Abstract
Endocytosis and autophagy are both membrane trafficking pathways vital for cell survival. Endocytosis, the primary means by which cells internalize material such as cell-surface receptors and their protein ligands, is essential for proper cell growth and communication. Autophagy is a catabolic process that degrades cargo ranging from organelles to protein aggregates to bacteria, and it is important for maintaining cellular homeostasis. Defects in both endosome and autophagosome maturation lead to an array of human diseases, including cancer; however, the molecular mechanisms underlying endosome and autophagosome maturation are not well characterized. In the case of endocytosis, small GTPases, key players in membrane organization, are required for endosome maturation. Specifically, activation of the small GTPase Rab7 is required for the initiation of the early-to-late endosome transition, although how this is regulated is largely unknown. Now recent findings from our laboratory show that Rubicon, a component of the PI3KC3 complex, inhibits endosome maturation by preventing activation of Rab7. Not only do our results clarify the molecular link between PI3KC3 and Rab7 function in endosome maturation, they lead us to propose new models for PI3KC3 involvement in membrane trafficking, particularly at the convergence between the endosome and autophagosome pathways.
Key words: endocytosis, autophagy, PI3KC3, Rubicon, Rab7
Background
Proteins entering the cell are enclosed within early endosomes, which then progress to different fates—recycling back to the plasma membrane, transport to the Golgi, or degradation in the lysosome.1 Endocytosis contributes to several levels of control of signal transduction: spatial control, temporal control and amplitude control.2 As such, defects in endocytosis have been linked to human disease, most notably cancer.
Two important regulators of intracellular membrane trafficking are phophatidylinositol 3-kinases (PI3K) and Rab GTPases. The class III PI3K (PI3KC3) phosphorylates the 3-hydroxyl group of phosphatidylinositol (PI3P). Membrane domains enriched in PI3P then recruit specific proteins with PI3P-binding motifs.3 Rab GTPases similarly localize to specific domains of organelle membranes and, via their “molecular switch” capabilities, bind to effectors to exert spatial and temporal regulation of membrane trafficking events.4,5
Early endosomes are characterized by PI3KC3 and Rab5, whereas late endosomes are characterized by Rab7.6,7 For early endosomes to mature, Rab7 must be activated and recruited to early endosomes, displacing Rab5.6 Active Rab7 then functions in vesicle tethering and fusion via interactions with effector proteins.8 How is the switch from Rab5 to Rab7 on the transitioning endosome membrane accomplished? Does PI3KC3 function in this switch? Understanding these mechanisms is key to understanding endosome maturation and other membrane trafficking pathways that involve the same proteins.
PI3KC3 also plays a crucial role in autophagy.9,10 Cytosolic double-membraned structures called autophagosomes engulf targets ranging from entire organelles to microorganisms to protein aggregates. The initiating autophagosome membrane is enriched in PI3P produced by PI3KC3;11–14 these PI3P-enriched regions constitute a docking site for downstream effectors including Atg5-Atg12-Atg16 and LC3.15–19 LC3 has a critical role in bringing in cargoes through interaction with distinct adaptors.20–24 The completed autophagosomes eventually fuse with lysosomes in a Rab7 dependent manner,25,26 resulting in the degradation of the autophagosomes' contents. Autophagy is a tightly regulated process, and this recycling of cellular components allows cells to survive under stress conditions, for example, nutrient deprivation, hypoxia or infection, and it is relevant to many human diseases.27 For example, the role of autophagy in cancer is an active area of research. Autophagy-regulator Beclin 1 has been found to have tumor suppressor properties, but autophagy may also be a mechanism cancer cells use to survive in the hypoxic environment of a tumor.28
Rubicon (Run domain protein as Beclin 1 interacting and cysteine-rich containing) is a protein found in complex with PI3KC3 on both endosomes and autophagosomes.12,13,29,30 In our recent publication, we find that it prevents Rab7 activation by sequestering UVRAG and preventing it from activating C-VPS/HOPS, the guanine nucleotide exchange factor for Rab7. As such, Rubicon acts as a negative regulator of endosome maturation. We discuss these results in more detail below and their implications for our understanding of both the endosome and autophagosome maturation.
Rubicon Inhibition of Endosome Maturation
We identified Rubicon as a component of the PI3KC3 holocomplex on endosomes, which also includes VPS34, p150, Beclin 1 and UVRAG. Rubicon contains a RUN domain, characteristic of a group of proteins interacting with small GTPases.31–33 We found that Rubicon specifically interacts with Rab7 but not Rab5, although not through the RUN domain. Further, immunoprecipitation experiments showed that it also interacts with UVRAG and Rab7 in a mutually exclusive manner.
Using constitutively active or inactive GTP- or GDP-bound mutants, we showed that Rubicon preferentially binds to GTP-bound Rab7. Overexpressing constitutively active GTP-bound Rab7 Q67L mutant competed with UVRAG for Rubicon binding much more efficiently than a GDP-bound Rab7 T22N mutant, further supporting the mutually exclusive binding of Rab7 and UVRAG with Rubicon, and indicating that this occurs in a nucleotide-dependent manner.
When in endosome maturation does Rubicon come into play? We investigated the subcellular localization of Rubicon in human osteosarcoma (U2OS) cells and found that Rubicon showed significant colocalization with Rab5. Further, in cells expressing a Rab5 GTP-bound Q79L mutant that arrests endosomes at the point of endosome maturation after acquiring Rab7, we found that Rubicon again showed marked colocalization with Rab5. These data indicate that Rubicon localizes to early and maturing endosomes.
Rab7 is activated by the guanine nucleotide exchange factor C-VPS/HOPS, and this complex is in turn activated by UVRAG.34 We propose that Rubicon interacts with UVRAG to block it from activating C-VPS/HOPS, which then blocks Rab7 activation. Indeed, in Rubicon RNA-i depleted cells, we observed increased interaction between UVRAG and Vps16, a component of C-VPS/HOPS. We also observed increased interaction between Rab7 and its effectors, including RILP and Vps41.35,36 Rubicon's inhibitory effect on endocytic degradation is physiologically relevant: degradation of epidermal growth factor was accelerated in cells depleted of Rubicon as compared to wild-type and slowed in cells that overexpressed Rubicon.
Molecular Connection between Rab7 and PI3KC3
It has long been speculated that there are cross-regulatory interactions between small GTPases and PI3KC3. On the one hand, PI3KC3 is required for Rab5 and Rab7 activation. On the other hand, Rab7 activates PI3KC3. The molecular mechanisms underlying this mutual regulation are not fully understood. It had previously been shown that the PI3KC3 VPS34 functions in endocytic transport, and it and its adaptor, p150, have been shown to colocalize with Rab7 on late endosomes.37 Overexpression of Rab7 stimulates VPS34 activity, and a previous model has proposed that this occurs via interaction with p150. However, p150 preferentially binds to the inactive, nucleotide-free form of Rab7, so this model may not fully explain the role of p150 in the activation of VPS34 by GTP-bound Rab7. On the basis of our results and previous evidence showing that UVRAG activates PI3KC3, we propose an alternate model: by competing with UVRAG for Rubicon binding, active Rab7 releases UVRAG to activate PI3KC3 and C-VPS/HOPS. C-VPS/HOPS in turn promotes the GDP to GTP transition on Rab7. This generates a feed-forward loop, in which activated, GTP-bound Rab7 facilitates activation of more Rab7, ensuring that the Rab5-to-Rab7 switch occurs and maturation can proceed.
Different from the competitive binding among Rab7, Rubicon, and UVRAG, Rubicon inhibition of PI3KC3 is executed within the same complex. Rubicon, in complex with UVRAG, Beclin-1, VPS34 and p150, has been previously shown to inhibit autophagy. Knockdown of Rubicon causes reduction in protein levels of autophagic substrates. Overexpression of Rubicon leads to the accumulation of these substrates, and also inhibits VPS34 kinase activity. When autophagosome maturation was tracked, it was found that cells overexpressing Rubicon showed accumulation of early autophagosomes, suggesting that Rubicon blocks autophagosome maturation, perhaps through modulating VPS34 activity. We have shown that Rubicon directly interacts with UVRAG and VPS34. However, the PI3KC3-UVRAG complex, without Rubicon, stimulates autophagosome maturation and autophagy.12,13 In the case of autophagy, PI3KC3 seems to exert its regulatory effect through two distinct complexes. On the basis of our results, we have shown that PI3KC3 can also regulate endosome maturation via a feed-forward loop.
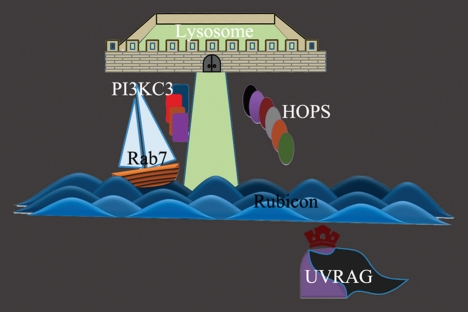
Simply put, during autophagosome and endosome maturation, both PI3KC3 and HOPS complex have to be activated. UVRAG is an activator for both enzymes but its activity is inhibited by Rubicon. Rab7 can release UVRAG from Rubicon sequestration, and therefore activate both PI3KC3 and HOPS and promotes lysosomal degradation (Fig. 1).
Figure 1.
A cartoon model of UVRAG-Rubicon in autophagosome and endosome maturation. UVRAG (the General) acts as a strong activator for both PI3KC3 and HOPS, but it is only active if it escapes the sequestration of Rubicon (by crossing the Rubicon river). Rab7 (the boat) allows UVRAG to sail through the Rubicon by competing for its binding with Rubicon, leading to PI3KC3 and HOPS activation and lysosomal degradation.
Endocytosis and Autophagy
The autophagy and endocytic pathways are intimately linked. Instead of directly fusing with lysosomes to form autolysosomes, autophagosomes may first fuse with endosomes to form amphisomes; in fact, five times as many amphisomes as autolysosomes are found in hepatocytes.38 Several groups of proteins are involved in both autophagy and endocytosis, including PI3K, coat protein complex I (COPI) and endosomal sorting complex required for transport (ESCRT).39–41 However, the exact sequence of fusion is not known—when does the autophagosome fuse with endosome, and does it fuse with an early or late endosome? Now our results lend support to the model in which autophagosomes first fuse with early endosomes. Previously, it was not known whether the PI3K VPS34 in complex with Rab5 and Rab7 could also form a complex with Beclin 1 and Beclin 1 interacting partners. Here we have shown that Rubicon, a component of the endosomal PI3K complex, is highly enriched on Rab5-decorated early endosomes and can form a complex with Rab7. PI3KC3 may thus serve to recruit autophagosome proteins to the early endosome, promoting fusion. In this way, the endosomal PI3KC3 complex might switch with autophagosomal PI3KC3 complex to promote fusion between autophagosomes and endosomes. In this model, we propose that the autophagy pathway is one of several pathways an early endosome can take. As the early endosome stage can be viewed as a “sorting station,” with endosomes proceeding towards different fates, we propose that fusing with autophagosomes may be one option.
Future Directions
Endosome and autophagosome maturation are not completely understood. Our studies show that PI3KC3 and Rab7 function in endosome maturation via a feed-forward loop: Rubicon prevents Rab7 activation by binding and sequestering UVRAG; when GTP-bound Rab7 is present, it competes with UVRAG to bind Rubicon, thus promoting its own activation and stimulating the early-to-late endosome transition. We contrast this with other mechanisms of PI3K action in autophagy, and suggest that our results support a model in which autophagosomes first fuse with early endosomes. In addition to elucidating the pathways of endosome and autophagosome maturation, these results may help us understand the role of the endocytosis and autophagy, and the PI3KC3 complex, in human diseases such as cancer. Beclin 1 and UVRAG have both been found to have tumor supressor activity. However, a recent study showed that microsatellite colon cancer carcinomas with monoallelic UVRAG mutations showed no inhibition of autophagy, suggesting that another mechanism may be responsible for the tumorigenicity.42 It is possible that these mutations in UVRAG affect endosome maturation rather than autophagosome maturation. Future studies will further delineate the relationship between endocytosis and autophagy and their role in human disease.
Acknowledgements
This work is supported by a New Investigator Award for Aging from the Ellison Medical Foundation, Hellman Family Foundation and NIH RO1 (CA133228) to Q. Z.
Extra View to: Sun Q, Westphal W, Wong KN, Tan I, Zhong Q. Rubicon controls endosome maturation as a Rab7 effector. Proc Natl Acad Sci USA. 2010;107:19338–19343. doi: 10.1073/pnas.1010554107.
References
- 1.Spang A. On the fate of early endosomes. Biol Chem. 2009;390:753–759. doi: 10.1515/BC.2009.056. [DOI] [PubMed] [Google Scholar]
- 2.Lanzetti L, Di Fiore PP. Endocytosis and cancer: an ‘insider’ network with dangerous liaisons. Traffic. 2008;9:2011–2021. doi: 10.1111/j.1600-0854.2008.00816.x. [DOI] [PubMed] [Google Scholar]
- 3.Lindmo K, Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci. 2006;119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- 4.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 5.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 6.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 7.Nottingham RM, Pfeffer SR. Defining the boundaries: Rab GEFs and GAPs. Proc Natl Acad Sci USA. 2009;106:14185–14186. doi: 10.1073/pnas.0907725106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markgraf DF, Peplowska K, Ungermann C. Rab cascades and tethering factors in the endomembrane system. FEBS Lett. 2007;581:2125–2130. doi: 10.1016/j.febslet.2007.01.090. [DOI] [PubMed] [Google Scholar]
- 9.Sun Q, Fan W, Zhong Q. Regulation of Beclin 1 in autophagy. Autophagy. 2009;5:713–716. doi: 10.4161/auto.5.5.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- 11.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol-3-kinase. Proc Natl Acad Sci USA. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 13.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol-3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350 kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- 17.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan W, Tang Z, Chen D, Moughon D, Ding X, Chen S, et al. Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy. Autophagy. 2010;6:614–621. doi: 10.4161/auto.6.5.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 23.Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 24.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 25.Gutierrez MG, Munafó DB, Berón W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 26.Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 27.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hippert MM, O'Toole PS, Thorburn A. Autophagy in cancer: good, bad or both? Cancer Res. 2006;66:9349–9351. doi: 10.1158/0008-5472.CAN-06-1597. [DOI] [PubMed] [Google Scholar]
- 29.Sun Q, Westphal W, Wong KN, Tan I, Zhong Q. Rubicon controls endosome maturation as a Rab7 effector. Proc Natl Acad Sci USA. 2010;107:19338–19343. doi: 10.1073/pnas.1010554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Q, Zhang J, Fan W, Wong KN, Ding X, Chen S, et al. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition and autophagy suppression. J Biol Chem. 2011;286:185–191. doi: 10.1074/jbc.M110.126425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Recacha R, Boulet A, Jollivet F, Monier S, Houdusse A, Goud B, et al. Structural basis for recruitment of Rab6-interacting protein 1 to Golgi via a RUN domain. Structure. 2009;17:21–30. doi: 10.1016/j.str.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 32.Kukimoto-Niino M, Takagi T, Akasaka R, Murayama K, Uchikubo-Kamo T, Terada T, et al. Crystal structure of the RUN domain of the RAP2-interacting protein x. J Biol Chem. 2006;281:31843–31853. doi: 10.1074/jbc.M604960200. [DOI] [PubMed] [Google Scholar]
- 33.Mari M, Macia E, Le Marchand-Brustel Y, Cormont M. Role of the FYVE finger and the RUN domain for the subcellular localization of Rabip4. J Biol Chem. 2001;276:42501–42508. doi: 10.1074/jbc.M104885200. [DOI] [PubMed] [Google Scholar]
- 34.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price A, Seals D, Wickner W, Ungermann C. The docking stage of yeast vacuole fusion requires the transfer of proteins from a cis-SNARE complex to a Rab/Ypt protein. J Cell Biol. 2000;148:1231–1238. doi: 10.1083/jcb.148.6.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stein MP, Feng Y, Cooper KL, Welford AM, Wandinger-Ness A. Human VPS34 and p150 are Rab7 interacting partners. Traffic. 2003;4:754–771. doi: 10.1034/j.1600-0854.2003.00133.x. [DOI] [PubMed] [Google Scholar]
- 38.Strømhaug PE, Berg TO, Fengsrud M, Seglen PO. Purification and characterization of autophagosomes from rat hepatocytes. Biochem J. 1998;335:217–224. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 40.Razi M, Chan EYW, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rusten TE, Stenmark H. How do ESCRT proteins control autophagy? J Cell Sci. 2009;122:2179–2183. doi: 10.1242/jcs.050021. [DOI] [PubMed] [Google Scholar]
- 42.Knævelsrud H, Ahlquist T, Merok MA, Nesbakken A, Stenmark H, Lothe RA, et al. UVRAG mutations associated with microsatellite unstable colon cancer do not affect autophagy. Autophagy. 2010;6:863–670. doi: 10.4161/auto.6.7.13033. [DOI] [PubMed] [Google Scholar]