Abstract
α1-Antitrypsin (AAT) secreted from hepatocytes is an inhibitor of neutrophil elastase. Its normal circulating concentration functions to maintain the elasticity of the lung by preventing the hydrolytic destruction of elastin fibers. Severely diminished circulating concentrations of AAT, resulting from the impaired secretion of genetic variants that exhibit distinct polypeptide folding defects, can function as an etiologic agent for the development of chronic obstructive pulmonary disease. In addition, the inappropriate accumulation of structurally aberrant AAT within the hepatocyte endoplasmic reticulum can contribute to the etiology of liver disease. This article focuses on the discovery and characterization of a biosynthetic quality control system that contributes to the secretion of AAT by first facilitating its proper structural maturation, and then by orchestrating the selective elimination of those molecules that fail to attain structural maturation. Mechanistic elucidation of these interconnected quality control events recently led to the identification of an underlying genetic modifier capable of accelerating the onset of end-stage liver disease by impairing the efficiency of an initial step in the protein disposal process.
Keywords: endoplasmic reticulum, glycoproteins, biosynthetic quality control, proteolysis, autophagy
A POST-TRANSLATIONAL PERSPECTIVE OF DISEASE PATHOGENESIS
It stands to reason that if most diseases are mechanism based, then the successful identification of biomarkers and development of therapeutic treatments will rely on a detailed understanding of the defective system that underlies each disorder (1). Because many genetic diseases are actually manifested at the level of aberrant protein structure (2), a current challenge is to elucidate how post-translational processes events (acting on the encoded proteins, rather than the genomic blueprint) might influence the molecular pathogenesis of numerous inherited disorders. This mechanistic pursuit is well designed to elucidate the underlying mechanisms that contribute to the development of numerous loss-of-function and gain-of-toxic-function disorders caused by failed protein deployment and the inappropriate accumulation of structurally aberrant molecules, respectively.
α1-Antitrypsin (AAT) is a member of the serine proteinase inhibitor (serpin) superfamily. One of its primary physiological roles is to inhibit the hydrolytic activity of elastase secreted from activated neutrophils, thereby maintaining the structural integrity of lung elastin fibers (3, 4). Importantly, a severely diminished circulating concentration of the inhibitor can result in the elastolytic destruction of lung connective tissue, which is a known risk factor for the development of chronic obstructive pulmonary disease (the predominant loss-of-function phenotype) (5–7). Importantly, hepatocytes function as the predominant site for AAT biosynthesis, and the inappropriate accumulation of structurally impaired molecules in the endoplasmic reticulum (ER) is known to function as a risk factor for the development of childhood liver disease (the primary gain-of-toxic-function disorder) (8). AAT deficiency has emerged as a paradigm to investigate the pathologic variability associated with conformational diseases, all of which are caused by cytotoxicity associated with the inappropriate accumulation of a structurally aberrant protein.
Several naturally occurring genetic variants of AAT have been identified that coincide with a diminished circulating concentration of the inhibitor, making it an ideal paradigm to initiate a mechanistic investigation of how cells facilitate the structural maturation and intracellular transport of secretory proteins. Intensive experimental investigation led to the discovery that correct conformational maturation of the newly synthesized polypeptide is a prerequisite for its productive transport along compartments of the secretory pathway. Subsequent discoveries included the identification of a biosynthetic quality control checkpoint that operates in the early secretory pathway, branches of which (1) initially facilitate the conformational maturation of newly synthesized AAT, and then (2) orchestrate the proteolytic elimination of structurally aberrant molecules. The mechanistic insights gained from the detailed investigation of the newly identified post-translational checkpoint are described subsequently here, as is the recent identification of a genetic modifier capable of accelerating the onset of end-stage liver disease.
SELECTIVE DEPLOYMENT OF FUNCTIONAL AAT
Genetic information is directly transformed into biological activity in response to the correct conformational maturation and deployment of the encoded proteins. The proper distribution of these molecules is of paramount importance to all living cells, and requires the services of numerous checkpoints. Arguably, this dual intention is best exemplified in the ER, which provides a physical route through which newly synthesized proteins are transported prior to their delivery to the eukaryotic cell surface (9). Therein, checkpoint systems operate to ensure the correct conformational maturation of translated proteins and the selective removal of those unable to attain this structural milestone.
Checkpoints systems are of interest to current biomedical research endeavors because one's ability to treat either the lung or liver diseases associated with AAT deficiency successfully might ultimately require detailed understanding of how the systems operate. Our early studies indicated that the productive transport of newly synthesized AAT requires prior adoption of native structure. Subsequent studies uncovered the existence of a checkpoint system composed of two opposing, but complementary, branches. Their binary nature is to initially facilitate AAT conformational maturation, and then orchestrate the selective degradation of misfolded molecules by 26S cytosolic proteasomes (10).
FACILITATED CONFORMATIONAL MATURATION
Identical to other proteins, the final three-dimensional conformation of AAT is dictated by the primary amino acid sequence. Nevertheless, the efficiency of the process is boosted in response to physical engagement with molecular chaperones, which help to facilitate the attainment of native structure by impeding nonproductive aggregation events (11). In particular, the ER contains a specialized “protein folding environment” where molecular chaperones and chaperone-related proteins engage newly synthesized polypeptides. A variety of molecular chaperones and accessory propteins that function in the ER have been discovered (10–12).
AAT undergoes asparagine-linked glycosylation at three specific sites on the polypeptide backbone during its translocation into the ER (13). The appendages not only help maintain AAT's solubility, but conveniently generate a scaffold by which a small ensemble of oligosaccharide-processing enzymes mediate the protein's entrance into a specialized glycoprotein folding pathway (Figure 1, step 2). Each appendage consists of branched, 14-unit oligosaccharide. Cotranslational removal of the two outer glucose units by glucosidases I and II generates a monoglycosylated asparagine-linked oligosaccharide (i.e., GlcMan9GlcNAc2) recognized by the lectin-like molecular chaperones, calnexin and calreticulin. Release from either lectin coincides with enzymatic removal of the remaining glucose unit by ER glucosidase II (14). UDP-glucose:glycoprotein glucosyltransferase functions as a glycoprotein-folding sensor that uses UDP-glucose as a donor to catalyze the transfer of a single glucose unit back to high mannose-type glycans attached to nonnative proteins (15, 16). The monoglucosylated oligosaccharides promote post-translational reassembly of nonnative glycoproteins with the glycoprotein-folding machinery (17, 18). Conformational maturation releases glycoproteins from the protein-folding machinery, coupling this process to productive transport beyond the ER (19) by entrance into coatamer protein complex II (COP II) vesicles en route to the Golgi complex (10). In contrast, proteins that remain unfolded, or eventually misfold, can persistently interact with the system that is used to promote folding, thereby preventing productive deployment.
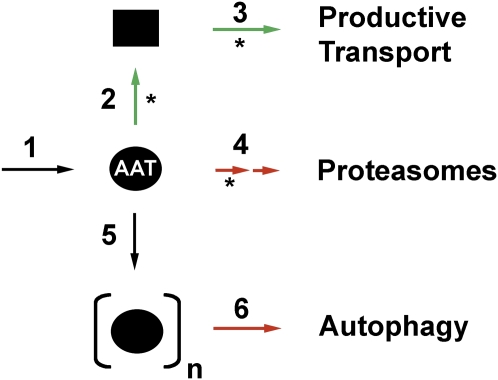
Figure 1.
Intracellular fates for newly synthesized α1-antitrypsin (AAT). After biosynthesis and translocation into the endoplasmic reticulum (ER) lumen (step 1), the modification of asparagine-linked oligosaccharides promotes physical interaction with the glycoprotein-folding machinery, which promotes conformational maturation (step 2) and productive transport (step 3). The removal of mannose units and polyubiquitinylation targets misfolded polypeptides for dislocation into the cytosol for proteolytic destruction by 26S proteasomes (step 4). The PI Z variant can undergo loop-sheet polymerization (step 6), and these are removed from cells by autophagy (step 6). Events that lead to correct folding and transport are depicted with green arrows. Events that selectively target AAT for intracellular degradation are depicted with red arrows. Events that are orchestrated through the modification and/or recognition of asparagine-linked oligosaccharides are shown with asterisks.
RETENTION AND DEGRADATION OF MISFOLDED AAT
A different fate (than that described previously here) awaits polypeptides unable to complete structural maturation and/or unassembled into larger macromolecular complexes. Rather than clogging the secretory pathway, these molecules are eliminated from the secretory pathway by a complex series of disposal systems, collectively coined “ER-associated degradation” (ERAD), which targets substrates into the cytosol for degradation by 26S proteasomes (12, 20). The exact combination of events by which any individual protein is selectively removed is still under investigation, but apparently depends on its location within the ER (lumen or membrane), and whether it has undergone asparagine-linked glycosylation (21), as described previously here.
Opportunistic removal of α1,2-linked mannose units in response to prolonged ER residence (22, 23) is diagnostic of the failure of a soluble secretory glycoprotein, such as AAT, to acquire native structure within a biologically relevant time frame. The intracellular degradation of mammalian glycoproteins, via ERAD, involves the removal of several mannose units by ER mannosidase I (ERManI) and perhaps additional members of the class 47 glycosylhydrolase family, especially in yeast (24–28). The modified glycans promote extraction of glycoproteins from the calnexin cycle (18), and, in combination with nonnative protein structure, are suspected of completing the formation of a proposed bipartite glycoprotein ERAD (GERAD) signal. The temporal manner in which the bipartite signal is formed is suspected of distinguishing chronically misfolded glycoproteins from nonnative wild-type folding intermediates, such that only the former population is degraded (29) (Figure 1, step 4). Subsequent dislocation of the tagged proteins into the cytosol for proteasomal destruction involves the cooperation of additional events and proteins, some of which are currently under investigation.
REGULATION OF GERAD SUBSTRATE SELECTION
The aforementioned findings support a model in which ER mannosidase I contributes to the operation of a post-translational checkpoint through the regulation of an important tempospatial decision that underlies a performance-based measurement of glycoprotein conformational maturation. That the glycosidase's intracellular concentration plays a central role in the establishment of an equitable glycoprotein quality control standard was recently substantiated by the capacity of experimentally overexpressed ERManI to selectively target nascent molecules of wild-type AAT and transferrin into GERAD (29).
Basal Checkpoint Regulation
The underlying mechanism by which GERAD substrate selection is controlled under basal conditions recently emerged. We recently reported (30) that the recombinant human and endogenous mouse ERManI orthologs are subject to rapid lysosomal down-regulation after biosynthesis (Figure 2). This fate is apparently controlled by signals that reside in the aminoterminal cytoplasmic tail, and that hypothesis is still under intense investigation. The simplest explanation as to why lysosomes, rather than proteasomes, are used in this process is to avoid the potential complications that might arise from controlling the glycosidase concentration by the system for which it functions. However, regardless of the explanation, the data imply that ERManI is subjected to proteolytically driven checkpoint control, as are other members of the ERAD system (31, 32). Proteolytic regulation of ERManI has not yet been demonstrated in budding yeast, suggesting that a distinct mechanism might operate.
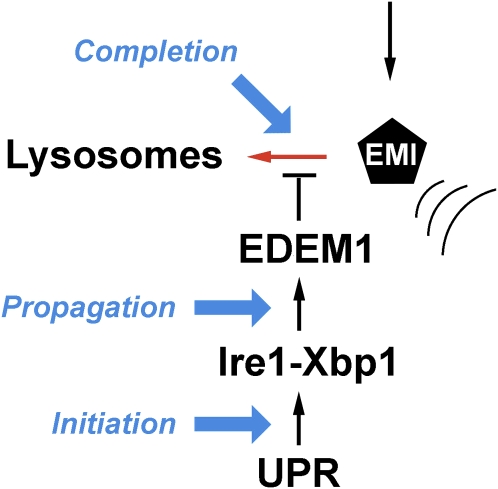
Figure 2.
Regulation of the intracellular endoplasmic reticulum (ER) mannosidase I (EMI) concentration. The newly synthesized protein is subjected to intracellular proteolysis under basal conditions (red arrow). Its concentration is elevated as a component of the unfolded protein response (UPR). Signal generation is initiated as part of the inositol-responsive element 1 - X box binding protein 1 (Ire1-Xbp1) branch, and is propagated through the elevated transcription of the gene that encodes an ER degradation–enhancing mannosidase-like (EDEM) protein that is translocated into the ER lumen. Completion of the signal circuit, which helps to boost the efficiency of glycoprotein ER-associated degradation (GERAD) substrate selection, involves a physical interaction with EDEM1, which suppresses the lysosomal down-regulation of EMI. The sequential stages of signaling are shown in blue.
Checkpoint Activation
The excessive accumulation of nonnative proteins in the ER can evoke a coordinated adaptive signaling network, designated the unfolded protein response (UPR) (33), that minimizes the consequences of ER stress through the action of several transcriptional programs.
ERAD is easily saturated, which can lead to the inappropriate accumulation of unfolded proteins and induction of the UPR (34). Therefore, the efficiency of ERAD is boosted by the UPR as one of several means to relieve ER stress. The accelerated rate at which misfolded proteins are selectively degraded is coordinated by the inositol-responsive element 1 - X box binding protein (Ire1-Xbp1) branch of the mammalian UPR (35). However, the mechanics by which linkage operates in higher eukaryotes has remained elusive.
The enhanced efficiency of ERAD coincides with the elevated transcription of ER degradation–enhancing mannosidase-like proteins (EDEMs), which are members of the class 47 glycosylhydrolase family (24). Although some EDEM orthologs in lower eukaryotes exhibit mannosidase activity (36), in vitro activity has not been detected for any of the mammalian orthologs (37, 38). Recent findings imply that the transcriptional elevation of EDEM1 boosts the efficiency of glycoprotein ERAD through the formation of a complex that suppresses the proteolytic down-regulation of ERManI (39). The new findings imply that ERManI, by functioning as a downstream effector target of EDEM1, represents a checkpoint activation paradigm by which the mammalian UPR coordinates the boosting of ERAD. The partnership is suspected to allow the rate of mannose removal to operate over a range of conditions and on a specific timescale, commensurate with the overall folding capacity of the ER. This mechanistic insight is in agreement with the recent prediction (40) that post-translational events will probably contribute to the completion of UPR signaling circuits (Figure 2). Again, this observation represents a paradigm shift in the transcriptional manner by which budding yeast coordinate the boosting of GERAD in response to ER stress. Importantly, the new knowledge sets the stage for exploiting this interface as a potential target for the therapeutic intervention of conformational diseases of the secretory pathway (1, 2).
LOOP-SHEET POLYMERIZATION OF AAT
To this point, the fate of misfolded AAT has been discussed. However, approximately 95% of patients that exhibit severe AAT deficiency are homozygous for the proteinase inhibitor Z (PI Z) variant (incidence of homozygosity is ∼1/1,800 live births). A single amino acid substitution (Glu394Lys) at the base of the reactive center loop diminishes the molecule's secretion in response to hindered folding kinetics (3, 41, 42), such that only a very small fraction of monomers are secreted from hepatocytes (41–43). The mutation promotes the specific conformational rearrangement of a late folding intermediate that favors reactive-center-loop insertion between molecules, resulting in subsequent polymerization (5, 44). In the generally accepted “loop-sheet polymerization” model, sequential insertion of the reactive center loop into the β sheet A of another molecule generates noncovalent PI Z polymers. The concomitant accumulation of a fraction of undegraded molecules leads to the formation of diastase-resistant inclusion bodies derived from the hepatocyte ER, which functions as an etiologic agent in the development of liver disease.
MECHANISMS OF CELLULAR INJURY
Accumulation of the PI Z variant induces ER stress. However, several reports have convincingly demonstrated that the UPR is not activated either in numerous cultured cell lines (15, 45, 46) or in transgenic mice (47). The exact mechanism(s) by which the protein-accumulated protein leads to cellular injury is still unknown, but is currently under intense investigation. A full explanation of the discoveries would exceed the page limitations for this article, but include the augmented transcription of nuclear factor–B (45). Moreover, a marked autophagic response has been detected that strongly correlates with the absolute amount of the mutant protein accumulated within the individual cell (48). This alone implies that the system is playing a pivotal role in endeavoring to remove the material in an attempt to protect cells from injury. Finally, stimulation of the apoptotic cascade with specific patterns of both mitochondrial autophagy and mitochondrial injury has been detected (49, 50).
IDENTIFICATION OF A DISEASE MODIFIER
The development of liver disease associated with the accumulation of PI Z polymers in the hepatocyte ER is highly variable and can occur at any age. Importantly, only a subset (10–15%) of patients who accumulate PI Z polymers in the hepatocyte ER actually develop clinically relevant liver damage (8).
The time of disease onset generally falls into three broad age categories (i.e., infants and toddlers, children, and adults). These clinical observations, plus the fact that transgenic mouse models that overexpress the human PI Z transgene develop liver injury, but not end-stage liver disease, implicates the contribution of genetic modifiers. To this end, a few groups (including our own) have attempted to unravel the mechanisms by which glycoprotein folding and degradation systems might function as a mediator of the liver disease.
The observation that PI Z polymers accumulate in the distended hepatocyte ER (4, 47, 51), rather than in the cytosol, implies that an early step in the disposal process may lack the ability to efficiently clear the secretion-impaired molecules. Notably, a delay in the degradation of the PI Z variant has been observed in transduced fibroblasts from patients who eventually underwent liver transplantation (29), implying that the degradation process might very well contribute to disease pathogenesis. Because synthesis of the PI Z variant does not activate the unfolded response, the hepatocytes of homozygous ZZ patients must rely on the low basal concentration of ERManI to target monomers into GERAD for the purpose of preventing the formation of polymers.
Presently, organ transplantation is most commonly used to alleviate the end-stage liver disease. Therefore, genomic DNA was extracted from these tissues as a means to take a candidate gene approach to determine whether variations in gene that encodes ERManI (MAN1B1) might influence the age at onset of end-stage liver disease in homozygous ZZ patients. In a small cohort of unrelated ZZ white subjects, it was discovered that homozygosity for a single-nucleotide polymorphism in the 3′ untranslated region of the ERManI mRNA induces the conditional suppression of ERManI translation in response to ER stress caused by the intracellular retention of variant PI Z (52). A model has been proposed in which the single-nucleotide polymorphism generates a conditional hypomorphic allele for ERManI that, when inherited in a homozygous fashion, can impairs the liver's capacity to efficiently degrade PI Z polymers, likely accelerating the rate at which polymers are formed, thereby inducing cytotoxicity (Figure 3).
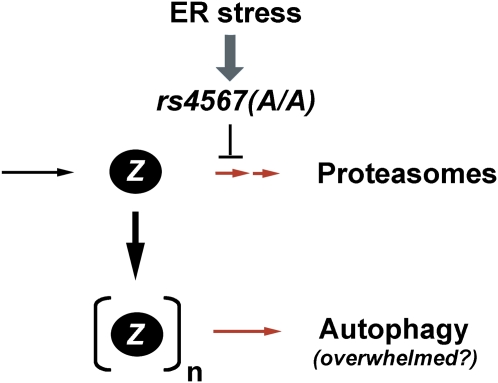
Figure 3.
Proposed mechanism by which a single-nucleotide polymorphism (rs4567(A/A)) suppresses the translation of endoplasmic reticulum (ER) mannosidase I in response to ER stress. The model proposes that impaired proteasomal degradation of the proteinase inhibitor Z (PI Z) monomers leads to an enhanced rate at which polymers are allowed to form, thereby overwhelming the functional capacity of the autophagic system.
CONCLUSIONS
Because the efficient, or inadequate, elimination of aberrant proteins underlies the molecular pathogenesis associated with numerous loss-of-function and gain-of-toxic-function disorders, quality control checkpoints warrant attention comparable to that given to other biological systems as a source for novel diagnostic marker sites for rational therapeutic intervention. This article has dealt with the underlying mechanistic principles that govern the operation of these systems with regard to their role in handling newly synthesized AAT. The chosen pursuit has established a disease paradigm in which a subtle defect in the multilevel regulation of gene expression can modify a classical gain-of-toxic-function disorder. It is, however, quite likely that subtle defects in autophagy, apoptosis, and/or mitochondrial integrity play a significant role in the molecular pathogenesis of the liver disease associated with the intracellular accumulation of PI Z polymers.
Acknowledgments
The authors acknowledge the Liver Tissue Cell Distribution System (no. N01-DK-7-0004/HHSN267200700004C) and the Alpha1-Foundation–University of Florida DNA and Tissue Bank for providing liver tissue samples and genomic DNA.
Supported by grants from the National Institutes of Health (National Heart, Lung, and Blood Institute and National Institute of Diabetes and Digestive and Kidney Diseases), the American Lung Foundation, the American Heart Association, the Moran Foundation, and the Alpha-1 Foundation.
Author Disclosure: R.N.S. received lecture fees from Boehringer-Ingelheim ($1,001–$5,000) and grant support from FoldRx Pharmaceuticals ($10,001–$50,000).
References
- 1.Molinari M. N-glycan structure dictates extension of protein folding or onset of disposal. Nat Chem Biol 2007;3:313–320. [DOI] [PubMed] [Google Scholar]
- 2.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science 2008;319:916–919. [DOI] [PubMed] [Google Scholar]
- 3.Lomas DA, Evans DLI, Finch JT, Carrell RW. The mechanism of Z alpha 1-antitrypsin accumulation in the liver. Nature 1992;357:605–607. [DOI] [PubMed] [Google Scholar]
- 4.Carrell RW, Lomas DA. Alpha1-antitrypsin deficiency—a model for conformational diseases. N Engl J Med 2002;346:45–53. [DOI] [PubMed] [Google Scholar]
- 5.Lomas DA, Mahadeva R. Alpha1-antitrypsin polymerization and the serpinopathies: pathobiology and prospects for therapy. J Clin Invest 2002;110:1585–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sifers RN. Heritable α1-antitrypsin deficiency. In: Zander DS, Popper HH, Jagirdar J, Haque AK, Cagle PT, Barrios R, editors. Molecular pathology of lung disease. New York, NY: Springer; 1998. pp. 541–548.
- 7.Perlmutter DH, Pierce JA. The alpha 1-antitrypsin gene and emphysema. Am J Physiol 1989;257:L147–L162. [DOI] [PubMed] [Google Scholar]
- 8.Ellgaard L, Helenius A. ER quality control: towards an understanding at the molecular level. Curr Opin Cell Biol 2001;13:431–437. [DOI] [PubMed] [Google Scholar]
- 9.Sifers RN, Finegold MJ, Woo SLC. Molecular biology and genetics of alpha 1-antitrypsin deficiency. Semin Liver Dis 1992;12:301–310. [DOI] [PubMed] [Google Scholar]
- 10.Fewell SW, Travers KJ, Weissman JS, Brodsky JL. The action of molecular chaperones in the early secretory pathway. Annu Rev Genet 2001;35:149–191. [DOI] [PubMed] [Google Scholar]
- 11.Gething MJ, Sambrook J. Protein folding in the cell. Nature 1992;355:33–45. [DOI] [PubMed] [Google Scholar]
- 12.Plemper RK, Wolf DH. Endoplasmic reticulum degradation: reverse protein transport and its end in the proteasome. Mol Biol Rep 1999;26:125–130. [DOI] [PubMed] [Google Scholar]
- 13.Sifers RN, Brashears-Macatee S, Kidd VJ, Muensch H, Woo SLC. A frameshift mutation results in a truncated alpha1-antitrypsin that is retained within the rough endoplasmic reticulum. J Biol Chem 1989;263:7330–7335. [PubMed] [Google Scholar]
- 14.Ellgaard L, Molinari M, Helenius A. Setting the standards: quality control in the secretory pathway. Science 1999;286:1882–1888. [DOI] [PubMed] [Google Scholar]
- 15.Cabral CM, Liu Y, Sifers RN. Dissecting glycoprotein quality control in the secretory pathway. Trends Biochem Sci 2001;26:619–624. [DOI] [PubMed] [Google Scholar]
- 16.Cabral CM, Liu Y, Moremen KW, Sifers RN. Organizational diversity among distinct glycoprotein ER-associated degradation programs. Mol Biol Cell 2002;13:2639–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trombetta SE, Ganan S, Parodi AJ. The UDP-Glc:glycoprotein glucosyltransferase is a soluble protein of the endoplasmic reticulum. Glycobiology 1991;1:155–161. [DOI] [PubMed] [Google Scholar]
- 18.Sousa MC, Ferro-Garcia MA, Parodi AJ. Recognition of the oligosaccharide and protein moieties of glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. Biochemistry 1992;31:97–105. [DOI] [PubMed] [Google Scholar]
- 19.Klausner RD, Sitia R. Protein degradation in the endoplasmic reticulum. Cell 1990;62:611–614. [DOI] [PubMed] [Google Scholar]
- 20.Bonifacino JS, Weissman AM. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu Rev Cell Dev Biol 1998;14:19–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernasconi R, Galli C, Calanca V, Nakajima T, Molinari M. Stringent requirement for HRD1, SEL1, and OS-9/XTP-3-B for disposal of ERAD-LS substrates. J Cell Biol 2010;188:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez DS, Karaveg K, Vandersall-Nairn AS, Lal A, Moremen KW. Identification, expression, and characterization of a cDNA encoding human endoplasmic reticulum mannosidase I, the enzyme that catalyzes the first mannose trimming step in mammalian Asn-linked oligosaccharide biosynthesis. J Biol Chem 1999;274:21375–21386. [DOI] [PubMed] [Google Scholar]
- 23.Tremblay LO, Herscovics A. Cloning and expression of a specific human alpha1,2-mannosidase that trims Man9GlcNAc2 to Man8GlcNAc2 isomer B during N-glycan biosynthesis. Glycobiology 1999;9:1073–1078. [DOI] [PubMed] [Google Scholar]
- 24.Olivari S, Galli C, Alanen H, Ruddock L, Molinari M. A novel stress-induced EDEM variant regulating endoplasmic reticulum-associated glycoprotein degradation. J Biol Chem 2005;280:2424–2428. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez DS, Jordan IK. The alpha-mannosidases: phylogeny and adaptive diversification. Mol Biol Evol 2000;17:292–300. [DOI] [PubMed] [Google Scholar]
- 26.Aebi M, Bernasconi R, Clerc S, Molinari M. N-glycan structures: recognition and processing in the ER. Trends Biochem Sci 2010;35:74. [DOI] [PubMed] [Google Scholar]
- 27.Hosokawa N, Kamiya Y, Kato K. The role of MRH domain–containing lectins in ERAD. Glycobiology 2010;20:651–660. [DOI] [PubMed] [Google Scholar]
- 28.Clerc S, Hirsch C, Oggier DM, Deprez P, Jakob C, Sommer T, Aebi M. Hrtm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. Biochem Biophys Res Commun 2006;349:1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y, Swulius MT, Moremen KW, Sifers RN. Elucidation of the molecular logic by which misfolded alpha1-antitrypsin is preferentially selected for intracellular degradation. Proc Natl Acad Sci USA 2003;100:8229–8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Y, Termine DJ, Swulius MT, Moremen KW, Sifers RN. Human endoplasmic reticulum mannosidase I is subject to regulated proteolysis. J Biol Chem 2007;282:4841–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cali T, Vanoni O, Molinari M. The endoplasmic reticulum crossroads for newly synthesized polypeptide chains. Prog Mol Biol Transl Sci 2008;83:135. [DOI] [PubMed] [Google Scholar]
- 32.Reggiori F, Monastyrska I, Verheije MH, Cali T, Ulasli M, Bianchi S, Bernasconi R, de Haan CA, Molinari M. Coronoviruses hijack the LC3-I-positive EDEMsomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 2010;7:500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007;8:519–529. [DOI] [PubMed] [Google Scholar]
- 34.Travers KJ, Patil CK, Wodicka L, Lockhardt DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 2000;101:249–258. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell 2003;4:265–271. [DOI] [PubMed] [Google Scholar]
- 36.Banerjee S, Vishwanath P, Cui J, Kelleher DJ, Gilmore R, Robbins PW, Samuelson J. The evolution of N-glycan–dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc Natl Acad Sci USA 2007;104:11676–11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosokawa N, Wada I, Hasegawa K, Yorihuzi T, Tremblay LO, Herscovics A, Nagata K. A novel ER alpha-mannosidase–like protein accelerates ER-associated degradation. EMBO Rep 2001;2:415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mast SW, Diekman K, Davis AW, Karaveg K, Sifers RN, Moremen KW. Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology 2005;15:421–436. [DOI] [PubMed] [Google Scholar]
- 39.Termine DJ, Moremen KW, Sifers RN. The mammalian UPR boosts glycoprotein ERAD by suppressing the proteolytic down-regulation of ER mannosidase I. J Cell Sci 2009;122:976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rutkowski DT, Kaufman RJ. That which does not kill me makes me stronger: adapting to chronic ER stress. Trends Biochem Sci 2007;32:469–476. [DOI] [PubMed] [Google Scholar]
- 41.Le A, Ferrell GA, Dishon DS, Le Q-Q, Sifers RN. Soluble aggregates of the human PI Z alpha1-antitrypsin variant are degraded within the endoplasmic reticulum by a mechanism sensitive to inhibitors of protein synthesis. J Biol Chem 1992;267:1072–1080. [PubMed] [Google Scholar]
- 42.Qu D, Teckman JH, Omura S, Perlmutter DH. Degradation of a mutant secretory protein, alpha1-antitrypsin Z, in the endoplasmic reticulum requires proteasome activity. J Biol Chem 1996;271:22791–22795. [DOI] [PubMed] [Google Scholar]
- 43.Teckman JH, Perlmutter DH. The endoplasmic reticulum degradation pathway for mutant secretory proteins alpha1-antitrypsin Z and S is distinct from that for an unassembled membrane protein. J Biol Chem 1996;271:13215–13220. [DOI] [PubMed] [Google Scholar]
- 44.Yu MH, Lee KN, Kim J. The Z type variation of human alpha 1-antitrypsin causes a protein folding defect. Nat Struct Biol 1995;2:363–367. [DOI] [PubMed] [Google Scholar]
- 45.Lawless MW, Greene CM, Mulgrew A, Taggart CC, O'Neill SJ, McElvaney NG. Activation of endoplasmic reticulum–specific stress responses associated with the conformational disease Z alpha 1-antitrypsin deficiency. J Immunol 2004;172:5722–5726. [DOI] [PubMed] [Google Scholar]
- 46.Hidvegi T, Schmidt BZ, Hale P, Perlmutter DH. Accumulation of mutant alpha1-antitrypsin Z in the endoplasmic reticulum activates caspases-4 and -12, NFkappaB, and BAP31 but not the unfolded protein response. J Biol Chem 2005;280:39002–39015. [DOI] [PubMed] [Google Scholar]
- 47.Graham KS, Le A, Sifers RN. Accumulation of the insoluble PiZ variant of human alpha 1-antitrypsin within the hepatic endoplasmic reticulum does not elevate the steady-state level of grp78/BiP. J Biol Chem 1990;265:20463–20468. [PubMed] [Google Scholar]
- 48.Lindblad D, Blomenkamp K, Teckman J. Alpha-1-antitrypsin mutant Z protein content in individual hepatocytes correlates with cell death in a mouse model. Hepatology 2007;46:1228–1235. [DOI] [PubMed] [Google Scholar]
- 49.Teckman JH, Perlmutter DH. Retention of mutant alpha(1)-antitrypsin Z in endoplasmic reticulum is associated with an autophagic response. Am J Physiol Gastrointest Liver Physiol 2000;279:G961–G974. [DOI] [PubMed] [Google Scholar]
- 50.Teckman JH, An JK, Blomenkamp K, Schmidt B, Perlmutter DH. Mitochondrial autophagy and injury in the liver in alpha1-antitrypsin deficiency. Am J Physiol Gastrointest Liver Physiol 2004;286:G851–G862. [DOI] [PubMed] [Google Scholar]
- 51.Volpert D, Molleston JP, Perlmutter DH. Alpha1-antitrypsin deficiency–associated liver disease progresses slowly in some children. J Pediatr Gastroenterol Nutr 2000;31:258–263. [DOI] [PubMed] [Google Scholar]
- 52.Pan S, Huang L, McPherson J, Muzny D, Rouhani F, Brantly M, Gibbs R, Sifers RN. Single nucleotide polymorphism–mediated translational suppression of endoplasmic reticulum mannosidase I modifies the onset of end-stage liver disease in alpha1-antitrypsin deficiency. Hepatology 2009;50:275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]