells (i.e., shRNA-expressing), together with uninfected cells (GFP-negative). Cell junctions are visualized using anti-VE-cadherin antibodies (red). Box (i) is enlarged to show a representative cell that has gaps in the VE-cadherin staining pattern (indicated by arrows). Box (ii) is enlarged to show a GFP-positive cell scored as having no gaps (cells indicated by asterisks). (B) Quantification of monolayer disruption using the criteria shown in (A) reveals that knockdown of Rap1A results in a significantly greater percentage of cells with gaps/discontinuities compared to both negative control shRNA or Rap1B knockdown. Combined knockdown of both Rap1A and Rap1B together was not additive. Data represents percentage of cells with gaps counted from 3 independent experiments and a total of 500 cells counted for each condition. Comparing knockdown of Rap1A to either Neg or 1B knockdown, **p < 0.01. Comparing 1B knockdown to the Neg control, *p = 0.046. Combined knockdown is not significantly different (NS) from 1A knockdown alone, p = 0.47. (C) Knockdown/rescue experiment: HUVEC were infected with shRNA virus to knockdown total Rap1 (both 1A and 1B) for 48 hrs, followed by 48 hr re-expression (“RE ”) of wildtype Rap1A, Rap1B, GFP only or nothing (knockdown only). The percentage of cells with gaps was quantified as indicated above. Re-expression of wt-Rap1A in 1A + 1B knockdown cells results in a significant reduction in the percentage of cells with gaps/discontinuities compared to both knockdown alone (*p = 0.014) or knockdown/re-expression of wt-Rap1B (*p = 0.002). Re-expression of wt-Rap1B does not significantly reduce the number of cells with gaps compared to KD alone or KD/RE GFP (NS). Data represents average ± SD from 2 independent experiments.
To confirm the specific requirement for Rap1A in the regulation of EC junctional integrity, and to rule-out off-target effects, knockdown/rescue experiments were also performed. HUVEC in which both Rap1A and Rap1B were knocked down (i.e., total Rap1), were treated with adenovirus to reexpress either nothing, GFP alone, wildtype Rap1A or wildtype Rap1B, and then monolayer gaps were quantified (Fig. 5C). Significant reduction of gaps (i.e., rescue) was seen only when Rap1A was re-expressed. Re-expression of Rap1B or GFP alone had no significant difference compared to knockdown only. Together, these results suggest that Rap1A is the more important isoform when it comes to EC junctional protein localization.
To determine which isoform is required for maintenance of junctional barrier function, we used an electrical impedance assay to quantify monolayer integrity.50,51 The RTCA (Real-Time Cell Analysis) system is an automated real-time method to monitor the impedance generated by a monolayer of cells grown on microelectrode sensors. ECs were preinfected with indicated shRNAs, trypsinized and replated at confluent cell density onto microelectrode coated dishes and analyzed for 24 hrs. A representative impedance trace (represented as Cell Index) is shown in Figure 6A. Single knockdown of Rap1A, but not Rap1B reduced the steadystate, plateau level of impedance after 24 hrs compared with negative control knockdown monolayers. Combined knockdown of both isoforms reduced the level of impedance similar to Rap1A knockdown alone. The relative Cell Index at the 24 hr time point (i.e., steady state) combined from 3 individual RTCA experiments is shown graphically in Figure 6B. At 24 hours, knockdown of Rap1A but not Rap1B results in a significant decrease in impedance relative to negative control cells (Rap1A knockdown, 73.6% ± 13.8 of control levels; Rap1B knockdown, 94.7% ± 5.2 of control). To confirm these results, a second shRNA designed to target a different region of Rap1A sequence was also used in separate experiments. Supplemental Figure 3 graph shows the relative Cell Index (impedance) after 24 hrs from 3 additional RTCA experiments. Both Rap1A#1 and Rap1A#2 shRNA treatment significantly decreased monolayer impedance compared to control. These results point to a more critical need for Rap1A, rather than Rap1B for acquisition of a strong EC monolayer barrier at steady state conditions. Interestingly, we found that not all cellular events differentially utilized Rap1 isoforms. In agreement with previously published work,36,42 we observed that loss of either isoform inhibited directed chemotactic migration of ECs (Sup. Fig. 4). Thus, it seems that some, but not all, physiological events are differentially impacted by Rap1A and Rap1B isoforms.
Figure 6.
Real-time Cell Impedance Assay (RTCA) shows that Rap1A but not Rap1B is required for steady-state acquisition of monolayer barrier function. HUVEC expressing indicated shRNAs were cultured on microelectrode-coated surfaces and monolayer integrity was monitored for 24 hrs using the Roche xCE LLigence system. (A) Representative trace of impedance (graphed as Cell Index) taken every 15 min; lower Cell Index values indicate decreased monolayer barrier function. Each data point is presented as average cell index of at least triplicate wells ±SD for each condition. (B) Graph showing relative cell index (impedance) of HUVEC monolayer having Rap1A, Rap1B, or both combined knockdown compared to negative control. Data is the average ± SD combined from 3 independent experiments. *p ≤ 0.015, comparing Rap1A and combined knockdown to Neg control.
Discussion
In this study, our aim was to identify unique and/or complementary roles of the closely related Rap1 isoforms, Rap1A and Rap1B, in endothelial cells. Our previous research identified a role for Rap GTPases during EC junctional regulation, in particular during leukocyte transendothelial migration.23 However, the relative contribution of each isoform was not identified in this earlier work and isoform-specific functional information is either lacking or contradictory. The significant differences in phenotypes of the isoform-specific knockout mice support the idea that Rap1A and Rap1B may serve non-redundant functions. To examine the individual contributions of each isoform we used an engineered microRNA-knockdown approach to selectively reduce expression of one or both isoforms. Surprisingly, given the extremely high amino acid sequence identity (95%) between the two isoforms, we found that the Rap1A isoform is preferentially required for ECs to acquire steady-state monolayer barrier The discontinuous localization of VE-cadherin and other junctional proteins along cell-cell contacts which we observed following Rap1A knockdown (Fig. 4 and Sup. Fig. 2) is reminiscent of what has been seen in other scenarios of EC barrier disruption. Expression of constitutively active RhoA or treatment with permeability-inducing agents such as thrombin52,53 and TNFα54 induce phenotypically similar gaps in the VE-cadherin staining pattern to what we see with loss of Rap1A expression. This phenomenon has also been seen in vivo; Adamson et al. observed that perfusion of platelet-activating factor resulted in frequent short gaps in the otherwise continuous junctional VE-cadherin staining pattern in mesenteric microvessels.55 Finally, several studies have observed that depletion of certain proteins can also cause gaps in VE-cadherin staining. For example, knockdown of caveolin1 in brain microvascular ECs causes gaps in the junctional localization of VE-cadherin, ZO-1 and B-catenin.56 More recently, knockdown of PKA-C, PDE4D and interestingly, the Rap1 GEF, Epac1, have all been shown to cause discontiguous VE-cadherin staining in ECs, correlating with increased vascular permeability.57
The observed mis-localization of junctional proteins after specific knockdown of Rap1A correlated with the increase in monolayer gaps in the Rap1A compared to 1B knockdown cells (Fig. 5). In addition, electrical impedance measurements confirmed that Rap1A is the isoform that plays the dominant role for EC barrier function (Fig. 6 and Sup. Fig. 3). Significantly, combined knockdown of both isoforms was not additive for junction disruption in either of these assays, which suggests that Rap1A is uniquely regulating EC barrier function under our conditions. Of note, the impedance trace of the combined knockdown cells lagged compared to Rap1A-only knockdown cells (Fig. 6A). Our interpretation of this is that combined knockdown is delaying the initial cell adhesion and spreading phase in these impedance experiments (i.e., at the early time points of <4 hrs). This would suggest that both Rap1 isoforms may play a role in mediating the well-described process of talin-dependent integrin activation (reviewed in ref. 58). However, upon reaching plateau levels of impedance, a readout for EC barrier properties (i.e., after 24 hrs), combined knockdown was not additive. The importance of Rap1A in EC junction formation is underscored by the knockdown/rescue experiment (Fig. 5C). Upon combined knockdown of both isoforms, re-expression of shRNA-resistant Rap1A, but not Rap1B, significantly reduced the amount of gaps within the monolayer, confirming specificity and ruling out the possibility that Rap1B can compensate for Rap1A in the context of junctional barrier formation.
It is interesting to note that not all assays we performed displayed such isoform-specific differences. Another often-studied function of Rap1 GTPases is the regulation of cell migration, which is essential for angiogenic processes (reviewed in ref. 59). In epithelial cells knockdown of Rap1A, but not 1B, inhibited cell migration in a wound healing assay, correlating with a decrease in β1 integrin expression.41 However, in endothelial cells (both microvascular ECs and HUVECs), the consensus seems to be that both isoforms are required for effective migration in wound healing assays,36,39,42 an observation that our migration data confirms (Sup. Fig. 4). These differences may reflect cell type-specific functions for Rap1A and Rap1B.
With respect to EC junctional regulation and permeability, some studies have shown that siRNA-mediated knockdown of either Rap1A or Rap1B decreased transendothelial electrical resistance (TER),42 or enhanced FITC-dextran permeability.60 We found that only knockdown of Rap1A had a significant effect on electrical impedance or localization of VE-cadherin to cell-cell junctions, which are both experimental readouts for EC barrier function. The discrepancy with our results could be due to any one of a number of factors, such as differences in assay type, or the use of microvascular endothelial cells compared to HUVEC in our study, as vascular cell subtype can influence response61 (and reviewed in refs. 62 and 63). Another possibility that cannot be ruled out given the published data is if the “smartpool” of siRNA used to silence Rap1B in those studies also contained sequences that targeted Rap1A. These studies relied solely on western blot analysis to confirm siRNA specificity; however, many of the commercially available antibodies do not distinguish between isoforms. Thus in our study, unlike these, in addition to performing western blot analysis of knockdown, we used RT-PCR to confirm specificity of knockdown at the mRNA level and to ensure that our shRNA target sequences did not cross react between Rap1A and Rap1B (Fig. 3B). Furthermore, the conclusions described in our paper are supported by multiple lines of evidence, i.e., both the monolayer gap quantification, including the knockdown/rescue experiment (Fig. 5) and the RTCA impedance data (Fig. 6) support our conclusion that Rap1A, and not Rap1B is the critical isoform for EC junctional formation and barrier function at steady state.
Differences in function of Rap1A versus Rap1B may be explained by divergent downstream signaling and/or binding to unique effectors. Although it is tempting to speculate, the effector residues (aa 20–45) including the core effector binding domain64 are identical between isoforms, making it unlikely that this is the mechanism that accounts for their divergent roles in ECs. Instead, differential localization may be responsible for creating functional specificity. The subcellular localization of Rap1A/B (isoforms not distinguished) has been described previously as perinuclear,65 in late endosomes/lysosomes,66 golgi-localized,67 at the plasma membrane,68 or even nuclear in squamous cell carcinomas,69 depending on cell type and culture conditions. In epithelial cells, GFP-Rap1 was found at sites of cell-cell contact, colocalizing with E-cadherin, however, which isoform of Rap1 used was not disclosed in this paper.70 Recently, GFP-Rap1 (isoform not specified) was found to be recruited to the leading edge during migration of Drosophila immune cells undergoing developmental migration71 and in endothelial cells during wound healing,39 indicating there may be a dynamic regulation of localization during certain cellular events. Here we explored whether the Rap1 isoforms localize to different subcellular regions, which might account for their functional specificity. Our immunofluorescence data suggests that Rap1A is relatively more localized to EC junctions compared to Rap1B (Fig. 2A and B). Quantitative analysis of the junctional intensity of each Rap1 isoform confirmed this observation (Fig. 2C). A potential caveat here is the use of GFP-tagged proteins for localization studies. Ideally one would examine the localization of endogenous protein using specific antibodies. As isoform-specific antibodies amenable for immunofluorescence applications are not currently available, this remains an experiment for future studies.
On the other hand, our observation that Rap1A is more concentrated at EC junctions is strengthened by the AF-6 co-immunoprecipitation experiment (Fig. 2D). Previous studies have shown that Rap1A can bind to the junctional scaffold protein AF-6 in vitro and is recruited to cell-cell contacts by binding to AF-6 in mammalian cells.46 Even more interesting is the observation that AF-6 specifically binds Rap1-GTP (isoform not specified),26 consistent with AF-6 being a Rap1 effector at junctions. We therefore tested whether AF-6 preferentially interacted with one isoform over the other. The co-immunoprecipitation data indicate that AF-6 interacts preferentially with Rap1A under our conditions (Fig. 2D). Longer exposure of the western blot shows that there is a small amount AF-6 in the Rap1B pulldown, therefore we cannot say that the interaction is isoform-specific. Rather, it suggests that Rap1A, being enriched at cell junctions, is at the right location in the cell to interact with AF-6.
Our finding that loss of Rap1A has the most dramatic effect on EC junctions is particularly interesting given the fact that as described for other cell types,40,72,73 Rap1A is expressed at a far lower level than Rap1B in these ECs (Fig. 3). Put another way, cells lacking Rap1A still have a large amount of Rap1B protein present, as it accounts for ∼90% of total cellular Rap1 protein, and yet EC barrier function is still compromised. This argues strongly for divergent functions for these closely related isoforms as the presence of Rap1B evidently does not fully compensate for the loss of Rap1A, at least in the context of EC junctional organization and barrier function. In conclusion, the data presented in this study suggest that there are isoform-specific functional differences for the closely related Rap1 GTPases, with Rap1A being the critical isoform during the formation and maintenance of EC junctions and barrier properties.
Materials and Methods
Cell culture.
Human umbilical vein endothelial cells (HUVECs) were obtained commercially (Lonza, CC-2519). Cells were grown as recommended in EGM-2 media (Lonza, CC-3162) and cultured in a 37°C incubator with 10% CO2. All experiments were performed with low passage HUVEC (i.e., p6 or lower). 293A cells (Invitrogen, R705-07) were grown in DMEM (high glucose) with 10% FBS, non-essential amino acid supplement and Pen/Strep (Gibco, 11995, 16000-044, 11140050 and 15140122).
Knockdown of Rap1A and Rap1B using miRNA adenovirus.
Generation of miRNA constructs. MicroRNA (miRNA) adenoviral constructs were engineered using the BLOCK-iT™ Pol II miR RNAi expression vector system (Invitrogen, K4941-00K4936-00) according to the manufacturer's protocol. In this system, virally encoded miRNA is processed by the endogenous cellular machinery where it functions like an shRNA to result in cleavage of the mRNA target. Double-stranded oligonucleotides were designed using Invitrogen's RNAi Designer (www.invitrogen.com/rnai) to form an engineered pre-miRNA sequence structure that targets unique sequences in human Rap1A or Rap1B. For some experiments, two independent and unique Rap1A targeting sequences were used:
“Rap1A#1”: 5′-TGC TGT ACA CCA CTG TCT TGC TAA ATG TTT TGG CCA CTG ACT GAC ATT TAG CAA CAG TGG TGT A-3′; “Rap1A#2”: 5′-TGC TGA ACC AAG GAC CAC TAG CTT GTG TTT TGG CCA CTG ACT GAC ACA AGC TAG GTC CTT GGT T-3′; and “Rap1B miR”: 5′-TGC TGA ACT AAT GCA AAT CCT TGT CCG TTT TGG CCA CTG ACT GAC GGA CAA GGT TGC ATT AGT T-3′ (21 bp antisense target sequences underlined).
Synthesized oligonucleotides were annealed and ligated into pcDNA 6.2-GW/EmGFP-miR. As a negative control (“Neg shRNA”), we used the pcDNA6.2-GW/± EmGFP-miR-neg control plasmid (Invitrogen, K4936-00), which contains an insert that is processed into a mature miRNA, but is predicted not to target any known vertebrate gene. The EmGFP-miRNA cassette from these constructs was subsequently shuttled through pDONR221 (also from Invitrogen, 12536017) by Gateway BP recombination and then into pAd-CMV-Dest Gateway vector by LR recombination.
Adenovirus production. Virus was produced in 293A packaging cell line with the ViraPower Adenoviral Expression System (Invitrogen, K4930-00) using the manufacturer's recommended protocol. Briefly, 293A cells were transfected with PacI-digested pAd-CMV-Dest vector containing the desired miRNA cassette, using Lipofectamine 2000 (Invitrogen, 11668-019). Mature viral particles were harvested by collecting the cells/media, and subjecting to multiple freeze thaw cycles then centrifugation. Co-cistronically expressed EmGFP serves as a marker for knockdown cells; viral infection efficiency approaches 100% in HUVEC (data not shown).
Infection of ECs with miRNA adenovirus and analysis of knockdown. ECs were infected with adenovirus: negative control shRNA, Rap1A#1 and #2 shRNA, Rap1B shRNA or Rap1A#1 plus Rap1B (in combination) for times indicated in figure legends. Typically virus was added to the culture medium, incubated overnight, with a media change the next day. Efficient knockdown was usually attained within 72 hrs and confirmed regularly by either western blot using a polyclonal antibody that recognizes total Rap1 (Santa Cruz Biotechnology, SC-65) or reverse-transcriptase PCR using isoform-specific primers. Total RNA was isolated using Trizol and cDNA was reverse-transcribed using the iScript cDNA isolation kit (Bio-Rad, 170-8890) according to the manufacturer's protocol. Rap1A and Rap1B-specific primers were subsequently used for PCR analysis. Human β2 microglobulin primers served as control.
Microscopy.
Expression of GFP-tagged Rap1 proteins. GFP-wt-Rap1A was kindly provided by Lawrence Quilliam (University of Indiana), and was subsequently subcloned into the adenoviral expression vector pAdCMV-Dest. The wt-Rap1B construct (RAP1B00000) was obtained through the Missouri S&T cDNA Resource Center (www.cdna.org). To obtain efficient expression of these GFP-tagged proteins, adenoviruses encoding these constructs were generated using the ViraPower Adenoviral Expression System as indicated above.
Immunofluorescence. HUVECs cultured on Matrigel (BD Biosciences, 354230)-coated coverslips were fixed and permeabilized with 3.7% formaldehyde (30 min, RT) and 0.2% Triton X-100/TBS (5 min, RT). The following primary antibodies were used: VE-cadherin (clone F-8, Santa Cruz Biotechnology, SC-9989), β-catenin (Sigma, C2206), ZO-1 (Invitrogen, 339100), CD31 (R & D systems, BBA7) and AF6/afadin (BD Transduction Labs, 610732). F-actin was detected by Texas Red-X-phalloidin or Alexa 350-palloidin (Molecular Probes, T7471 and A22281). The following secondary antibodies were also used: Alexa 594-conjugated anti-mouse and anti-rabbit (Molecular Probes, A11005 and A11012).
Image acquisition and analysis. Fluorescence images were obtained with a Zeiss axiovert 200M microscope equipped with a Hamamatsu ORCA-ERAG digital camera and acquired using Metamorph Workstation (Universal Imaging Corp., META-40002). Adjustments of brightness/contrast and color balance were performed using ImageJ software. Any adjustments made fairly represent the original images, following accepted guidelines for digital image presentation.74 Figures were prepared using Adobe Photoshop software.
Quantification of junctional intensity of Rap1 isoforms. To quantify junctional localization of GFP-Rap1A vs. GFP-Rap1B, images were analyzed using ImageJ. VE-cadherin co-staining was used as a reference marker for cell-cell junctions. For each cell quantified, the “junctional proportion” was quantified as the ratio of junctional GFP integrated pixel intensity to the total cellular GFP intensity. This normalizes for any slight cell-cell variation of GFP-Rap expression levels. The data were graphed as the average “junctional proportion of GFP” in cells expressing either GFP-1A or GFP-1B.
Co-immunoprecipitation experiments.
To detect association between AF6 and Rap1A or 1B, GFP-tagged proteins (GFP-constitutively active Rap1A, -1B or GFP alone) were expressed in HUVEC, followed by immunoprecipitation with an anti-GFP antibody (Roche, 11814460001) coupled to Protein G-sepharose beads (GE Healthcare, 17-0618-01). Total cell lysates and IP samples were western blotted with anti-AF6 (Novus Biologicals, ab11338) and anti-total Rap1 (Santa Cruz, SC-65) antibodies.
Monolayer gap assay.
Knockdown only gap quantification. As a gauge of monolayer integrity, we quantified the amount of “gaps” in a given monolayer, using anti-VE-cadherin antibody (clone F-8, Santa Cruz, SC-9989) as a marker for EC junctions, together with Alexa 594-conjugated anti-mouse IgG secondary (Molecular Probes, A11005). To simplify the analysis and to more accurately compare the extent of monolayer disruption for loss of one isoform over the other, shRNA-expressing cells were co-cultured at a 1:4 ratio with uninfected HUVEC to ensure single-cell distribution of knockdown cells. Knockdown cells (GFP-positive) were scored as positive for having incomplete VE-cadherin junctional staining if there was at least one gap or discontinuity per cell. One hundred cells were counted (in a blinded manner) per condition, per experiment. Results were expressed as percent knockdown cells having gaps, comparing co-cultured single knockdown or combined Rap1A/1B knockdown to negative control shRNA-expressing cells.
Knockdown/Rescue experiments. For knockdown/rescue experiments, total cellular Rap1 (both 1A and 1B isoforms) was first knocked down for 48 hrs, followed by addition of shRNA-resistant GFP-Rap1A, GFP-Rap1B or GFP only re-expression virus for an additional 24 hrs. Knockdown/re-expression cells were then cocultured on coverslips with uninfected HUVEC, and monolayer gaps were quantified as described in previous section. Equal re-expression of Rap1A and Rap1B was confirmed by western blot (data not shown).
Real-time cell analysis (RTCA ) experiments.
Barrier function assays. The xCELLigence Real-Time Cell Analyzer (RTCA) system (Acea Biosciences/Roche Applied Science 05-469-759-001) was used to measure electrical impedance, as a readout of monolayer barrier function. This method uses electrical impedance signals to monitor the status of cells grown directly on micro-electrode coated surfaces. Changes in impedance reflect changes in barrier function and permeability.50 HUVEC preinfected with the indicated shRNA for 48–72 hrs were counted with an automated cell counter (Nexcelom Bioscience), then plated (in quadruplicate) at a confluent density directly onto a microelectrode-surface within the wells of an E-Plate 16 (Roche Applied Science, 05-469-813-001). Impedance readings were taken automatically every 15 min for another 24 hrs and plotted as Cell Index ± SD. Confirmation of equal seeding density was obtained for every experiment by plating in parallel an equivalent number of cells into a 24 well dish and staining and counting nuclei at the end of the experiment (data not shown).
Cell migration assays. The xCELLigence RTCA system can also be used for quantification of EC chemotactic migration when the CIM-Plate16 (Cellular Invasion/Migration) (Roche Applied Science, 05-665-817-001) dishes are used. These Transwell®-like plates have microelectrode sensors integrated onto the underside of an 8 µm pore-size membrane which separates an upper chamber (where ECs are seeded) and a lower chamber (containing chemoattractant). Increasing impedance values correlate to increasing numbers of migrated cells contacting and adhering to these sensors. Equal number of HUVEC cells pre-infected with indicated shRNA adenovirus were seeded into the upper chamber of a CIM-Plate 16 and allowed to migrate towards 25 ng/ml FGF-2 (R&D Systems, 234-FSE-025) present in the lower well. Cell Impedance was recorded automatically every 5 minutes for at least 12 hours.
Statistical analysis.
Statistical significance was determined by Student's t-test (one-tail, homoscedastic) using the average values obtained from 3 independent experiments. A p-value of less than 0.05 was considered statistically significant.
Acknowledgements
This work has been supported by NIH grants PO1 HL-080166, HL-45100 and 3-R01-GM029860-28S (awarded to K.B.). E.S.W. is currently supported by an American Heart Association Scientist Development Grant (10SDG3430042). A.A. received support from an NIH Medical Scientist Training Program Grant (T32 GM008719) and an NIH NRSA (F30HL094063). We thank members of the Burridge lab for insightful discussion and Lisa Sharek for technical assistance.
Abbreviations
- ECs
endothelial cells
- GAP
GTPase activating protein
- GEF
guanine nucleotide exchange factor
- HUVEC
human umbilical vein endothelial cell
- miR
microRNA
- RTCA
real-time cell analysis
- RTCA-CIM
real-time cell analysis-cell invasion and migration
- shRNA
short hairpin RNA
- VE-cadherin
vascular endothelial cadherin
Supplementary Material
References
- 1.Fanning AS, Jameson BJ, Jesaitis LA, Anderson JM. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J Biol Chem. 1998;273:29745–29753. doi: 10.1074/jbc.273.45.29745. [DOI] [PubMed] [Google Scholar]
- 2.Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci USA. 1995;92:8813–8817. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wittchen ES, Haskins J, Stevenson BR. Protein interactions at the tight junction. Actin has multiple binding partners and ZO-1 forms independent complexes with ZO-2 and ZO-3. J Biol Chem. 1999;274:35179–35185. doi: 10.1074/jbc.274.49.35179. [DOI] [PubMed] [Google Scholar]
- 4.Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukuhara S, Sakurai A, Sano H, Yamagishi A, Somekawa S, Takakura N, et al. Cyclic AMP potentiates vascular endothelial cadherin-mediated cell-cell contact to enhance endothelial barrier function through an Epac-Rap1 signaling pathway. Mol Cell Biol. 2005;25:136–146. doi: 10.1128/MCB.25.1.136-146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noda K, Zhang J, Fukuhara S, Kunimoto S, Yoshimura M, Mochizuki N. Vascular endothelial-cadherin stabilizes at cell-cell junctions by anchoring to circumferential actin bundles through alpha- and beta-catenins in cyclic AMP-Epac-Rap1 signal-activated endothelial cells. Mol Biol Cell. 2010;21:584–596. doi: 10.1091/mbc.E09-07-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Millán J, Cain RJ, Reglero-Real N, Bigarella C, Marcos-Ramiro B, Fernandez-Martin L, et al. Adherens junctions connect stress fibres between adjacent endothelial cells. BMC Biol. 2010;8:11. doi: 10.1186/1741-7007-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall A. Rho GTPases and the Actin Cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. 8. [DOI] [PubMed] [Google Scholar]
- 9.Wójciak-Stothard B, Ridley AJ. Rho GTPases and the regulation of endothelial permeability. Vascul Pharmacol. 2002;39:187–199. doi: 10.1016/S1537-1891(03)00008-9. [DOI] [PubMed] [Google Scholar]
- 10.Etienne S, Adamson P, Greenwood J, Strosberg AD, Cazaubon S, Couraud PO. ICAM-1 signaling pathways associated with Rho activation in microvascular brain endothelial cells. J Immunol. 1998;161:5755–5761. [PubMed] [Google Scholar]
- 11.Thompson PW, Randi AM, Ridley AJ. Intercellular adhesion molecule (ICAM)-1, but not ICAM-2, activates RhoA and stimulates c-fos and rhoA transcription in endothelial cells. J Immunol. 2002;169:1007–1013. doi: 10.4049/jimmunol.169.2.1007. 11. [DOI] [PubMed] [Google Scholar]
- 12.Adamson P, Etienne S, Couraud PO, Calder V, Greenwood J. Lymphocyte migration through brain endothelial cell monolayers involves signaling through endothelial ICAM-1 via a rho-dependent pathway. J Immunol. 1999;162:2964–2973. [PubMed] [Google Scholar]
- 13.Bogatcheva NV, Garcia JG, Verin AD. Molecular mechanisms of thrombin-induced endothelial cell permeability. Biochemistry (Mosc) 2002;67:75–84. doi: 10.1023/A:1013904231324. [DOI] [PubMed] [Google Scholar]
- 14.Garcia JG, Liu F, Verin AD, Birukova A, Dechert MA, Gerthoffer WT, et al. Sphingosine 1-phosphate promotes endothelial cell barrier integrity by Edg-dependent cytoskeletal rearrangement. J Clin Invest. 2001;108:689–701. doi: 10.1172/JCI12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Wetering S, van Buul JD, Quik S, Mul FP, Anthony EC, ten Klooster JP, et al. Reactive oxygen species mediate Rac-induced loss of cell-cell adhesion in primary human endothelial cells. J Cell Sci. 2002;115:1837–1846. doi: 10.1242/jcs.115.9.1837. [DOI] [PubMed] [Google Scholar]
- 16.Wójciak-Stothard B, Potempa S, Eichholtz T, Ridley AJ. Rho and Rac but not Cdc42 regulate endothelial cell permeability. J Cell Sci. 2001;114:1343–1355. doi: 10.1242/jcs.114.7.1343. [DOI] [PubMed] [Google Scholar]
- 17.Schnittler HJ. Structural and functional aspects of intercellular junctions in vascular endothelium. Basic Res Cardiol. 1998;93:s30–s39. doi: 10.1007/s003950050205. [DOI] [PubMed] [Google Scholar]
- 18.Cullere X, Shaw SK, Andersson L, Hirahashi J, Luscinskas FW, Mayadas TN. Regulation of vascular endothelial barrier function by Epac, a cAMP-activated exchange factor for Rap GTPase. Blood. 2005;105:1950–1955. doi: 10.1182/blood-2004-05-1987. [DOI] [PubMed] [Google Scholar]
- 19.Fukuhra S, Sakurai A, Yamagishi A, Sako K, Mochizuki N. Vascular endothelial cadherin-mediated cell-cell adhesion regulated by a small GTPase, Rap1. J Biochem Mol Biol. 2006;39:132–139. doi: 10.5483/BMBRep.2006.39.2.132. [DOI] [PubMed] [Google Scholar]
- 20.Kooistra MR, Corada M, Dejana E, Bos JL. Epac1 regulates integrity of endothelial cell junctions through VE-cadherin. FEBS Lett. 2005;579:4966–4972. doi: 10.1016/j.febslet.2005.07.080. [DOI] [PubMed] [Google Scholar]
- 21.Sakurai A, Fukuhara S, Yamagishi A, Sako K, Kamioka Y, Masuda M, et al. MAGI-1 is required for Rap1 activation upon cell-cell contact and for enhancement of vascular endothelial cadherin-mediated cell adhesion. Mol Biol Cell. 2006;17:966–976. doi: 10.1091/mbc.E05-07-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sehrawat S, Cullere X, Patel S, Italiano J, Jr, Mayadas TN. Role of epac1, an exchange factor for rap GTPases, in endothelial microtubule dynamics and barrier function. Mol Biol Cell. 2008;19:1261–1270. doi: 10.1091/mbc.E06-10-0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wittchen ES, Worthylake RA, Kelly P, Casey PJ, Quilliam LA, Burridge K. Rap1 GTPase inhibits leukocyte transmigration by promoting endothelial barrier function. J Biol Chem. 2005;280:11675–11682. doi: 10.1074/jbc.M412595200. [DOI] [PubMed] [Google Scholar]
- 24.Birukova AA, Zagranichnaya T, Alekseeva E, Bokoch GM, Birukov KG. Epac/Rap and PKA are novel mechanisms of ANP-induced Rac-mediated pulmonary endothelial barrier protection. J Cell Physiol. 2008;215:715–724. doi: 10.1002/jcp.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsukamoto N, Hattori M, Yang H, Bos JL, Minato N. Rap1 GTPase-activating protein SPA-1 negatively regulates cell adhesion. J Biol Chem. 1999;274:18463–18469. doi: 10.1074/jbc.274.26.18463. [DOI] [PubMed] [Google Scholar]
- 26.Su L, Hattori M, Moriyama M, Murata N, Harazaki M, Kaibuchi K, et al. AF-6 controls integrin-mediated cell adhesion by regulating Rap1 activation through the specific recruitment of Rap1GTP and SPA-1. J Biol Chem. 2003;278:15232–15238. doi: 10.1074/jbc.M211888200. [DOI] [PubMed] [Google Scholar]
- 27.Mandai K, Nakanishi H, Satoh A, Obaishi H, Wada M, Nishioka H, et al. Afadin: A novel actin filamentbinding protein with one PDZ domain localized at cadherin-based cell-to-cell adherens junction. J Cell Biol. 1997;139:517–528. doi: 10.1083/jcb.139.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glading A, Han J, Stockton RA, Ginsberg MH. KRIT-1/CCM1 is a Rap1 effector that regulates endothelial cell cell junctions. J Cell Biol. 2007;179:247–254. doi: 10.1083/jcb.200705175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tawa H, Rikitake Y, Takahashi M, Amano H, Miyata M, Satomi-Kobayashi S, et al. Role of Afadin in Vascular Endothelial Growth Factor- and Sphingosine 1-Phosphate-Induced Angiogenesis. Circ Res. 2010;106:1731–1742. doi: 10.1161/CIRCRESAHA.110.216747. [DOI] [PubMed] [Google Scholar]
- 30.Kim S, Mizoguchi A, Kikuchi A, Takai Y. Tissue and subcellular distributions of the smg-21/rap1/Krev-1 proteins which are partly distinct from those of c-ras p21s. Mol Cell Biol. 1990;10:2645–2652. doi: 10.1128/mcb.10.6.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quilliam LA, Der CJ, Clark R, O'Rourke EC, Zhang K, McCormick F, et al. Biochemical characterization of baculovirus-expressed rap1A/Krev-1 and its regulation by GTPase-activating proteins. Mol Cell Biol. 1990;10:2901–2908. doi: 10.1128/mcb.10.6.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pizon V, Lerosey I, Chardin P, Tavitian A. Nucleotide sequence of a human cDNA encoding a ras-related protein (rap1B) Nucleic Acids Res. 1988;16:7719. doi: 10.1093/nar/16.15.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawata M, Matsui Y, Kondo J, Hishida T, Teranishi Y, Takai Y. A novel small molecular weight GTP-binding protein with the same putative effector domain as the ras proteins in bovine brain membranes. Purification, determination of primary structure and characterization. J Biol Chem. 1988;263:18965–18971. [PubMed] [Google Scholar]
- 34.Enserink JM, Christensen AE, de Rooij J, van Triest M, Schwede F, Genieser HG, et al. A novel Epac-specific cAMP analogue demonstrates independent regulation of Rap1 and ERK. Nat Cell Biol. 2002;4:901–906. doi: 10.1038/ncb874. [DOI] [PubMed] [Google Scholar]
- 35.Janoueix-Lerosey I, Polakis P, Tavitian A, de Gunzburg J. Regulation of the GTPase activity of the ras-related rap2 protein. Biochem Biophys Res Commun. 1992;189:455–464. doi: 10.1016/0006-291X(92)91580-J. [DOI] [PubMed] [Google Scholar]
- 36.Chrzanowska-Wodnicka M, Kraus AE, Gale D, White GC, 2nd, Vansluys J. Defective angiogenesis, endothelial migration, proliferation and MAPK signaling in Rap1b-deficient mice. Blood. 2008;111:2647–2656. doi: 10.1182/blood-2007-08-109710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Yan J, De P, Chang HC, Yamauchi A, Christopherson KW, 2nd, et al. Rap1a null mice have altered myeloid cell functions suggesting distinct roles for the closely related Rap1a and 1b proteins. J Immunol. 2007;179:8322–8331. doi: 10.4049/jimmunol.179.12.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duchniewicz M, Zemojtel T, Kolanczyk M, Grossmann S, Scheele JS, Zwartkruis FJ. Rap1A-deficient T and B cells show impaired integrin-mediated cell adhesion. Mol Cell Biol. 2006;26:643–653. doi: 10.1128/MCB.26.2.643-653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carmona G, Gottig S, Orlandi A, Scheele J, Bauerle T, Jugold M, et al. Role of the small GTPase Rap1 for integrin activity regulation in endothelial cells and angiogenesis. Blood. 2009;113:488–497. doi: 10.1182/blood-2008-02-138438. [DOI] [PubMed] [Google Scholar]
- 40.Chrzanowska-Wodnicka M, Smyth SS, Schoenwaelder SM, Fischer TH, White GC., 2nd Rap1b is required for normal platelet function and hemostasis in mice. J Clin Invest. 2005;115:680–687. doi: 10.1172/JCI22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Severson EA, Lee WY, Capaldo CT, Nusrat A, Parkos CA. Junctional adhesion molecule A interacts with Afadin and PDZ-GEF2 to activate Rap1A, regulate beta1 integrin levels and enhance cell migration. Mol Biol Cell. 2009;20:1916–1925. doi: 10.1091/mbc.E08-10-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan J, Li F, Ingram DA, Quilliam LA. Rap1a is a key regulator of fibroblast growth factor 2-induced angiogenesis and together with Rap1b controls human endothelial cell functions. Mol Cell Biol. 2008;28:5803–5810. doi: 10.1128/MCB.00393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-O. [DOI] [PubMed] [Google Scholar]
- 44.Michaelson D, Silletti J, Murphy G, D'Eustachio P, Rush M, Philips MR. Differential Localization of Rho Gtpases in Live Cells: Regulation by Hypervariable Regions and RhoGDI Binding. J Cell Biol. 2001;152:111–126. doi: 10.1083/jcb.152.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams CL. The polybasic region of Ras and Rho family small GTPases: a regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cellular Signalling. 2003;15:1071–1080. doi: 10.1016/S0898-6568(03)00098-6. [DOI] [PubMed] [Google Scholar]
- 46.Boettner B, Govek EE, Cross J, Van Aelst L. The junctional multidomain protein AF-6 is a binding partner of the Rap1A GTPase and associates with the actin cytoskeletal regulator profilin. Proc Natl Acad Sci USA. 2000;97:9064–9069. doi: 10.1073/pnas.97.16.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dejana E, Orsenigo F, Lampugnani MG. The role of adherens junctions and VE-cadherin in the control of vascular permeability. J Cell Sci. 2008;121:2115–2122. doi: 10.1242/jcs.017897. [DOI] [PubMed] [Google Scholar]
- 48.Vestweber D, Broermann A, Schulte D. Control of endothelial barrier function by regulating vascular endothelial-cadherin. Curr Opin Hematol. 2010;17:230–236. doi: 10.1097/MOH.0b013e328338664b. [DOI] [PubMed] [Google Scholar]
- 49.Aghajanian A, Wittchen ES, Allingham MJ, Garrett TA, Burridge K. Endothelial cell junctions and the regulation of vascular permeability and leukocyte transmigration. J Thromb Haemost. 2008;6:1453–1460. doi: 10.1111/j.1538-7836.2008.03087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Atienza JM, Yu N, Kirstein SL, Xi B, Wang X, Xu X, et al. Dynamic and Label-Free Cell-Based Assays Using the Real-Time Cell Electronic Sensing System. Assay Drug Dev Technol. 2006;4:597–607. doi: 10.1089/adt.2006.4.597. [DOI] [PubMed] [Google Scholar]
- 51.Li HB, Ge YK, Zhang L, Zheng XX. Astragaloside IV improved barrier dysfunction induced by acute high glucose in human umbilical vein endothelial cells. Life Sciences. 2006;79:1186–1193. doi: 10.1016/j.lfs.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 52.Moy AB, Blackwell K, Wang N, Haxhinasto K, Kasiske MK, Bodmer J, et al. Phorbol ester-mediated pulmonary artery endothelial barrier dysfunction through regulation of actin cytoskeletal mechanics. Am J Physiol Lung Cell Mol Physiol. 2004;287:L153–L167. doi: 10.1152/ajplung.00292.2003. [DOI] [PubMed] [Google Scholar]
- 53.Rabiet MJ, Plantier JL, Rival Y, Genoux Y, Lampugnani MG, Dejana E. Thrombin-induced increase in endothelial permeability is associated with changes in cell-to-cell junction organization. Arterioscler Thromb Vasc Biol. 1996;16:488–496. doi: 10.1161/01.atv.16.3.488. [DOI] [PubMed] [Google Scholar]
- 54.Wójciak-Stothard B, Entwistle A, Garg R, Ridley AJ. Regulation of TNFalpha-induced reorganization of the actin cytoskeleton and cell-cell junctions by Rho, Rac and Cdc42 in human endothelial cells. J Cell Physiol. 1998;176:150–165. doi: 10.1002/(SICI)1097-4652(199807)176:1<150::AID°CP17>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 55.Adamson RH, Ly JC, Sarai RK, Lenz JF, Altangerel A, Drenckhahn D, et al. Epac/Rap1 pathway regulates microvascular hyperpermeability induced by PAF in rat mesentery. Am J Physiol Heart Circ Physiol. 2008;294:H1188–H1196. doi: 10.1152/ajpheart.00937.2007. [DOI] [PubMed] [Google Scholar]
- 56.Song L, Ge S, Pachter JS. Caveolin-1 regulates expression of junction-associated proteins in brain microvascular endothelial cells. Blood. 2007;109:1515–1523. doi: 10.1182/blood-2006-07-034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rampersad SN, Ovens JD, Huston E, Umana MB, Wilson LS, Netherton SJ, et al. Cyclic AMP phosphodiesterase 4D (PDE4D) Tethers EPAC1 in a vascular endothelial cadherin (VE-Cad)-based signaling complex and controls cAMP-mediated vascular permeability. J Biol Chem. 2010;285:33614–33622. doi: 10.1074/jbc.M110.140004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boettner B, Van Aelst L. Control of cell adhesion dynamics by Rap1 signaling. Curr Opin Cell Biol. 2009;21:684–693. doi: 10.1016/j.ceb.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chrzanowska-Wodnicka M. Regulation of angiogenesis by a small GTPase Rap1. Vascul Pharmacol. 2010;53:1–10. doi: 10.1016/j.vph.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 60.Li X, Stankovic M, Lee BP, Aurrand-Lions M, Hahn CN, Lu Y, et al. JAM-C induces endothelial cell permeability through its association and regulation of {beta}3 integrins. Arterioscler Thromb Vasc Biol. 2009;29:1200–1206. doi: 10.1161/ATVBAHA.109.189217. [DOI] [PubMed] [Google Scholar]
- 61.Wang Q, Pfeiffer GR, 2nd, Stevens T, Doerschuk CM. Lung microvascular and arterial endothelial cells differ in their responses to intercellular adhesion molecule-1 ligation. Am J Respir Crit Care Med. 2002;166:872–877. doi: 10.1164/rccm.2201007. [DOI] [PubMed] [Google Scholar]
- 62.Aird WC. Phenotypic heterogeneity of the endothelium: I. Structure, function and mechanisms. Circ Res. 2007;100:158–173. doi: 10.1161/01.RES.0000255691.76142.4a. [DOI] [PubMed] [Google Scholar]
- 63.Pries AR, Kuebler WM. Normal endothelium. Handb Exp Pharmacol. 2006;(176 Pt 1):1–40. doi: 10.1007/3-540-32967-6_1. [DOI] [PubMed] [Google Scholar]
- 64.Wittinghofer A, Herrmann C. Ras-effector interactions, the problem of specificity. FEBS Lett. 1995;369:52–56. doi: 10.1016/0014-5793(95)00667-X. [DOI] [PubMed] [Google Scholar]
- 65.Mochizuki N, Yamashita S, Kurokawa K, Ohba Y, Nagai T, Miyawaki A, et al. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–8. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- 66.Pizon V, Desjardins M, Bucci C, Parton RG, Zerial M. Association of Rap1a and Rap1b proteins with late endocytic/phagocytic compartments and Rap2a with the Golgi complex. J Cell Sci. 1994;107:1661–1670. doi: 10.1242/jcs.107.6.1661. [DOI] [PubMed] [Google Scholar]
- 67.Béranger F, Goud B, Tavitian A, de Gunzburg J. Association of the Ras-antagonistic Rap1/Krev-1 proteins with the Golgi complex. Proc Natl Acad Sci USA. 1991;88:1606–1610. doi: 10.1073/pnas.88.5.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bivona TG, Wiener HH, Ahearn IM, Silletti J, Chiu VK, Philips MR. Rap1 upregulation and activation on plasma membrane regulates T cell adhesion. J Cell Biol. 2004;164:461–470. doi: 10.1083/jcb.200311093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mitra RS, Zhang Z, Henson BS, Kurnit DM, Carey TE, D'Silva NJ. Rap1A and rap1B ras-family proteins are prominently expressed in the nucleus of squamous carcinomas: nuclear translocation of GTP-bound active form. Oncogene. 2003;22:6243–6256. doi: 10.1038/sj.onc.1206534. [DOI] [PubMed] [Google Scholar]
- 70.Hogan C, Serpente N, Cogram P, Hosking CR, Bialucha CU, Feller SM, et al. Rap1 regulates the formation of E-cadherin-based cell-cell contacts. Mol Cell Biol. 2004;24:6690–6700. doi: 10.1128/MCB.24.15.6690-6700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Siekhaus D, Haesemeyer M, Moffitt O, Lehmann R. RhoL controls invasion and Rap1 localization during immune cell transmigration in Drosophila. Nat Cell Biol. 2010;12:605–610. doi: 10.1038/ncb2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Awasthi A, Samarakoon A, Chu H, Kamalakannan R, Quilliam LA, Chrzanowska-Wodnicka M, et al. Rap1b facilitates NK cell functions via IQGAP1-mediated signalosomes. J Exp Med. 2010;207:1923–1938. doi: 10.1084/jem.20100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chu H, Awasthi A, White GC, 2nd, Chrzanowska-Wodnicka M, Malarkannan S. Rap1b regulates B cell development, homing and T cell-dependent humoral immunity. J Immunol. 2008;181:3373–3383. doi: 10.4049/jimmunol.181.5.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rossner M, Yamada KM. What's in a picture? The temptation of image manipulation. J Cell Biol. 2004;166:11–15. doi: 10.1083/jcb.200406019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.