Abstract
Macroautophagy (autophagy hereafter) is a catabolic process by which cells degrade intracellular components in lysosomes. This cellular garbage disposal and intracellular recycling provided by autophagy serves to maintain cellular homeostasis by eliminating superfluous or damaged proteins and organelles, and invading microbes, or to provide substrates for energy generation and biosynthesis in stress. Thus, autophagy promotes the health of cells and animals and is critical for development, differentiation and maintenance of cell function and for the host defense against pathogens. Deregulation of autophagy is linked to susceptibility to various disorders including degenerative diseases, metabolic syndrome, aging, infectious diseases and cancer. Autophagic activity emerges as a critical factor in development and progression of diseases that are associated with increased cancer risk as well as in different stages of cancer. Given that cancer is a complex process and autophagy exerts its effect in multiple ways, role of autophagy in tumorigenesis is context-dependent. As a cytoprotective survival pathway, autophagy prevents chronic tissue damage and cell death that can lead to cancer initiation and progression. As such, stimulation or restoration of autophagy may prevent cancer. By contrast, once cancer occurs, cancer cells may utilize autophagy to enhance fitness to survive with altered metabolism and in the hostile tumor microenvironment. In this setting autophagy inhibition would instead become a strategy for therapy of established cancers.
Keywords: autophagy, metabolism, homeostasis, inflammation, cancer prevention
Tumorigenesis is a complex multistage process. It includes tumor initiation, promotion, progression to malignancy and metastasis. This process involves profound alteration of cells in terms of metabolism, growth, proliferation, stress tolerance and survival, and interaction with the microenvironment where they grow. Genetic and epigenetic changes initiate and facilitate progression of normal cells toward malignancy, and chronic tissue damage provides a pro-mutagenic environment to accelerate this process. Chronic inflammation can create a cancer-promoting environment to support survival and proliferation of abnormal cells and hastens progression.
Autophagy plays an important role, not only in different stages of tumorigenesis, but also in the disease states that give rise to a microenvironment that promotes tumorigenesis in the first place. The role of autophagy in disease states associated with a higher risk of cancer, such as chronic liver disease, obesity, and inflammatory bowel disease is increasingly clearer. Pharmacological manipulation of autophagy with the intention of preventing a microenvironment rife for tumor initiation in these disease states may in fact require an opposite approach than pharmacological manipulation of autophagy to limit tumor progression once premalignant cells have been established. In this review we will highlight the regulation of autophagy machinery with specific emphasis on drugable targets, the role of autophagy in physiology and the links of autophagy with the disease states listed above associated with increased cancer risk, and the role of autophagy within the cells destined to become cancers. Finally we will discuss how induction of autophagy may limit the promotion of a hospitable tumor microenvironment, whereas inhibition of autophagy may limit tumor growth and metastases in established tumors.
Regulation of the autophagy machinery
Autophagy involves the engulfment of intracellular components into double membrane vesicles, termed autophagosomes, which fuse with lysosomes to form autolysosomes, where the autophagic contents are degraded. The availability of nutrients, growth factors and hormones as well as stress, regulate autophagy. The mammalian target of rapamycin complex 1 (mTORC1) is a major negative regulator of autophagy. mTORC1 promotes protein synthesis, cell division and metabolism in response to the nutrient, growth factor and hormone availability, while suppressing autophagy. It is not uncommon that tumor cells take advantage of the growth-promoting function of mTOR by acquiring activating mutations upstream; therefore, suppression of autophagy may occur in some tumors. Stressors such as amino acid depletion and hypoxia also signal through mTORC1 and induce autophagy (1, 2). Autophagy can also be regulated by pathways independent of mTORC1, including hypoxia-inducible factors (HIFs), LKB1-AMP-activated protein kinase (AMPK) and Protein kinase C (3–10).
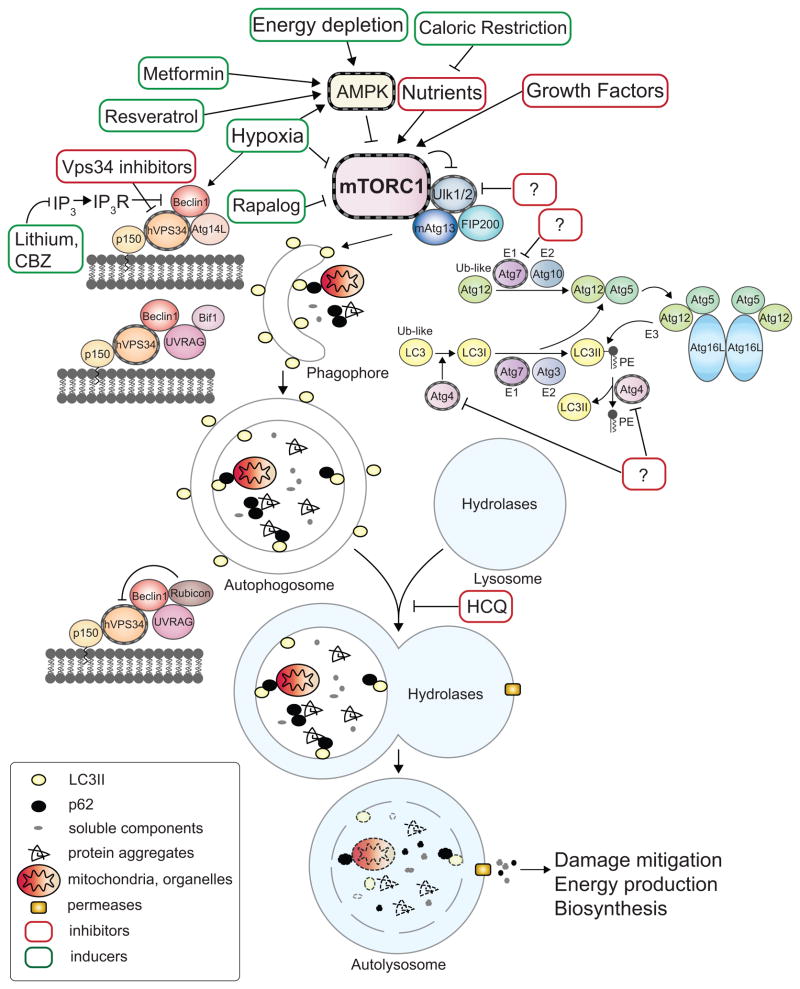
The central machinery of autophagy includes a series of complexes comprised of autophagy-related (Atg) proteins, executing the assembly of autophagosomes. The Ulk1/2 (mammalian Atg1) complex receives signals from mTORC1 and AMPK. AMPK regulates the Ulk1 complex by phosphorylating Ulk1 directly as well as the mTORC1 component Raptor (9–12). Several Beclin1/Vps34 (class III phosphatidylinositol-3 kinase)-containing complexes dictate the sequential steps of autophagosome formation and promote fusion of autophagosomes with lysosomes. Two interconnected ubiquitin-like conjugation systems act in expansion of autophagosome membranes. For detailed molecular mechanism, refer to (5). Proteins that can be targeted to modulate autophagy are highlighted in Fig. 1. Rapamycin and its analogs, metformin, resveratrol, lithium and carbamazepine (CBZ) stimulate autophagy (13–15). Vps34 inhibitors can potentially suppress autophagy induction while chloroquine (CQ) and its analog hydroxychloroquine (HCQ) prevent lysosomal acidification and impede the degradation of autophagic cargo and clearance of autolysosomes (16).
Figure 1. Machinery and small molecule modulators of autophagy.
The events of autophagosome formation- nucleation, expansion and maturation are depicted along with molecular machinery that regulates this process. The major negative regulator of autophagy, mTORC1, which integrates stimuli including availability of nutrients or growth factors, energy depletion or hypoxia, is also shown. Drugable protein targets are highlighted with striated outlines. Green or Red boxes indicate autophagy stimulators or inhibitors respectively. Question marks represent inhibitors aiming at potential targets, the kinase Ulk1, the cysteine protease Atg4 and the E1-like ubiquitination enzyme Atg7. AMPK: AMP-activated protein kinase. PE: phosphatidylethanolamine. IP3: inositol-1,4,5-triphosphate. IP3R: inositol-1,4,5-triphosphate receptor. CBZ: carbamazepine. HCQ: hydroxychloroquine.
Distinct from proteasomal degradation, which only proteolyzes individual, soluble proteins inside the proteasomal barrels, autophagy is the only cellular mechanism for degrading large cellular components such as protein aggregates and entire organelles. Autophagic substrates may include cytoplasm, organelles, proteins and protein aggregates as well as the autophagy components that associate with the autophagosomal inner membrane (Fig. 1). Autophagy can function in the non-selective bulk degradation of cytoplasmic contents or in the selective turnover and elimination of specific cellular components. To selectively target substrates to autophagosomes, protein and organelle substrates are conjugated with ubiquitin, while selective autophagic degradation of lipid droplets may use other signals (17, 18).
Autophagy cargo receptors such as p62 and NBR1, interact with both the ubiquitin on cargo and with the autophagy machinery through LC3-interacting (LIR) domains. This enables the cargo receptors to recognize and deliver cargo to autophagosomes (19–22) (Fig. 2). The Bcl-2 family BH3-only proteins BNIP3 and NIX (BNIP3L) are also critical for selective elimination of mitochondria (23). Evidence suggests that NIX, which localizes to the mitochondrial outer membrane, interacts with LC3 family proteins such as γ-aminobutyric acid receptor-associated protein (GABARAP) (24, 25) and likely serves as a cargo receptor for mitophagy in erythroid maturation (26). NIX may also promote mitochondrial loss of membrane potential (27), facilitating selective autophagy. Whether BNIP3 acts as a cargo receptor is still unknown. Cargo receptors are degraded along with the cargo and their accumulation, often in large aggregates in the case of p62, is symptomatic of autophagy inhibition.
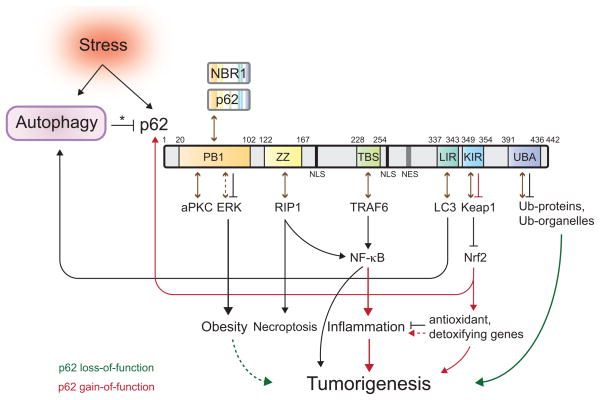
Figure 2. Role of autophagic regulation of p62 in cell signaling.
Stress upregulates autophagy and also induces the accumulation of the autophagic cargo receptor protein p62. Autophagy regulates p62 turnover and its protein levels in cells (denoted by an asterisk). Domains of p62 are depicted diagrammatically. PB1: Phox and Bem1p-1 oligomerization domain. ZZ: zinc finger. TBS: TRAF6-binding sequence. LIR: LC3-interacting region. KIR: Keap1-interacting region. UBA: ubiquitin-associated domain. Numbers represent the corresponding amino acid positions in mouse p62. NLS: nuclear localization signal. NES: nuclear export signal (40, 43, 76). Brown arrows indicate p62-protein interactions. The dash indicates a putative interaction between p62 and ERK (77). Accumulation of p62 activates Nrf2, while p62 itself is a transcriptional target of Nrf2, creating a positive feedback loop (45). Green or red depicts signals occurring when p62 loses or gains function, respectively. The dashed green line shows a putative role of obesity in tumorigenesis (see Fig. 3). The dashed red line shows a putative consequence of hyperactive Nrf2.
Autophagy in cellular refreshment
Autophagy-mediated protein quality control
Autophagy constitutively degrades excess or damaged proteins and organelles through its basal activity, which is critical to maintain cellular homeostasis and function. This is especially important for post-mitotic cells, which cannot dilute cellular waste products through cell division. Impaired autophagy in mice causes quiescent cells such as neurons and hepatocytes, to accumulate ubiquitin-and p62-positive protein inclusions, aberrant membranous structures and deformed mitochondria, accompanied by neuronal degeneration and liver injury (28, 29). p62- and ubiquitin-containing inclusions (Mallory-Denk bodies) have been associated with a variety of liver diseases including hepatitis and hepatocellular carcinoma (HCC) (30). This accumulated p62 causes hepatotoxicity in liver, since genetic ablation of p62 partially suppresses the formation of protein inclusions and liver injury in autophagy-deficient mouse cells (19). It will be of interest to see if genetic impairment of autophagy is the cause of some liver disease in humans and if stimulation of autophagy mitigates disease progression.
In neuronal cells, the accumulation of p62 does not account for the neurodegeneration observed in autophagy-deficient mice, since the ablation of p62 did not prevent disease, indicating that role of autophagy in disease development is context-dependent. Failure of autophagy to remove dysfunctional mitochondria (mitophagy) has been mechanistically linked to the neurodegenerative Parkinson’s disease (20). Additionally, autophagy deficiency accelerates the progression of neurodegenerative diseases precipitated by expression of aggregate-prone mutant proteins, which rely on autophagy for clearance. Enhancement of autophagy has been suggested as an approach to mitigate neurodegeneration caused by pathogenic proteins such as mutant Huntingtin in Huntington’s disease (13, 31).
Autophagy deficit may also contribute to Alzheimer’s disease (AD). It is reported recently that presenilin1 (PS1), which is commonly mutated in early-onset familial AD, is required for the acidification of lysosomes/autolysosomes and the clearance of autophagosomes and cargo (32). This provides a possibility that defective autophagy causes AD and that estoration of autophagic degradation may slow the progression of AD.
Development of liver disease in α1-antitrypsin (AT)-deficient patients exemplifies another case of the significance of autophagy in cellular and organismal well-being by discarding aggregate-prone proteins and links the autophagy-mediated garbage disposal with chronic liver disease that is associated with increased risk of cancer. A point mutation in the liver-derived secretory AT protein (ATZ) renders it aggregate-prone, causing its accumulation in the endoplasmic reticulum (ER). ATZ aggregates cause hepatotoxicity, liver injury, inflammation and carcinogenesis. However, only a subgroup of afflicted homozygous patients develop liver fibrosis and eventually hepatocellular carcinoma (33). This discrepancy has been attributed to the interaction of ATZ with genetic or environment determinants. The autophagy-stimulating drug carbamazepine (CBZ) reduces the ATZ load and alleviates the associated symptoms in mice. This suggests that autophagy impairment may predispose ATZ homozygous patients to the ATZ-associated progressive liver disease and that enhancement of autophagy may be beneficial for disease prevention and therapy (34) (Fig. 3).
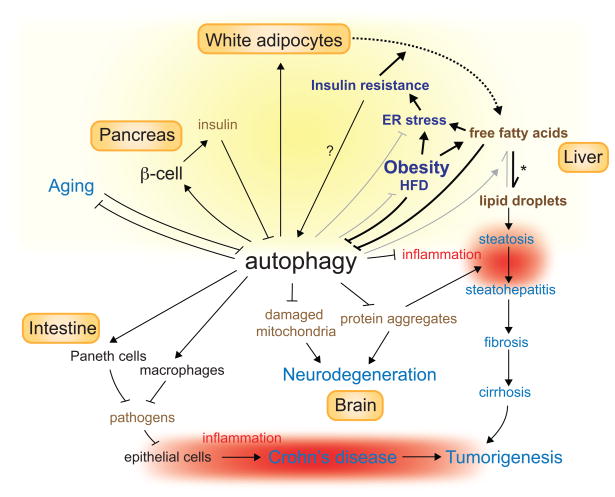
Figure 3. Role of autophagy in health and disease progression.
Autophagy is required for cellular and organismal heath and is involved in the pathogenesis of a variety of diseases. Disorders are shown in blue. Lines with an arrowhead or a cross short line represent positive or negative regulation of the pathway, respectively. Bold or gray lines represent pathways upregulated or downregulated respectively during lipid homeostasis collapse. The dashed line depicts flux of fatty acids from adipocytes to liver. The asterisk indicates the abnormal deposit of fatty acids in liver when lipid homeostasis is disrupted. The question mark denotes a putative regulation. Yellow shading shows where autophagy is involved in the regulation of lipid homeostasis. Red shading shows inflammation, which promotes tumorigenesis and is suppressed by autophagy. HFD: high-fat diet.
Autophagy-mediated organelle quality control
Autophagy eliminates damaged organelles and ensures their quality control, thereby maintaining organelle function and preventing the harmful consequences of organelle damage. Autophagy may sustain cellular metabolism through mitochondrial quality control, while impaired autophagy may lead to compromised or altered metabolism in part through the accumulation of dysfunctional mitochondria. Skeletal muscle-specific atg7 deficiency in mice causes reduced mitochondrial function, revealing the significance of autophagy for preservation of the functional mitochondrial pool (35). This mitochondrial function preserved by autophagy is particularly important for cells that need mitochondrial β-oxidation and oxidative phosphorylation for efficient energy production. Moreover, autophagy-mediated turnover of peroxisomes, which also function in fatty acid β-oxidation, might contribute to normal metabolism as well.
The autophagic elimination of damaged proteins and organelles, particularly mitochondria and peroxisomes, also removes potential sources of reactive oxygen species (ROS). ROS production in autophagy-defective mice is associated with tissue damage, chronic cell death and inflammation in the liver (19, 36). In mouse skeletal muscle, autophagy deficiency causes accumulation of abnormal mitochondria and elevated oxidative stress in myofibers accompanied by muscle degeneration with age (37). Studies in yeast suggest that the peroxisomal autophagy also restricts intracellular ROS. In methylotrophic yeast, autophagy constitutively degrades peroxisomes. Mutations in atg1, the yeast homolog of mammalian Ulk1, cause peroxisome accumulation and decreased activity of the peroxisomal enzyme catalase, which functions in detoxification and converts H2O2 to H2O, and increase ROS levels. Senescent human cells also show accumulation of peroxisomes with reduced capacity to import catalase and increased load of ROS (38). The autophagy status was not assessed in these aging cells; nevertheless, it is possible that autophagy also preserves the functional integrity of peroxisomes and limits ROS production from peroxisomes in mammals (38). Thus, failure of organelle quality control by autophagy can lead to toxic ROS production and disease.
The importance of controlling p62 levels by autophagy
The ability of autophagy to selectively eliminate specific proteins has an important role in cellular function. The autophagic substrate receptor p62, which is induced by stress to facilitate selective autophagic degradation, itself is a substrate of autophagy (39). p62 contains oligomerization and protein interaction domains and facilitates protein aggregate formation (19, 40) (Fig. 2). Failure to clear p62 due to impaired autophagy causes liver damage in mice and promotes tumorigenesis of allografts (19, 36). Thus, autophagic degradation of a specific protein, p62, has a role in preventing disease development and progression.
p62 regulates the NF-κB signaling
p62 is required for oncogenic Ras-driven NF-κB activation and lung adenocarcinoma in mice (41). By contrast, aberrant p62 accumulation in autophagy-defective cells abrogates NF-κB signaling that may promote tumorigenesis in the liver (36). NF-κB activates prosurvival and proinflammatory gene transcription. The role of NF-κB in tumorigenesis in the liver is becoming clear. Inhibition of NF-κB activation in hepatocytes in the liver causes hepatocyte cell death. Dying cells activate and recruit immune cells (such as Kupffer cells), causing cytokine and growth factor production and inflammation, which leads to compensatory proliferation and tumorigenesis. By contrast, inhibition of NF-κB in Kupffer cells prevents expression of tumor-promoting cytokines and inflammation and suppresses tumor development (42). Therefore, the role of NF-κB in tumorigenesis is context dependent. It will be interesting to delineate the interplay between autophagy deficiency-dependent p62 accumulation and NF-κB signaling in tumorigenesis.
p62 accumulation activates Nrf2-mediated transcription
Recent studies indicate that the autophagy deficiency-dependent p62 accumulation alters the regulation of another signaling pathway, transcription by NF-E2-related factor 2 (Nrf2). Nrf2 is a transcription factor mediating transcription of antioxidant and detoxifying genes. p62 interacts with Keap1, an adaptor for the Cullin-based E3-ligase that promotes Nrf2 degradation. By sequestrating Keap1, p62 accumulation causes activation of the Nrf2-target genes (43–45). The enzymes encoded by these genes metabolize and detoxify environmental carcinogens or endogenous mutagens such as ROS, protecting cells from damage and carcinogenesis. Counter-intuitively, simultaneous ablation of Nrf2, and the cytoprotective antioxidant/detoxifying pathway, in mouse liver alleviates the autophagy deficiency-dependent liver injury (43). Whether this is directly related to the function of Nrf2-targeted genes remains to be investigated. Constitutive activation of Nrf2 is associated with increased cancer incidence (46). It will be of interest to see if the autophagy-dependent accumulation of p62 promotes tumorigenesis through activating both Nrf2 and NF-κB.
Autophagy in metabolic homeostasis
Recent findings have indicated multiple roles of autophagy in lipid homeostasis. Epidemiological studies have linked disruption of lipid homeostasis manifested as obesity to increased risk for several types of cancer (47), suggesting the relevance of autophagy in cancer developed under this condition. Fatty liver disease and systemic metabolic disorders are associated with disturbed lipid homeostasis. Fatty liver ranges from simple steatosis to steatohepatitis, cirrhosis and ultimately can lead to HCC. Dysregulation of lipid metabolism or hepatic lipotoxicity is thought to trigger inflammation and fibrogenesis, which are associated with development of aggressive disease (48). Accumulation of fatty acids and the resulting fatty acid metabolites in liver renders hepatocytes more susceptible to injury, leading to cell death and activation of the inflammatory responses (49). This is similar to the phenotypes of autophagy-deficient mice; allelic loss of beclin1 causes steatosis, steatohepatitis, and spontaneous HCC (50). Although it is still unclear how exactly autophagy deficiency contributes to initiation or progression of disease, failure of lipid homeostasis leading to lipotoxicity and chronic inflammation, in addition to p62 accumulation, is a likely possibility.
Autophagy is important for accessing lipid stores through lipophagy, promoting lipid breakdown and preventing deposition in liver (18). In liver, lipid droplets continually undergo hydrolysis and the resulting free fatty acids are used forβ-oxidation or synthesis of very low-density lipoprotein (VLDL) for export, or are re-esterified back to triglycerides. Autophagy constitutively degrades lipid droplets (lipophagy) and inhibition of autophagy causes accumulation of lipid droplets in cultured hepatocytes and in mouse liver (18). Thus, autophagy facilitates the disposal of fatty acids and prevents fatty acid accumulation, which causes lipotoxicity, in liver.
Lipid homeostasis also involves the whole body storage and mobilization of fatty acids, and autophagy contributes to this systemic aspect. Recent data demonstrates that autophagy is required for differentiation of white adipose tissue, where most lipids in the body are stored. Adipocyte-specific atg7 deficiency in mice causes decreased numbers of white adipocytes, lower fatty acid levels in plasma, an imbalanced adipokine secretion profile, and higher insulin responsiveness. These mice are lean even when subjected to a high-fat diet, indicative of altered lipid metabolism (51, 52). Moreover, autophagy is also important for the integrity and function of the pancreaticβ-cells, which secrete insulin, a hormone regulating the storage and utilization of glucose and fat. β-cell mass and insulin secretion is also reciprocally regulated by fatty acids. Autophagy is again indispensible for the compensatory β-cell mass augmentation in response to a high-fat diet (53, 54). Therefore, autophagy is involved in local hepatocyte lipid metabolism as well as the systemic regulation of lipid homeostasis. To delineate the impact of autophagy defects in each aspect on pathology of lipid imbalance, inducible conditional knockout mouse models will be useful.
Autophagy is critical for lipid homeostasis. Unfortunately, factors that disrupt lipid homeostasis commonly suppress autophagy, leading to a vicious cycle. To achieve homeostasis, lipophagy should be upregulated during fasting to facilitate the mobilization of fatty acids and β-oxidation in hepatocytes in response to fluctuation in lipid levels. However, a long-term high-fat diet impairs the selectivity of autophagic degradation toward lipid droplets in liver when the high-fat fed mice were starved (18). Obesity also suppresses autophagy. In liver, inhibition of autophagy is likely responsible for the obesity-induced ER stress and insulin resistance (55). Insulin resistance, which mimics starvation, causes flux of fatty acids from white adipose tissue to liver, where it deposited. Impaired lipid autophagy impedes access of lipid. Autophagy inhibition-caused insulin resistance further augments the accumulation of lipids in liver. Inhibition of autophagy by a high-fat diet or obesity, therefore, exacerbates the imbalanced homeostasis (Fig. 3). In this regard, enhancement of autophagy would help restore lipid homeostasis.
Autophagy declines with age (56), correlating with altered metabolism manifested as ectopic fat deposition and intracellular garbage accumulation. This suggests that the imbalance of lipid homeostasis as well as waste accumulation and cellular functional degeneration due to suppressed autophagy may accelerate aging. In worms, autophagy is required for lifespan extension provided by dietary restriction (56). Resveratrol, which mimics starvation and induces autophagy, increases survival of mice on a high-fat diet and reverses age-related syndromes (57). Pharmacological inhibition of mTOR by rapamycin also extends lifespan in mice (58), providing a possibility that autophagy improves fitness and mediates longevity in mammals. Rapamycin prolongs the lifespan of mice, even late in their life (58), suggesting that enhancement of autophagy may possibly interrupt aging and related syndromes including increased cancer incidence (Fig. 3).
Autophagy confronts stress and environmental insults
Autophagy is upregulated in response to stress, including growth factor and nutrient limitation, energy depletion and hypoxia. In yeast, starvation induces autophagy, which recycles intracellular constituents to support metabolism, leading to adaptation and survival. In mammals, this self-cannibalistic function is conserved, and autophagy deficient mice cannot survive the neonatal starvation period and show indications of energy depletion (59).
The capability of autophagy to degrade proteins, lipids, glycogen and nucleic acids provides cells the flexibility to utilize intracellular components or various stores of nutrients for energy production and biosynthesis under stress (4, 60). Autophagy generates substrates such as nucleosides, amino acids, fatty acids and sugars from the breakdown of intracellular components. Metabolism of different substrates can produce unequal redox equivalents such as NADPH, which can support lipid biosynthesis and maintain cytosolic redox equilibrium (61, 62). Therefore, through selective autophagy, central metabolism may be supported by different substrates to restore metabolic and energy homeostasis, redox balance and biosynthesis. In this way autophagy can enable stress adaptation, maintenance of cellular fitness, and survival. Considering the nature of autophagy: inducible, selective or non-selective bulk degradation within a defined, membrane-enclosed area, autophagy is an efficient way to reallocate intracellular “macromolecular stores” to support bioenergetics and for use as building blocks for biosynthetic pathways under stress conditions.
Autophagy and host defense
Autophagy can also prevent disease by destroying microbial invaders (xenophagy). The autophagic endomembrane system delivers viral nucleic acids or microbial antigens to endosomes/lysosomes for induction of interferon or antigen presentation, activating immune responses (56, 63, 64). Recent findings indicate that impairment of the host defense function of autophagy cooperates with environmental factors such as infection to affect development of the chronic inflammatory bowel disease (IBD) Crohn’s disease. IBD increases the risk of developing cancer (65).
The etiology of Crohn’s disease has been attributed to the interaction of pathogens and genetic factors. A genome-wide study in patients of Crohn’s disease identified the association of variants of autophagy genes ATG16L1 and IRGM with disease susceptibility (64). ATG16L1 is recruited to the bacterial entry site, triggering autophagic degradation of pathogens and antigen presentation. This prevents persistence of pathogens and chronic inflammation (66, 67). Moreover, mice engineered to have a hypomorphic mutation in ATG16L1 develop disease that mimics Crohn’s disease. Hypomorphic ATG16L1 causes an abnormal response to murine norovirus infection in the intestine. In the mutant mice, Paneth cells, the epithelial cells in the small intestine that secrete antimicrobial peptides to protect the intestine, show abnormal granule packaging and secretion and altered transcription profiles upon infection (Fig. 3). If additionally challenged by chemical-induced injury, these mice manifest aberrant cytokine production and inflammation in intestine. These phenotypes resemble what has been seen in patients with Crohn’s disease (68). It will be interesting to see if these Atg16L mutant mice are also tumor prone. Role of autophagy or autophagy-related process in this type of disease, therefore, is multifaceted (63, 64).
Autophagy and early tumor cells
Autophagy suppresses tumor initiation by limiting genome mutation
The housekeeping function of autophagy maintains turnover of proteins and organelles and ensures homeostasis and cellular health, preventing disease conditions. In response to stress, autophagy eliminates damaged proteins and organelles. This damage mitigation of autophagy can be important for survival of tumor cells, which are commonly subjected to metabolic stress due to insufficient vascularization. Autophagy-deficient murine cells, although more susceptible to metabolic stress, have evident DNA damage response activation and have an increased frequency of chromosome gains and losses (69). Autophagy deficient mice are tumor-prone and liver tumors there from, as well as human liver tumors accumulate p62-containing protein aggregates, ER chaperones and activate the DNA damage response (36). Thus, by taking out the garbage, autophagy may suppress genomic instability to limit tumor initiation and progression.
Autophagy may also hinder proliferation of cells with cancer mutations, in addition to limiting genomic mutations in cells. Inhibition of autophagy delays oncogene-induced senescence (70). Senescence is believed to be a tumor suppressive mechanism attributed to induction of cell cycle exit. By facilitating senescence, autophagy can limit propagation of oncogenic mutations thereby suppressing tumorigenesis.
Autophagy suppresses tumor initiation and progression by limiting chronic inflammation
Autophagy maintains homeostasis by removing excess or damaged intracellular components and microbial invaders as well as by regulating lipid metabolism. This not only restrains damage, including genome instability, but also the subsequent inflammation. In autophagy-suppressive conditions, persistence of unresolved damage leads to chronic cell death and inflammation. In liver, and probably other tissues, this can provide a cancer-promoting environment.
Autophagy provides internal resource to support metabolism and mitigates damage, allowing cells to survive stress. Autophagy-deficient murine tumor cells in a background of defective apoptosis, which commonly occurs in tumors, undergo necrosis when subjected to metabolic stress (71). Necrosis also occurs in tumor allografts when apoptosis and autophagy are inhibited concurrently and is associated with inflammation. Therefore, autophagy limits genetic instability and inflammation that can predispose to tumor initiation, promotion and progression. In this regard, autophagy stimulation may prevent tumorigenesis.
Pharmacological manipulation of autophagy for cancer prevention
Autophagy induction with rapamycin or metformin may reduce the likelihood of malignant transformation
Epidemiological studies have linked metformin treatment with a reduced cancer risk in type 2 diabetic patients. In vitro and animal models also indicate that metformin suppresses tumorigenesis of various types of cancer. This anticancer activity of metformin has been attributed to activation of AMPK and the subsequent metabolic responses including reduced insulin resistance, insulin/insulin like growth factor level and mTOR signaling (15). It is therefore reasonable to speculate that autophagy activation with metformin, which is involved in these altered metabolic responses, will lower cancer risk. Retrospective analysis in renal-transplant recipients reveals that rapamycin suppresses tumorigenesis. Animal models also suggest the delayed tumor development by a rapalog (72). Given the role of autophagy in physiology discussed above, stimulation of autophagy with metformin or a rapalog may benefit patients with metabolic syndrome, chronic hepatitis or IBD.
Autophagy inhibition sensitizes transformed cells to cell death
Since autophagy is a stress survival pathway, aggressive cancer cells may become more sensitive to autophagy inhibition. The autophagy inhibitor CQ, which induces lysosomal stress and prevents autophagic degradation, preferentially kills Myc-expressing mouse cells in vitro in a p53-dependent manner. CQ impairs spontaneous lymphomagenesis in Myc-transgenic mice that model Burkitt’s lymphoma and in atm-deficient mice that model ataxia telangiectasia also dependent on p53 (73). Thus lysosomal stress and/or autophagy inhibition may serve as a stress to enhance tumor suppression pathways to harness tumorigenesis when cancer mutations exist.
In a p53-deficient, Myc-induced mouse model of lymphoma, inhibition of autophagy promotes cell death and delays the recurrence of tumors following p53 reactivation or alkylating agent treatment (74). This suggests that once cancer occurs, tumor cells may rely on the function of autophagy to survive the metabolic demand resulting from proliferation, but also survival to therapeutic stress, and in the tumor microenvironment during latency. Taking advantage of the dependency of tumor cells on autophagy to survive, many clinical trials HCQ are underway for advanced disease. (75).
Conclusion
As a cell-refreshing and metabolism-regulating pathway, autophagy is required for normal operation of cellular and organismal physiology. Autophagy-deficient models in mice reveal the role of autophagy in preventing progression of chronic diseases. In this perspective, autophagy stimulation/restoration underscores a strategy to prevent disease initiation and progression by sustaining normal physiology and handling stress properly. Of note, since autophagy exerts its effects in multiple aspects, effects of autophagy enhancement are highly cellular context-dependent. In addition to benefits, the risk of autophagy stimulation should be taken into account (75). For example, the magnitude and duration of autophagy activation may lead to different consequences. Over-induction may lead to perturbation of homeostasis, an autophagy traffic jam or autophagic cell death. Moreover, in contrast to efforts to exploit the cytoprotective nature of autophagy to prevent tumor initiation, inhibition of autophagy may enhance tumor suppression responses and promote cell death of existing cancers. Currently, our understanding of the role of autophagy in physiology and pathology is still in an early stage. Further investigation of the role of autophagy in different contexts would warrant development of tractable regimens for modulation of autophagy in cancer prevention.
Acknowledgments
We thank members of the White laboratory for helpful comments and discussions and Tami Sharkey for assistance with preparation of the manuscript. E. W. acknowledges support from the NIH (R37 CA53370, RO1 CA130893, RC1 CA147961, and the New Jersey Commission on Cancer Research (09-1083-CCR-EO) and the DoD (W81XWH06-1-0514 and W81XWH05).
References
- 1.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y, Wang Y, Kim E, Beemiller P, Wang CY, Swanson J, et al. Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with Rheb. J Biol Chem. 2007;282:35803–13. doi: 10.1074/jbc.M705231200. [DOI] [PubMed] [Google Scholar]
- 3.Mazure NM, Pouyssegur J. Hypoxia-induced autophagy: cell death or cell survival? Curr Opin Cell Biol. 2010;22:177–80. doi: 10.1016/j.ceb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 4.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–8. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–31. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–24. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 7.Sakaki K, Wu J, Kaufman RJ. Protein kinase Ctheta is required for autophagy in response to stress in the endoplasmic reticulum. J Biol Chem. 2008;283:15370–80. doi: 10.1074/jbc.M710209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 2010;22:157–68. doi: 10.1016/j.ceb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011 doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-Activated Protein Kinase Connects Energy Sensing to Mitophagy. Science. 2011;331:456–61. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JW, Park S, Takahashi Y, Wang HG. The association of AMPK with ULK1 regulates autophagy. PLoS One. 2010;5:e15394. doi: 10.1371/journal.pone.0015394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–26. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renna M, Jimenez-Sanchez M, Sarkar S, Rubinsztein DC. Chemical inducers of autophagy that enhance the clearance of mutant proteins in neurodegenerative diseases. J Biol Chem. 2010;285:11061–7. doi: 10.1074/jbc.R109.072181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morselli E, Galluzzi L, Kepp O, Criollo A, Maiuri MC, Tavernarakis N, et al. Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol. Aging (Albany NY) 2009;1:961–70. doi: 10.18632/aging.100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ben Sahra I, Le Marchand-Brustel Y, Tanti JF, Bost F. Metformin in cancer therapy: a new perspective for an old antidiabetic drug? Mol Cancer Ther. 2010;9:1092–9. doi: 10.1158/1535-7163.MCT-09-1186. [DOI] [PubMed] [Google Scholar]
- 16.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–12. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 17.Oku M, Sakai Y. Peroxisomes as dynamic organelles: autophagic degradation. FEBS J. 2010;277:3289–94. doi: 10.1111/j.1742-4658.2010.07741.x. [DOI] [PubMed] [Google Scholar]
- 18.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–5. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 20.Wild P, Dikic I. Mitochondria get a Parkin' ticket. Nat Cell Biol. 2010;12:104–6. doi: 10.1038/ncb0210-104. [DOI] [PubMed] [Google Scholar]
- 21.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–16. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 22.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–46. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarten M, Mohrluder J, Ma P, Stoldt M, Thielmann Y, Stangler T, et al. Nix directly binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy. 2009;5:690–8. doi: 10.4161/auto.5.5.8494. [DOI] [PubMed] [Google Scholar]
- 25.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–5. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–5. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 30.Zatloukal K, French SW, Stumptner C, Strnad P, Harada M, Toivola DM, et al. From Mallory to Mallory-Denk bodies: what, how and why? Exp Cell Res. 2007;313:2033–49. doi: 10.1016/j.yexcr.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 31.Rose C, Menzies FM, Renna M, Acevedo-Arozena A, Corrochano S, Sadiq O, et al. Rilmenidine attenuates toxicity of polyglutamine expansions in a mouse model of Huntington's disease. Hum Mol Genet. 2010;19:2144–53. doi: 10.1093/hmg/ddq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–58. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perlmutter DH. Autophagic disposal of the aggregation-prone protein that causes liver inflammation and carcinogenesis in alpha-1-antitrypsin deficiency. Cell Death Differ. 2009;16:39–45. doi: 10.1038/cdd.2008.103. [DOI] [PubMed] [Google Scholar]
- 34.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–32. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 35.Wu JJ, Quijano C, Chen E, Liu H, Cao L, Fergusson MM, et al. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging (Albany NY) 2009;1:425–37. doi: 10.18632/aging.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–15. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 38.Manjithaya R, Nazarko TY, Farre JC, Subramani S. Molecular mechanism and physiological role of pexophagy. FEBS Lett. 2010;584:1367–73. doi: 10.1016/j.febslet.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moscat J, Diaz-Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci. 2007;32:95–100. doi: 10.1016/j.tibs.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, et al. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13:343–54. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–90. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 43.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–23. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 44.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–85. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–91. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kensler TW, Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis. 2010;31:90–9. doi: 10.1093/carcin/bgp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer. 2004;4:579–91. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- 48.Trauner M, Arrese M, Wagner M. Fatty liver and lipotoxicity. Biochim Biophys Acta. 2010;1801:299–310. doi: 10.1016/j.bbalip.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 49.Neuschwander-Tetri BA. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: the central role of nontriglyceride fatty acid metabolites. Hepatology. 2010;52:774–88. doi: 10.1002/hep.23719. [DOI] [PubMed] [Google Scholar]
- 50.White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol. 2010;22:212–7. doi: 10.1016/j.ceb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106:19860–5. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–39. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–24. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 54.Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–32. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 55.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–78. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–5. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 60.Kuma A, Mizushima N. Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Semin Cell Dev Biol. 2010;21:683–90. doi: 10.1016/j.semcdb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 61.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noguchi Y, Young JD, Aleman JO, Hansen ME, Kelleher JK, Stephanopoulos G. Effect of anaplerotic fluxes and amino acid availability on hepatic lipoapoptosis. J Biol Chem. 2009;284:33425–36. doi: 10.1074/jbc.M109.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galluzzi L, Kepp O, Zitvogel L, Kroemer G. Bacterial invasion: linking autophagy and innate immunity. Curr Biol. 2010;20:R106–8. doi: 10.1016/j.cub.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 64.Netea MG, Joosten LA. A NOD for autophagy. Nat Med. 2010;16:28–30. doi: 10.1038/nm0110-28. [DOI] [PubMed] [Google Scholar]
- 65.Danese S, Mantovani A. Inflammatory bowel disease and intestinal cancer: a paradigm of the Yin-Yang interplay between inflammation and cancer. Oncogene. 2010;29:3313–23. doi: 10.1038/onc.2010.109. [DOI] [PubMed] [Google Scholar]
- 66.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–7. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 67.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 68.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–45. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blagosklonny MV. Prevention of cancer by inhibiting aging. Cancer Biol Ther. 2008;7:1520–4. doi: 10.4161/cbt.7.10.6663. [DOI] [PubMed] [Google Scholar]
- 73.Maclean KH, Dorsey FC, Cleveland JL, Kastan MB. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest. 2008;118:79–88. doi: 10.1172/JCI33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.White E, Dipaola RS. The Double-Edged Sword of Autophagy Modulation in Cancer. Clin Cancer Res. 2009;15:5308–17. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wooten MW, Geetha T, Seibenhener ML, Babu JR, Diaz-Meco MT, Moscat J. The p62 scaffold regulates nerve growth factor-induced NF-kappaB activation by influencing TRAF6 polyubiquitination. J Biol Chem. 2005;280:35625–9. doi: 10.1074/jbc.C500237200. [DOI] [PubMed] [Google Scholar]
- 77.Lee SJ, Pfluger PT, Kim JY, Nogueiras R, Duran A, Pages G, et al. A functional role for the p62-ERK1 axis in the control of energy homeostasis and adipogenesis. EMBO Rep. 2010;11:226–32. doi: 10.1038/embor.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]