Abstract
We recently showed that palmitoylated Ras proteins (H-Ras and N-Ras) localize intracellularly at recycling endosomes (REs) and that REs act as a way-station for Ras proteins as they move along the post-Golgi exocytic pathway to the plasma membrane (PM). Palmitoylation is essential for H-Ras/N-Ras targeting to REs. H-Ras requires two palmitoyl groups for RE targeting. A lack of either or both palmitoyl groups causes H-Ras to be mislocalized to the endoplasmic reticulum (ER), the Golgi apparatus, or the PM. In this commentary, we summarize recent progress about the Ras trafficking cycle between the endomembranes (endosomes/ER/Golgi) and the PM. We further discuss (1) the critical determinants of RE targeting of lipidated proteins and (2) possible Ras-mediated signaling pathways that originate from REs.
Key words: H-Ras, N-Ras, K-Ras, Rap2, palmitoylation, recycling endosomes, COS-1
Ras Traffic to the PM
Ras proteins are small GTPases that regulate cell growth, death and differentiation. The three ubiquitously expressed Ras isoforms, H-, N- and K-Ras, are anchored to the inner leaflet of the membrane by two motifs contained in their C-terminal hypervariable domain.1 The first motif, which is common to all Ras proteins, is a C-terminal CAAX motif that undergoes posttranslational modification by sequential farnesylation, proteolysis and carboxyl methylation. The second motif varies between Ras isoforms and is comprised of a polybasic domain of 6 lysine residues for K-Ras, and either one or two palmitoylation sites for N-Ras (C181) and H-Ras (C181 and C184). H- and N-Ras acquire these lipid modifications at ER/Golgi while transiting through the exocytic pathway leading to the PM.2,3 K-Ras, by virtue of its C-terminal polybasic domain, is sorted out of the conventional exocytic pathway and takes an undefined pathway to the PM that bypasses the Golgi.2
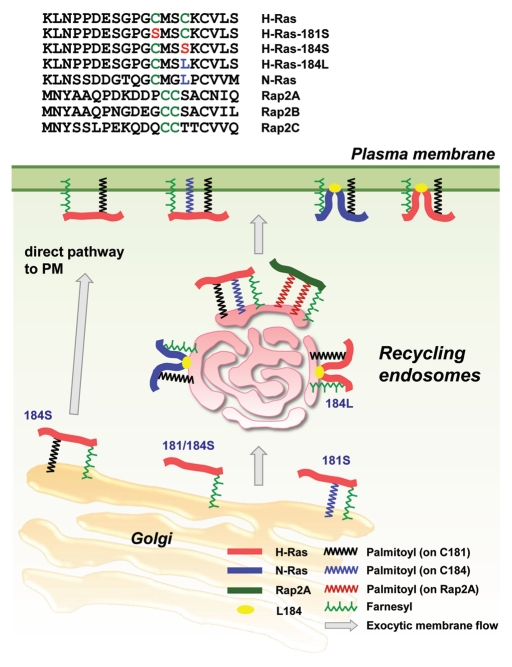
REs are endosomes that are responsible for recycling internalized proteins and lipids to the PM.4 We showed that REs are also involved in exocytic membrane traffic.5 In many mammalian cells, Golgi and REs are intermixed around the perinuclear region. COS-1 cells (green monkey kidney cells) have a unique spatial organization of organelles: REs are exclusively confined within the ring-shaped structure of Golgi (“Golgi ring”), and the organelles associated with degradation [early endosomes, late endosomes and lysosomes] are excluded from inside the Golgi ring.6 By exploiting this feature of COS-1 cells, we recently showed that palmitoylated Ras proteins (H-Ras and N-Ras) localize intracellularly at REs and that REs act as a way-station for Ras proteins as they move along the post-Golgi exocytic pathway to the PM7 (Fig. 1).
Figure 1.
Exocytic pathway of palmitoylated Ras proteins through REs to PM. The sequences of C-terminal 20-amino acid stretch of Ras proteins, palmitoylation deficient mutants and Rap2 proteins are shown. After the exit from Golgi, H-Ras is first transported to REs, then to PM. A palmitoyl group on C181 (black) is required to exit Golgi. Without the palmitoyl group on C184 (blue) upon the exit, mutant H-Ras (H-Ras-C184S) is directly transported to PM. L184 (yellow) can substitute the palmitoyl group on C184 (H-Ras-C184L can access REs), which explains N-Ras residency on REs. Rap2 proteins with two palmitoyl groups on C176/C177 are localized at REs.
Ras Translocation from PM to Endomembranes
Ras proteins at the PM can be translocated to endomembranes (endosomes/ER/Golgi) by two routes. One involves endocytosis with membrane carriers. The biological significance of H-Ras endocytosis is indicated by the observation that Raf-1 activation is sensitive to the function of dynamin (a GTPase responsible for clathrin- and caveolae-mediated endocytosis) and Rab5 (a member of Rab small GTPases that regulate membrane traffic).8 The putative endocytic membrane carrier for H-Ras was shown to harbor ADP-ribosylation factor 6 (Arf6), a member of Arf small GTPases.9 H-Ras and N-Ras can be ubiquitinated, which promotes their interaction with endosomal membranes and suppresses Ras-dependent ERK activation.10 Rabex-5 (also known as RabGEF1) was recently identified as an E3 ubiquitin ligase for H-Ras.11 Intriguingly, the study also found that the Ras effector RIN1 is required for Rabex-5-dependent ubiquitination, suggesting a feedback mechanism by which Ras activation can be coupled to its ubiquitination. K-Ras was shown to be internalized in a clathrin-dependent fashion and to be transported along early endosomes, late endosomes and eventually into lysosomes.12
The other translocation mechanism does not involve membrane carriers. H-Ras and N-Ras at the PM can be depalmitoylated.13,14 Depalmitoylation releases the proteins back to the cytosol, then the proteins are subjected to repalmitoylation at the ER/Golgi. The repalmitoylated Ras proteins at the ER/Golgi can follow the exocytic pathway leading to the PM. K-Ras can also be detached from the PM by phosphorylation of S181 within the polybasic domain.15 The K-Ras released into the cytosol is recruited to the outer mitochondrial membrane through its association with Bcl-XL, which induces apoptosis.
Critical Determinants for RE Targeting
During our Ras study,7 we found that a 20-amino acid stretch at the C terminus of H-Ras or N-Ras that contains all of the lipid modifications (farnesylation and palmitoylation) (Fig. 1) was sufficient for RE targeting. H-Ras has palmitoyl groups at two cysteine residues (C181 and C184). By analysis of the subcellular localization of palmitoyl-deficient mutant H-Ras by introducing serine point mutations, we found that both palmitoyl groups are essential for the correct H-Ras targeting to REs. A monopalmitoylation mutant, H-Ras C181S, localized exclusively at the Golgi, whereas a monopalmitoylation mutant, H-Ras C184S, localized at the Golgi and the PM. A null palmitoylation mutant, H-Ras C181/184S, accumulated mostly at the Golgi and a small amount localized at the ER. We further showed that palmitoylation on C184 can be functionally replaced with L184, since a mono-palmitoylation mutant, H-Ras C184L, localized at REs and the PM, in contrast to the localization of H-Ras C184S. Because L184 on N-Ras was found to be involved in N-Ras membrane binding,16,17 palmitoyl groups and specific amino acid residues buried in membranes can function as the RE localization determinant. M182 may also be important because it (1) is conserved in H-Ras and N-Ras and (2) was also shown to be involved in N-Ras membrane binding.16,17
We previously showed that three other small GTPases (Rap2A, Rap2B and Rap2C) also localize at REs.18 Rap2B is geranylgeranylated and Rap2A is farnesylated,19 and Rap2C is assumed to be farnesylated. All Rap2 proteins have two cysteines (C176 and C177) upstream of the CAAX cysteine (C180), and in Rap2B, C176/C177 were demonstrated as the sites of palmitoylation (Fig. 1).20 The palmitoyl-null mutants of Rap2A, 2B and 2C do not localize to REs,18 which indicates that palmitoylation of Rap2 is required for RE targeting, as is the case of H-Ras and N-Ras. Knowing the three-dimensional structure of the C-terminus of Rap2 proteins and how it binds to membranes might help to fully understand the RE targeting determinants of Rap2.
A notable characteristic of REs is that they have a unique lipid composition.21 REs purified from MDCK cells are enriched in the raft lipids sphingomyelin and cholesterol as well as in the raft-associated proteins caveolin-1 and flotillin-1. A possible mechanism for the localization of palmitoylated Ras and Rap2 to REs is that their C-termini preferentially bind to these raft lipids.
Possible Ras-Mediated Signaling from REs
By using Ras-binding domain of Raf1 that binds only to the active form of Ras, we showed that H-Ras at REs is active.7 This leads to the question: what downstream signals does Ras activate at REs? Ras can activate small GTPases, Ral (Ras-like GTPase) through the action of Ral GEFs that are direct Ras effectors.22,23 Two Ral proteins are known, RalA and RalB. RalA localizes intracellularly at REs and together with the exocyst complex regulates membrane traffic through REs to the PM.24,25 The exocyst complex consists of eight subunits (Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70 and Exo84),26,27 and several of which localize at REs.28,29 Sec10 interacts with GTP-bound Arf6,28 that also regulate recycling membrane traffic through REs to the PM.30–32 Another Ras effector, Rho small GTPase Cdc42, is involved in membrane traffic/actin regulation,33,34 and is found mainly at REs.7 Therefore, it is tempting to speculate that Ras-mediated signaling from REs is tightly connected to a variety of membrane trafficking pathways through REs.
Extra View to: Misaki R, Morimatsu M, Uemura T, Waguri S, Miyoshi E, Taniguchi N, et al. Palmitoylated Ras proteins traffic through recycling endosomes to the plasma membrane during exocytosis. J Cell Biol. 2010;191:23–29. doi: 10.1083/jcb.200911143.
References
- 1.Hancock JF. Ras proteins: different signals from different locations. Nat Rev Mol Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 2.Apolloni A, Prior IA, Lindsay M, Parton RG, Hancock JF. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol Cell Biol. 2000;20:2475–2487. doi: 10.1128/MCB.20.7.2475-87.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, et al. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- 4.Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 5.Ang AL, Taguchi T, Francis S, Folsch H, Murrells LJ, Pypaert M, et al. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J Cell Biol. 2004;167:531–543. doi: 10.1083/jcb.200408165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Misaki R, Nakagawa T, Fukuda M, Taniguchi N, Taguchi T. Spatial segregation of degradation- and recycling-trafficking pathways in COS-1 cells. Biochem Biophys Res Commun. 2007;360:580–585. doi: 10.1016/j.bbrc.2007.06.101. [DOI] [PubMed] [Google Scholar]
- 7.Misaki R, Morimatsu M, Uemura T, Waguri S, Miyoshi E, Taniguchi N, et al. Palmitoylated Ras proteins traffic through recycling endosomes to the plasma membrane during exocytosis. J Cell Biol. 2010;191:23–29. doi: 10.1083/jcb.200911143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy S, Wyse B, Hancock JF. H-Ras signaling and K-Ras signaling are differentially dependent on endocytosis. Mol Cell Biol. 2002;22:5128–5140. doi: 10.1128/MCB.22.14.5128-40.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKay J, Wang X, Ding J, Buss JE, Ambrosio L. H-ras resides on clathrin-independent ARF6 vesicles that harbor little RAF-1, but not on clathrin-dependent endosomes. Biochim Biophys Acta. 2011;1813:298–307. doi: 10.1016/j.bbamcr.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 10.Jura N, Scotto-Lavino E, Sobczyk A, Bar-Sagi D. Differential modification of Ras proteins by ubiquitination. Mol Cell. 2006;21:679–687. doi: 10.1016/j.molcel.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Xu L, Lubkov V, Taylor LJ, Bar-Sagi D. Feedback regulation of Ras signaling by Rabex-5-mediated ubiquitination. Curr Biol. 2010;20:1372–1377. doi: 10.1016/j.cub.2010.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu A, Tebar F, Alvarez-Moya B, Lopez-Alcala C, Calvo M, Enrich C, et al. A clathrin-dependent pathway leads to KRas signaling on late endosomes en route to lysosomes. J Cell Biol. 2009;184:863–879. doi: 10.1083/jcb.200807186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodwin JS, Drake KR, Rogers C, Wright L, Lippincott-Schwartz J, Philips MR, et al. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J Cell Biol. 2005;170:261–272. doi: 10.1083/jcb.200502063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 15.Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell. 2006;21:481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 16.Huster D, Vogel A, Katzka C, Scheidt HA, Binder H, Dante S, et al. Membrane insertion of a lipidated ras peptide studied by FTIR, solid-state NMR and neutron diffraction spectroscopy. J Am Chem Soc. 2003;125:4070–4079. doi: 10.1021/ja0289245. [DOI] [PubMed] [Google Scholar]
- 17.Gorfe AA, Pellarin R, Caflisch A. Membrane localization and flexibility of a lipidated ras peptide studied by molecular dynamics simulations. J Am Chem Soc. 2004;126:15277–15286. doi: 10.1021/ja046607n. [DOI] [PubMed] [Google Scholar]
- 18.Uechi Y, Bayarjargal M, Umikawa M, Oshiro M, Takei K, Yamashiro Y, et al. Rap2 function requires palmitoylation and recycling endosome localization. Biochem Biophys Res Commun. 2009;378:732–737. doi: 10.1016/j.bbrc.2008.11.107. [DOI] [PubMed] [Google Scholar]
- 19.Farrell FX, Yamamoto K, Lapetina EG. Prenyl group identification of rap2 proteins: a ras superfamily member other than ras that is farnesylated. Biochem J. 1993;289:349–355. doi: 10.1042/bj2890349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canobbio I, Trionfini P, Guidetti GF, Balduini C, Torti M. Targeting of the small GTPase Rap2b, but not Rap1b, to lipid rafts is promoted by palmitoylation at Cys176 and Cys177 and is required for efficient protein activation in human platelets. Cell Signal. 2008;20:1662–1670. doi: 10.1016/j.cellsig.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 21.Gagescu R, Demaurex N, Parton RG, Hunziker W, Huber LA, Gruenberg J. The recycling endosome of Madin-Darby canine kidney cells is a mildly acidic compartment rich in raft components. Mol Biol Cell. 2000;11:2775–2791. doi: 10.1091/mbc.11.8.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chardin P, Tavitian A. The ral gene: a new ras related gene isolated by the use of a synthetic probe. EMBO J. 1986;5:2203–2208. doi: 10.1002/j.1460-2075.1986.tb04485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolthuis RM, Zwartkruis F, Moen TC, Bos JL. Ras-dependent activation of the small GTPase Ral. Curr Biol. 1998;8:471–474. doi: 10.1016/S0960-9822(98)70183-6. [DOI] [PubMed] [Google Scholar]
- 24.Chen XW, Inoue M, Hsu SC, Saltiel AR. RalA-exocyst-dependent recycling endosome trafficking is required for the completion of cytokinesis. J Biol Chem. 2006;281:38609–38616. doi: 10.1074/jbc.M512847200. [DOI] [PubMed] [Google Scholar]
- 25.Balasubramanian N, Meier JA, Scott DW, Norambuena A, White MA, Schwartz MA. RalA-exocyst complex regulates integrin-dependent membrane raft exocytosis and growth signaling. Curr Biol. 2010;20:75–79. doi: 10.1016/j.cub.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.TerBush DR, Maurice T, Roth D, Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu SC, Ting AE, Hazuka CD, Davanger S, Kenny JW, Kee Y, et al. The mammalian brain rsec6/8 complex. Neuron. 1996;17:1209–1219. doi: 10.1016/S0896-6273(00)80251-2. [DOI] [PubMed] [Google Scholar]
- 28.Prigent M, Dubois T, Raposo G, Derrien V, Tenza D, Rosse C, et al. ARF6 controls post-endocytic recycling through its downstream exocyst complex effector. J Cell Biol. 2003;163:1111–1121. doi: 10.1083/jcb.200305029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Folsch H, Pypaert M, Maday S, Pelletier L, Mellman I. The AP-1A and AP-1B clathrin adaptor complexes define biochemically and functionally distinct membrane domains. J Cell Biol. 2003;163:351–362. doi: 10.1083/jcb.200309020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radhakrishna H, Donaldson JG. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J Cell Biol. 1997;139:49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D'Souza-Schorey C, van Donselaar E, Hsu VW, Yang C, Stahl PD, Peters PJ. ARF6 targets recycling vesicles to the plasma membrane: insights from an ultrastructural investigation. J Cell Biol. 1998;140:603–616. doi: 10.1083/jcb.140.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balasubramanian N, Scott DW, Castle JD, Casanova JE, Schwartz MA. Arf6 and microtubules in adhesion-dependent trafficking of lipid rafts. Nat Cell Biol. 2007;9:1381–1391. doi: 10.1038/ncb1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kroschewski R, Hall A, Mellman I. Cdc42 controls secretory and endocytic transport to the basolateral plasma membrane of MDCK cells. Nat Cell Biol. 1999;1:8–13. doi: 10.1038/8977. [DOI] [PubMed] [Google Scholar]
- 34.Ang AL, Folsch H, Koivisto UM, Pypaert M, Mellman I. The Rab8 GTPase selectively regulates AP-1B-dependent basolateral transport in polarized Madin-Darby canine kidney cells. J Cell Biol. 2003;163:339–350. doi: 10.1083/jcb.200307046. [DOI] [PMC free article] [PubMed] [Google Scholar]