Abstract
Transforming growth factor-β (TGFβ) signaling pathways regulate a wide array of cellular activities that are crucial for cell proliferation, apoptosis, migration and differentiation. TGFβ signaling pathways are initiated by ligand-activated TGFβ receptors, with type I TGFβ receptors (TβR-I) kinase being essential for phosphorylation of downstream targets. Until now, a prevalent view was that the TGFβ intracellular signaling targets would regulate transcription. Recently, we uncovered a novel TGFβ signaling pathway that exerts a direct regulatory effect on mRNA translation and protein synthesis. Eukaryotic elongation factor eEF1A1 is a GTP-binding protein that plays a central role in protein synthesis. By using a screening method for kinase substrate that was developed in our laboratory, we identified eEF1A1 as a novel substrate of TβR-I. This shed a new light on the convergence of TGFβ signaling and protein synthesis. We also showed phosphorylation of eEF1A1 at Ser300 by TβR-I prevents aa-tRNA binding to eEF1A1. As a consequence, TGFβ-dependent phosphorylation of eEF1A1 has an inhibitory effect on protein synthesis and cell proliferation. Therefore, we unveiled a novel regulatory mechanism of cellular proliferation by TGFβ at the translational level. Here we discuss this finding in the context of its potential role in the multiplicity of TGFβ signaling, and in the regulation of fundamental cellular functions, such as proliferation.
Key words: TGFβ, elongation, protein, translation, eEF1A1, cancer
eEF1A1 and Cancer
Protein synthesis is the fundamental mechanism essential for any living cell, and is involved in normal physiology and disease development. Protein synthesis is a multiple step process that depends on the coordinated action of hundred of proteins and different RNAs. The protein synthesis consists of three phases: initiation, elongation and termination. Eukaryotic elongation factor eEF1A is a GTP-binding protein that plays a crucial role in translational elongation process. There are two eEF1A isoforms, eEF1A1 and eEF1A2, that are expressed in a tissue-specific manner.1,2
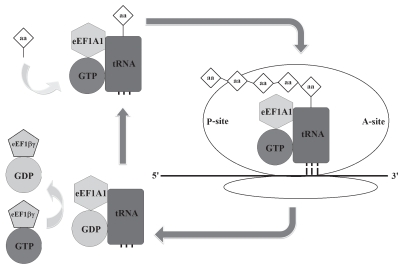
During protein translation, following the codon/anticodon matching, GTPase activity of eEF1A catalyzes the binding of aa-tRNA to the A site of ribosome. The formation of peptide bond between the amino acid and the growing peptide chain is catalyzed by ribosomal peptidyl-transferase. Following hydrolysis of GTP, eEF1A-GDP leaves the ribosome. In order for additional translocation events to occur, the GDP must be exchanged for GTP, which is carried out by eEF-1βγ (Fig. 1).3
Figure 1.
eEF1A1 in protein synthesis. During protein-synthesis codon/anticodon matching leads to the binding of eEF1A1-aminoacyl-tRNA-GTP complex to the A-site of ribosome which contains the growing polypeptide chain at the P-site of ribosome. Following hydrolysis of GTP, eEF1A-tRNA-GDP complex leaves the ribosome. In order for additional translocation events to take place, the GDP must be exchanged for GTP, which is carried out by eEF1βγ.
eEF1A is a key regulator of various physiological processes, such as embryogenesis, aging, proliferation, apoptosis, protein degradation and cytoskeletal rearrangement.4–6 It is believed that the role of eEF1A in protein synthesis is of the key importance in these activities. However, whether these are non-canonical functions of eEF1A or part of an efficient channelling of protein synthesis components is currently under debate.
eEF1A is also involved in the tumorigenesis. The increased expression of eEF1A1 was observed in hepatocellular carcinoma,7,8 and prostate carcinoma.9 eEF1A increased metastatic potential of mammary adenocarcinoma.10 The overexpression of eEF1A2 was found in over 30% of ovarian tumor11 and 83% of pancreatic cancers.12 Targeting eEF1A, as a strategy to combat apoptotic-resistant melanoma, has also been reported in reference 13.
Several reports implicated that post-translational modifications (PTMs) of eEF1A are associated with its regulatory function. As examples, phosphorylation of eEF1A1 is involved in stimulation of GDP/GTP-exchange rate in rabbit reticulocytes,14 and reduces binding to F-actin in rat liver cells.15 Methylation of eEF1A was associated with the SV40-dependent transformation of mouse 3T3B cells.16 As PTMs play a significant role in a wide array of cellular processes, uncovering novel PTMs in eEF1A enable us to understand how eEF1A functions.
TGFβ Signaling
Transforming growth factor-β (TGFβ) exerts diverse effects on a wide variety of cellular responses, including proliferation, differentiation, migration, invasion, cell cycle and apoptosis. It acts as a growth inhibitor at the early stage of tumorigenesis, but as a tumor promoter at the later stage.17,18 TGFβ intracellular signaling is initiated by binding of the ligand to the heterotetrameric complex of TGFβ receptors type II and type I. Activated type II receptor kinase subsequently phosphorylated and activated type I receptor. The activated type I receptor phosphorylates Smad2 and Smad3 proteins, and initiates the TGFβ intracellular signaling.
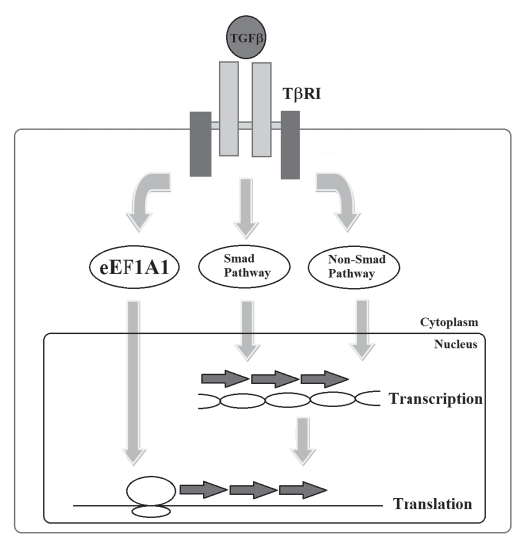
In addition to the canonical Smad-dependent TGFβ pathway, there are numbers of TGFβ signaling pathway that do not involve direct activation of Smad proteins.19 These non-Smad pathways include MAPK, Rho-like GTPase and PI3K/Akt. These pathways are activated by receptor-interacting proteins, and also by non-Smad substrates of the receptor kinases. Among which ShcA and Par6 were shown to be directly phosphorylated by TGFβ receptor kinases.20,21 Recently, our study uncovered that eEF1A1 is phosphorylated by type I TGFβ receptor kinase (Fig. 2).22
Figure 2.
Targeting of eEF1A1 is a novel mechanism in TGFβ signaling. Canonical TGFβ signaling pathway involves activation of Smad2 and Smad3 proteins (Smad pathway). There are numbers of TGFβ signaling pathways that do not involve direct activation of Smads (non-Smad pathway). These non-Smad pathways include MAPK, Rho-like GTPase and PI3K/Akt, as examples. Activation of Smad and non-Smad signaling cascade ultimately leads to the transcription activation of target genes. Recently, our study uncovered that eEF1A1 is phosphorylated by type I TGFβ receptor kinase, which leads to the direct regulation of protein synthesis, without involvement of transcriptional regulation.
More than 100 proteins associated with TGFβ receptor complex were identified, and many of these interactions are dynamically dependent on the activation of the receptor complex.23 These suggested that, in addition Smad2 and Smad3, and Smad-independent signaling substrate (i.e., ShcA, Par6 and eEF1A1), TGFβ receptors might have more substrates to be uncovered. Since the kinetics of phosphorylation is very rapid by its nature, and the interaction between a kinase and its substrate is transient, identification of substrates of TGFβ receptors remains a daunting task. Undoubtedly, characterization of the novel substrates might reveal novel molecular mechanisms of TGFβ signaling.
Phosphorylation of eEF1A1 by TβR-I
In the past few years, considerable efforts have been made toward understanding the cross-talk between TGFβ signaling and other pathway. In our recent article, we described a screening method for novel substrates of type I TGFβ receptor (TβR-I) using a protein expression library, and identified eEF1A1 as a novel substrate of TβR-I. We showed that phosphorylation of eEF1A1 at Serine 300 (Ser300) by TβR-I exerted a direct inhibitory effect of TGFβ on protein synthesis. Our study shed a new light on the links between TGFβ and protein synthesis machinery.22
In the presence of TGFβ1, TβR-I phosphorylated eEF1A1 at Ser300. Ser300 is located in the domain II of eEF1A1, which is a binding site for aa-tRNA. This implies that Ser300 phosphorylation could interfere with aa-tRNA binding which lead to the inhibition of the protein synthesis, as confirmed in our study.22 This phenomenon was observed as correlated with the potent growth inhibitory activity of TGFβ. Therefore, Ser300 phosphorylation may be the mechanism to render the cells into the lower rate of proliferation.
Our study showed that enhanced expression of the wild-type eEF1A1 promoted proliferation of cells and exhibited TGFβ-dependent inhibition of cell proliferation. Mutations of the Ser300 residue in eEF1A1 resulted in the inhibition of protein synthesis and abrogation of TGFβ responsiveness, which was due to the inhibition of aa-tRNA loading onto eEF1A1. The similar outcome was shown after a long-term treatment of cells with TGFβ in anchorage-independent growth assay. Notably, the enhanced expression of the wild-type eEF1A1 promoted formation of mammospheres and preserved TGFβ responsiveness. Mutation of Ser300 in eEF1A1 inhibited mammospheres formation and abrogated TGFβ responsiveness. This suggests that TβR-I-dependent phosphorylation of eEF1A1 at Ser300 is a part of the TGFβ signaling that regulates cell proliferation. The growth inhibition is probably due to the decrease in synthesis of a number of proteins involved in the cell proliferation. However, the exact cause and detailed mechanism remains unexplored. Further investigation of Ser300 phosphorylation in eEF1A1 may lead to the further understanding of the molecular mechanism coupling TGFβ signaling, protein synthesis and cell proliferation.
Since TGFβ and eEF1A1 are involved in tumorigenesis, we also explored the phosphorylation status of Ser300 of eEF1A1 in normal human breast tissue and breast tumors. The result showed decreased Ser300 phosphorylation in human breast tumors, indicating that this mechanism is affected in tumorigenesis.22 This can be explained by the fact that decreased phosphorylation at Ser300 correlated with higher proliferation rate of cells and interrupted TGFβ intracellular signaling. Therefore, phosphorylation status of Ser300 can be a potential marker of tumorigenesis, and further investigation may prove its usefulness.
Regulatory Functions of TGFβ-eEF1A1 Pathway
TGFβ utilizes a multitude of intracellular signaling pathway, both Smad-dependent and Smad-independent pathways, to regulate various cellular responses, including cell growth.18 It was known that both Smad-dependent and Smad-independent pathways are activated by ligand binding to receptors, followed by cascade of signaling events which lead to the transcriptional regulation of target genes. However, TGFβ effect on eEF1A1 renders a direct impact on the mRNA translation without involvement of transcriptional activation of genes.22 This growth regulatory pathway may function in coordination with other TGFβ pathways, including known Smad-dependent and Smad-independent pathways. The multiplicity of TGFβ-initiated pathways in regulation of transcription and translation activities is the feature which ensures robustness of TGFβ action to cope with various conditions, e.g., different status of regulatory components in different types of cells.
Adaptation of the cells to the dynamic microenvironment is one of the mechanisms to ensure cells survival. Fluctuation of energy source is one of the major challenges. Protein synthesis is energy-dependent activity, in which GTP represents the major source of energy. Previous study showed that mutation in the GTP binding site in eEF1A1 resulted in the dramatic reduction in translational fidelity.24–26 In addition, structural analysis of bacterial homologue of eEF1A, EF1A showed that the inactive, GDP-bound EF1A have markedly different conformation to that of the active, GTP-bound EF1A.27,28 These observations suggest that the eEF1A1 activity and activation may be affected by GTP abundance in cells. Thus, we speculate that GTP binding to eEF1A1 might affect the capability of the aa-tRNA binding in domain II, where phosphorylation of Ser300 occurred. In the low energy supplied cells, we propose that decrease GTP binding to eEF1A1 leads to the conformational change of eEF1A1, thereby promotes the phosphorylation of Ser300 by TβRI. This is followed by inhibition of protein synthesis, which leads to the lower proliferation rate of the cells. On the other hand, increased GTP binding to eEF1A1 may reduce the capability of TβR-I to phosphorylate Ser300, thereby increase of protein synthesis and proliferation of the cells. As such, we speculate that the phosphorylation status of Ser300 acts as a switch between low and high proliferation rates, which is crucial for the cells to survive in different condition.
Direct impact of TGFβ on the protein synthesis, without involving transcription, suggests that the phosphorylation at Ser300 may be of importance for coordinated and fast slowing-down of the cell cycle. Inhibition of the cell proliferation may occur by accumulation of cells in a specific phase of the cell cycle. We observed modulation of the TGFβ-dependent regulation of the cell cycle by eEF1A1, and that Ser300 phosphorylation may play a major role in this process.29 We showed that eEF1A1 promoted transition of cells through the S- and G2/M-phases, and TGFβ-dependent accumulation of cells in G0/G1 phase. The phosphorylation of Ser300 in eEF1A1 slowed down transition through the S-phase, and inhibited transition of the cells through the cell cycle. We speculate that phosphorylation of Ser300 on eEF1A1 represent a mechanism for inhibition of the cell cycle as a relatively fast response to TGFβ. It has been suggested that the cell proliferation is regulated predominantly at the transcriptional level. However, transcription regulation is a slow response due to involvement of the cascade of events, from initiation of signaling to transcriptional activation of target gene, synthesis of proteins and execution of their regulatory functions. Ser300 phosphorylation of eEF1A1 may act as an immediate switch that contributes to turning the active proliferating cells into quiescent cells. This mechanism does not require the involvement of transcriptional activity.
Our study identified eEF1A1 as a substrate of TβR-I, and unveiled a novel TGFβ-eEF1A1 pathway linking TGFβ signaling and protein synthesis machinery.22 We observed that this phosphorylation of eEF1A1 had an impact on protein synthesis, transition through the cell cycle phases, cell proliferation and was deregulated in human breast tumors. However, the cooperativity between TGFβ-eEF1A1 pathway and other TGFβ signaling pathways, both Smad-dependent and Smad-independent pathways, remains a major goal for further investigation. Understanding their functional relationship may provide a hint in unveiling the complexity of TGFβ signaling and may pave the way to clinical applications with modulation of TGFβ signaling for benefit of patients.
Abbreviations
- TGFβ
transforming growth factor-β
- TβR-I
type I TGFβ receptors
- PTM
post-translational modification
Extra View to: Lin KW, Yakymovych I, Jia M, Yakymovych M, Souchelnytskyi S. Phosphorylation of eukaryotic elongation factor eEF1A at Ser300 by type I transforming growth factor-β-receptor results in inhibition of mRNA translation. Curr Biol. 2010;20:1615–1625. doi: 10.1016/j.cub.2010.08.017.
References
- 1.Knudsen SM, Frydenberg J, Clark BF, Leffers H. Tissue-dependent variation in the expression of elongation factor-1 alpha isoforms: isolation and characterisation of a cDNA encoding a novel variant of human elongation-factor 1alpha. Eur J Biochem. 1993;215:549–554. doi: 10.1111/j.1432-1033.1993.tb18064.x. [DOI] [PubMed] [Google Scholar]
- 2.Lee S, Francoeur AM, Liu S, Wang E. Tissue-specific expression in mammalian brain, heart and muscle of S1, a member of the elongation factor-1alpha gene family. J Biol Chem. 1992;267:24064–24068. [PubMed] [Google Scholar]
- 3.Moldave K. Eukaryotic protein synthesis. Annu Rev Biochem. 1985;54:1109–1149. doi: 10.1146/annurev.bi.54.070185.005333. [DOI] [PubMed] [Google Scholar]
- 4.Condeelis J. Elongation factor 1alpha, translation and the cytoskeleton. Trends Biochem Sci. 1995;20:169–170. doi: 10.1016/S0968-0004(00)88998-88997. [DOI] [PubMed] [Google Scholar]
- 5.Kato MV, Sato H, Nagayoshi M, Ikawa Y. Upregulation of the elongation factor-1alpha gene by p53 in association with death of an erythroleukemic cell line. Blood. 1997;90:1373–1378. [PubMed] [Google Scholar]
- 6.Lamberti A, Caraglia M, Longo O, Marra M, Abbruzzese A, Arcari P. The translation elongation factor 1A in tumorigenesis, signal transduction and apoptosis: review article. Amino Acids. 2004;26:443–448. doi: 10.1007/s00726-004-0088-2. [DOI] [PubMed] [Google Scholar]
- 7.Grassi G, Scaggiante B, Farra R, Dapas B, Agostini F, Baiz D, et al. The expression levels of the translational factors eEF1A 1/2 correlate with cell growth but not apoptosis in hepatocellular carcinoma cell lines with different differentiation grade. Biochimie. 2007;89:1544–1552. doi: 10.1016/j.biochi.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Guo H, Mi Z, Gao C, Bhattacharya S, Li J, Kuo PC. EF1A1-actin interactions alter mRNA stability to determine differential osteopontin expression in HepG2 and Hep3B cells. Exp Cell Res. 2009;315:304–312. doi: 10.1016/j.yexcr.2008.10.042. [DOI] [PubMed] [Google Scholar]
- 9.Liu H, Ding J, Chen F, Fan B, Gao N, Yang Z, Qi L. Increased expression of elongation factor-1alpha is significantly correlated with poor prognosis of human prostate cancer. Scand J Urol Nephro. 2010;44:277–283. doi: 10.3109/00365599.2010.492787. [DOI] [PubMed] [Google Scholar]
- 10.Edmonds BT, Wyckoff J, Yeung YG, Wang Y, Stanley ER, Jones J, et al. Elongation factor-1alpha is an overexpressed actin binding protein in metastatic rat mammary adenocarcinoma. J Cell Sci. 1996;109:2705–2714. doi: 10.1242/jcs.109.11.2705. [DOI] [PubMed] [Google Scholar]
- 11.Anand N, Murthy S, Amann G, Wernick M, Porter LA, Cukier IH, et al. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nat Genet. 2002;31:301–315. doi: 10.1038/ng904. [DOI] [PubMed] [Google Scholar]
- 12.Cao H, Zhu Q, Huang J, Li B, Zhang S, Yao W, Zhang Y. Regulation and functional role of eEF1A2 in pancreatic carcinoma. Biochem Biophys Res Commun. 2009;380:11–16. doi: 10.1016/j.bbrc.2008.12.171. [DOI] [PubMed] [Google Scholar]
- 13.Van Goietsenoven G, Hutton J, Becker JP, Lallemand B, Robert F, Lefranc F, et al. Targeting of eEF1A with Amaryllidaceae isocarbostyrils as a strategy to combat melanomas. FASEB J. 2010;24:4575–4584. doi: 10.1096/fj.10-162263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peters HI, Chang YW, Traugh JA. Phosphorylation of elongation factor 1 (EF-1) by protein kinase C stimulates GDP/GTP-exchange activity. Eur J Biochem. 1995;234:550–556. doi: 10.1111/j.1432-1033.1995.550_b.x. [DOI] [PubMed] [Google Scholar]
- 15.Izawa T, Fukata Y, Kimura T, Iwamatsu A, Dohi K, Kaibuchi K. Elongation factor-1alpha is a novel substrate of rho-associated kinase. Biochem Biophys Res Commun. 2000;278:72–78. doi: 10.1006/bbrc.2000.3772. [DOI] [PubMed] [Google Scholar]
- 16.Coppard NJ, Clark BF, Cramer F. Methylation of elongation factor 1alpha in mouse 3T3B and 3T3B/SV40 cells. FEBS Lett. 1983;164:330–334. doi: 10.1016/0014-5793(83)80311-1. [DOI] [PubMed] [Google Scholar]
- 17.Massagué J. New concepts in tissue-specific metastases. Clin Adv Hematol Oncol. 2003;1:576–577. [PubMed] [Google Scholar]
- 18.Miyazono K, Suzuki H, Imamura T. Regulation of TGFbeta signaling and its roles in progression of tumors. Cancer Sci. 2003;94:230–234. doi: 10.1111/j.1349-7006.2003.tb01425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang YE. Non-Smad pathways in TGFbeta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee MK, Pardoux C, Hall MC, Lee PS, Warburton D, Qing J, et al. TGFbeta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J. 2007;26:3957–3967. doi: 10.1038/sj.emboj.7601818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307:1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- 22.Lin KW, Yakymovych I, Jia M, Yakymovych M, Souchelnytskyi S. Phosphorylation of eEF1A1 at Ser300 by TbetaR-I results in inhibition of mRNA translation. Curr Biol. 2010;20:1615–1625. doi: 10.1016/j.cub.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 23.Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 24.Carr-Schmid A, Durko N, Cavallius J, Merrick WC, Kinzy TG. Mutations in a GTP-binding motif of eukaryotic elongation factor 1A reduce both translational fidelity and the requirement for nucleotide exchange. J Biol Chem. 1999;274:30297–30302. doi: 10.1074/jbc.274.42.30297. [DOI] [PubMed] [Google Scholar]
- 25.Carr-Schmid A, Valente L, Loik VI, Williams T, Starita LM, Kinzy TG. Mutations in elongation factor 1beta, a guanine nucleotide exchange factor, enhance translational fidelity. Mol Cell Biol. 1999;19:5257–5266. doi: 10.1128/mcb.19.8.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soares DC, Barlow PN, Newbery HJ, Porteous DJ, Abbott CM. Structural models of human eEF1A1 and eEF1A2 reveal two distinct surface clusters of sequence variation and potential differences in phosphorylation. PLoS One. 2009;4:e6315. doi: 10.1371/journal.pone.0006315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen GR, Nissen P, Nyborg J. Elongation factors in protein biosynthesis. Trends Biochem Sci. 2003;28:434–441. doi: 10.1016/S0968-0004(03)00162-2. [DOI] [PubMed] [Google Scholar]
- 28.Andersen GR, Pedersen L, Valente L, Chatterjee I, Kinzy TG, Kjeldgaard M, Nyborg J. Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Balpha. Mol Cell. 2000;6:1261–1266. doi: 10.1016/S1097-2765(00)00122-2. [DOI] [PubMed] [Google Scholar]
- 29.Lin KW, Souchelnytskyi S. Eukaryotic elongation factor eEF1A1 promotes and Ser300 mutants of eEF1A1 inhibit transition through the S and G2/M phases of the cell cycle. J Cell Mol Biol. 2010;8:125–130. [Google Scholar]