Abstract
Rationale: Previously, we demonstrated a candidate region for susceptibility to airspace enlargement on mouse chromosome 5. However, the specific candidate genes within this region accounting for emphysema-like changes remain unrecognized. c-Kit is a receptor tyrosine kinase within this candidate gene region that has previously been recognized to contribute to the survival, proliferation, and differentiation of hematopoietic stem cells. Increases in the percentage of cells expressing c-Kit have previously been associated with protection against injury-induced emphysema.
Objectives: Determine whether genetic variants of c-Kit are associated with spontaneous airspace enlargement.
Methods: Perform single-nucleotide polymorphism association studies in the mouse strains at the extremes of airspace enlargement phenotype for variants in c-Kit tyrosine kinase. Characterize mice bearing functional variants of c-Kit compared with wild-type controls for the development of spontaneous airspace enlargement. Epithelial cell proliferation was measured in culture.
Measurements and Main Results: Upstream regulatory single-nucleotide polymorphisms in the divergent mouse strains were associated with the lung compliance difference observed between the extreme strains. c-Kit mutant mice (KitW-sh/W-sh), when compared with genetic controls, developed altered lung histology, increased total lung capacity, increased residual volume, and increased lung compliance that persist into adulthood. c-Kit inhibition with imatinib attenuated in vitro proliferation of cells expressing epithelial cell adhesion molecule.
Conclusions: Our findings indicate that c-Kit sustains and/or maintains normal alveolar architecture in the lungs of mice. In vitro data suggest that c-Kit can regulate epithelial cell clonal expansion. The precise mechanisms that c-Kit contributes to the development of airspace enlargement and increased lung compliance remain unclear and warrants further investigation.
Keywords: genetic, tyrosine kinase, SASH, chronic obstructive pulmonary disease, aging
AT A GLANCE COMMENTARY.
Scientific Knowledge on the Subject
Previous data suggest that bone marrow–derived cells can preserve alveolar structure in various models of lung injury. c-Kit is a receptor tyrosine kinase that contributes to the survival, proliferation, and differentiation of hematopoietic stem cells. The role of c-Kit in maintaining normal lung structure remains unknown.
What This Study Adds to the Field
Forward genetics identified a region within mouse chromosome 5 associated with the development of spontaneous airspace enlargement. In silico mapping of divergent mouse strains and characterization of c-Kit mutant mice indicate that genetic variation upstream of c-Kit tyrosine kinase is associated with premature emphysematous changes of the lung parenchyma of mice. Furthermore, our observations suggest a role of c-Kit in lung epithelial cell clonal expansion.
The delicate airway and parenchymal tissues of the lung follow a blueprint for structural development that relies on tightly controlled temporal and spatial interactions between many diverse cell types (1–4). In humans as in mice, the lung continues to develop after birth until the end of early adulthood (2, 3). However, because lung development proceeds more rapidly in the mouse (12–14 wk) than in the human (23–25 yr), mouse models allow for rapid testing of genetic and molecular determinants of lung structure and function. Delays and/or interruptions of normal postnatal lung growth together with normal aging of the lung may have serious functional consequences later in life (5) and can lead to reduced quality of life (6) and increased risk of mortality (7–9). Knowledge of the underlying genetic determinates of lung development could provide an unbiased view of the critical genes (10) and pathways (11) that contribute to declines in lung function during aging.
Previously we identified several quantitative trait loci (QTL) in mice that are associated with altered mouse lung function including increased static lung compliance (Cl) (12), which is a pathognomonic feature of chronic obstructive pulmonary disease (COPD). We observed a region on chromosome 5 that was associated with alterations in Cl and harbored several candidate genes including v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (Kit, also known as mast/stem cell growth factor receptor, c-Kit oncogene, or CD117) (12). The KIT protein encoded by this gene is a tyrosine kinase receptor that is expressed on the surface of hematopoietic stem cells (HSCs). Binding of KIT ligand (KITLG, also known as stem cell factor or Steel factor) to KIT activates a signaling pathway important to HSC survival, proliferation, and differentiation (13, 14).
Although KIT is strongly expressed at critical points in embryonic mouse lung development (15), the role that KIT plays in both lung development and repair has not been fully explored (16, 17). In humans, KIT-immunoreactive interstitial cells have been observed in the alveolar septa of normal newborn lungs, but were absent in lungs of patients with congenital alveolar capillary dysplasia (18). In mice, HSCs may contribute to lung regeneration after injury as demonstrated after lung injury induced by lipopolysaccharide (19), elastase (20), irradiation, and combined elastase/irradiation (21). In addition, repair after elastase-induced lung injury was enhanced by intranasal hepatocyte growth factor instillation and was associated with an increase in the percentage of KIT-expressing circulating mononuclear cells (22). In the current study we assess lung function in select genetically divergent inbred mouse strains and used a mutant strain to determine how Kit, a candidate gene associated with lung function, may provide novel insights into the later decline in lung function.
METHODS
Mice
Mice were obtained from commercial vendors. Lung function measurements were performed in the following 10 mouse strains: AKR/J (00648), BTBR T+ tf/J (002282), CAST/EiJ (000928), CZECHII/EiJ (001144), DBA/2J (000671), KK/HIJ (002106), MOLF/EiJ (000550), NOD/ShiLtJ (001976), NZW/LacJ (001058), and WSB/EiJ (001145) and supplemented by nine strains previously phenotyped: 129S1/SvImJ, A/J, BALB/cByJ, C3H/HeJ, C57BL/6J, FVB/NJ, JF1/Ms, PWD/PhJ, and SWR/J (12,23,24). Single-nucleotide polymorphisms (SNPs) in Kit were identified using the Mouse Phenome Database (2010) and the population was interrogated for the phenotypic difference observed in mice with either allele compared with the phenotypic difference of the extreme mouse strains (CZECHII/EiJ and NZW/LacJ).
To determine whether mice with mutant Kit have altered lung structure and function, B6.Cg-KitW-sh/HNihrJaeBsmJ mice (referred to here as SASH mice) were compared with C57BL/6J. The SASH mouse is homozygous for a KitW-sh allele (25), which has been backcrossed onto C57BL/6J for more than 10 generations. The KitW-sh mutation prevents KIT mRNA expression in mast cells and mesenchymal cells in the lung and digestive tract on Embryonic Day 13 (E13) and in adult lung, whereas expression in other tissues is normal (15, 26).
Lung Function
In the inbred mouse strains, invasive measurements of lung function measurements of total lung capacity (TLC), static lung compliance (Cl), dead space volume (Vd), diffusion capacity (Dco), alveolar volume (Va), and specific lung function values (TLC/body weight [BW]; Vd/TLC; Cl/TLC, Dco/Va) were determined and statistics performed as described previously (27). In SASH and additional C57BL/6J mice, lung function was measured with a commercial system (flexiVent; SCIREQ, Montreal, PQ, Canada).
Morphometric Analysis of Histology
Sections (5 μm thick) of paraffin-embedded, formalin-fixed mouse lung were stained with hematoxylin and eosin (H&E stain; AML Laboratories, Rosedale, MD). Five digital images of parenchymal architecture from each mouse were captured at ×40 magnification by an investigator blinded to the experimental groups. Images were captured from anatomically comparable regions of the lung.
Alveolar volume density (AVD) and alveolar surface density (ASD) were calculated as previously described (28). Briefly, five random fields were chosen (omitting large vascular and bronchiolar structures) and images were overlaid with an 11 × 11 point grid (Metamorph; Universal Imaging, West Chester, PA) for point (P) and intercept (I) counting of the alveolar septa. Alveolar volume density was calculated from Palveoli/Pparenchyma and alveolar surface density from 2Ialveoli/LT, where LT was the test line length within the lung parenchyma. For mean linear intercept (MLI), images were analyzed as previously described (29). Briefly, a series of grid lines was laid over each photomicrograph to determine the number of times those lines were intercepted by alveolar tissue, with Lm = L/Li, where L is the total length of the lines in the grid field and Li is the total number of times those lines are intercepted. Data from each group are expressed as means ± SE.
Lung Epithelial Cell Assays
Flow cytometry was used to characterize populations of airway epithelial cells. Primary lung epithelial cells expressing epithelial cell adhesion molecule (EpCAM+ cells) were isolated and cultured on fibroblasts ± imatinib. Number and size of colonies were quantified after culture for 28 days. Similarly, coculture experiments were performed ± stem cell factor (SCF) at doses of 0–100 ng/ml, and the number and size of colonies were quantified at 14 days.
RESULTS
Lung Function Varies among Mouse Strains
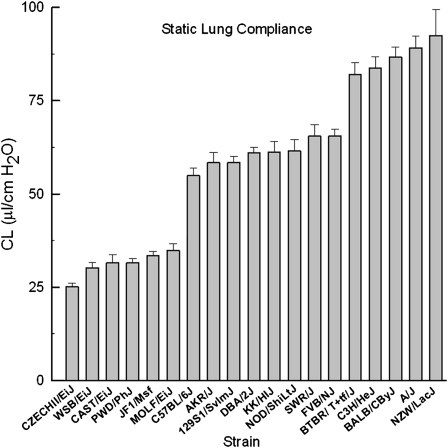
Lung function measurements varied significantly among the additional age- and sex-matched (12–14 wk, female) mouse strains tested (Table 1). For example, total lung capacity (TLC) (mean ± SE) varied from 679 ± 9 μl for CZECHII/EiJ (n = 12) to 1,409 ± 105 μl for BTBR T+ tf/J (n = 8) mouse strains. The CZECHII/EiJ strain mice have the smallest TLC we have noted in the combined 19 strains tested (12, 23, 24). In the nine strains previously phenotyped, the smallest TLC was 874 ± 17 μl for the sex- and age-matched JF1/Ms mouse strain (12). The largest TLC was 1,457 ± 82 for the BALB/cByJ mouse strain (24), which is only slightly smaller than observed in the BTBR T+ tf/J mouse strain. Thus, the strains with extreme phenotypes (CZECHII/EiJ and the BALB/cByJ) varied about twofold in TLC. This analysis also uncovered a large strain difference (4.7-fold) in Dco (CZECHII/EiJ Dco = 3.0 ± 0.1 μmol/minute/hPa; BTBR T+ tf/J Dco = 17.4 ± 0.3 μmol/minute/hPa), whereas the previous greatest difference was between JF1/Ms Dco (8.3 ± 0.4 μmol/minute/hPa) and FVB/NJ Dco (15.7 ± 1.8 μmol/minute/hPa).
TABLE 1.
LUNG FUNCTION MEASUREMENTS OF 10 INBRED MOUSE STRAINS
| Strain |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | AKR/J | BTBR/T+ tf/J | CAST/EiJ | CZECHII/EiJ | DBA/2J | KK/HIJ | MOLF/EiJ | NOD/ShiLtJ | NZW/LacJ | WSB/EiJ |
| BW, g | 31 (1.2) | 32 (0.6) | 13 (0.3) | 12.0 (0.9) | 24 (0.8) | 35 (1.3) | 13 (0.2) | 22 (0.6) | 27 (0.8) | 14 (0.03) |
| Age, d | 96 (0.4) | 84 (0.2) | 96 (0.2) | 94 (0.6) | 93 (0.2) | 84 (0.2) | 93 (2.3) | 91 (0.2) | 86 (0.2) | 89 (0.9) |
| Number | 8 | 8 | 6 | 12 | 8 | 7 | 14 | 8 | 8 | 11 |
| Lung volume | ||||||||||
| TLC, μl | 1,176 (32) | 1,409 (26) | 751 (9) | 679 (9) | 1,054 (17) | 1,304 (29) | 855 (14) | 1,012 (25) | 1,218 (26) | 744 (27) |
| TLC/BW, μl/g | 38.7 (1.4) | 44.4 (1.5) | 57.7 (0.8) | 56.6 (0.9) | 43.8 (1.7) | 37.5 (1.4) | 64.8 (1.2) | 46.4 (1.6) | 44.5 (0.6) | 51.5 (1.7) |
| FRC, μl | 304 (2) | 343 (9) | 272 (3) | 272 (4) | 289 (11) | 304 (9) | 281 (4) | 303 (6) | 310 (5) | 273 (6) |
| FRC/TLC | 0.26 (0.01) | 0.24 (0.01) | 0.36 (0.01) | 0.40 (0.01) | 0.27 (0.01) | 0.23 (0.01) | 0.33 (0.01) | 0.30 (0.01) | 0.25 (0.01) | 0.37 (0.01) |
| Vd, μl | 231 (4) | 251 (3) | 179 (5) | 177 (4) | 232 (3) | 246 (2) | 217 (5) | 222 (2) | 250 (4) | 201 (4) |
| Vd/TLC | 0.20 (0.01) | 0.18 (0.01) | 0.24 (0.01) | 0.26 (0.01) | 0.22 (0.01) | 0.19 (0.01) | 0.25 (0.01) | 0.22 (0.01) | 0.21 (0.01) | 0.27 (0.01) |
| Compliance | ||||||||||
| Cl, μl/cm H2O | 58.3 (2.8) | 82.1 (3.0) | 31.6 (2.2) | 25.2 (0.9) | 61.1 (1.5) | 61.2 (2.7) | 34.8 (1.9) | 61.5 (3.1) | 92.4 (7.0) | 30.2 (1.4) |
| Cl/TLC, 1/cm H2O | 49.8 (2.7) | 58.2 (1.7) | 46.1 (3.2) | 36.3 (1.1) | 58.0 (1.3) | 47.0 (1.9) | 40.7 (2.0) | 60.7 (2.4) | 75.9 (5.5) | 40.8 (1.6) |
| Cdyn, μl/cm H2O | 41.1 (1.7) | 57.7 (2.4) | 22.6 (1.5) | 19.9 (0.8) | 38.4 (0.8) | 46.5 (1.3) | 24.6 (0.6) | 43.5 (1.9) | 58.9 (3.4) | 21.1 (0.6) |
| Diffusion capacity | ||||||||||
| Dco, μmol/min/hPa | 13.9 (0.4) | 17.4 (0.3) | 4.7 (0.03) | 3.0 (0.1) | 9.5 (0.3) | 11.4 (0.7) | 5.6 (0.4) | 10.3 (0.4) | 11.4 (0.5) | 5.1 (0.4) |
| Dco/Va, μmol/min/hPa/ml | 12.4 (0.03) | 13.1 (0.2) | 6.4 (0.04) | 4.7 (0.2) | 10.1 (0.1) | 9.3 (0.3) | 7.4 (0.3) | 11.2 (0.2) | 10.4 (0.2) | 7.5 (0.3) |
Definition of abbreviations: BW = body weight; Cdyn = dynamic lung compliance; Cl = static lung compliance; Dco = diffusion capacity; FRC = functional residual capacity; TLC = total lung capacity; Va = alveolar volume; Vd = dead space volume.
Values represent means (SE).
Divergent Mouse Strains and Association between Polymorphisms in Kit and Static Lung Compliance
Previously, we identified on mouse chromosome 5 a QTL that was associated with lung compliance and that contained several candidate genes including Kit. We measured static lung compliance (Cl) for 19 mouse strains (213 mice). The range in the difference between the lowest and highest strains was about threefold (CZECHII/EiJ Cl = 25.2 ± 0.9 μl/cm H2O; NZW/LacJ Cl = 92.4 ± 7.0 μl/cm H2O) (Figure 1). Using a high-density SNP map for these 19 mouse strains (including the 5′ untranslated region [UTR] and 3′ UTR of the gene), we identified 534 SNPs within the Kit locus that included eleven in the 5′ UTR (“promoter”), one nonsynonymous SNP, and fifteen 3′ UTR SNPs.
Figure 1.
Strain distribution pattern for static lung compliance (Cl) in 19 inbred mouse strains. Mouse lung Cl obtained from the transpulmonary pressure–volume curve varied between strains (n = 6–16 mice per strain, female, 12–14 wk). The range in the difference between the lowest and highest strains was about threefold (CZECHII/EiJ Cl, 25.2 ± 0.9 μl/cm H2O; NZW/LacJ Cl, 92.4 ± 7.0 μl/cm H2O). Values represent means ± standard error.
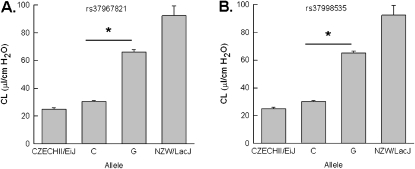
We interrogated possible functional consequences of the 11 promoter SNPs, using the Transcription Element Search System (30). Seven of the 11 SNPs result in a change that could alter the transcription factor DNA–binding efficiency (Table 2). Of these, two were particular noteworthy. The first, rs37967821, is a C-to-T conversion that would convert the conserved sequence for DNA binding of Deformed (Dfd), which has homology to homeobox B4 (HOXB4), to a DNA-binding site for v-myb myeloblastosis viral oncogene homolog (MYB1). The C allele had a frequency of 0.21 among the 19 strains phenotyped and could explain 53.2% of the Cl difference observed in the extreme mouse strains (Figure 2A). The second, rs37998535, is a C-to-G conversion that would form a conversed sequence to NF1 and thereby replaces a CCAAT/enhancer-binding protein (C/EBP)-α (CEBPA) binding site. The C allele had a frequency of 0.19 among the 19 mouse strains phenotyped and could explain 51.6% of the Cl difference observed between the extreme strains (Figure 2B). While the functional consequences of the genetic variations associated with Cl remain unknown, the nonsynonymous SNP (rs36972615) could produce an aspartic acid-to-glutamic acid conversion at position 927. It is noteworthy that the CZECHII/EiJ and JF1/Ms mouse strains share the same genotype for these three SNPs whereas the NZW/LacJ and C3H/HeJ mouse strains share the opposing genotype for these SNPs. Of the fifteen 3′ UTR SNPs, seven were present in the strains tested and six were A-to-G conversions. We did not detect differences in mRNA expression of c-Kit from adult (6 wk) whole lungs between the CZECHII/EiJ and NZW/LacJ mice. However, the timing and distribution of c-Kit expression required for normal lung maintenance remain unknown.
TABLE 2.
SINGLE-NUCLEOTIDE POLYMORPHISMS IN MOUSE KIT
| SNP | Location (Mbp) | Observed SNP | Allele 1 Frequency | Percentage of Phenotype | Region | Allele 1 | Allele 2 |
|---|---|---|---|---|---|---|---|
| rs37546426 | 75.969357 | C/T | 0.19 | 51.6 | 5′ L | IRF1/2 | YY1 |
| rs36684666 | 75.969567 | C/T | 0.38 | 35.7 | 5′ L | Zeste | POU1F1a |
| rs37967821 | 75.969778 | C/T | 0.21 | 53.2 | 5′ L | MYB1 | Dfb (Hoxb4) |
| rs38970347 | 75.970285 | T/A | 0.21 | 53.2 | 5′ L | No RAF | RAF |
| rs36956723 | 75.970368 | C/T | 0.38 | 35.7 | 5′ L | ETF, Sp1 | CACCC-binding factor |
| rs37998535 | 75.970867 | C/G | 0.19 | 51.6 | 5′ L | NF-1 | C/EBPα |
| rs36929749 | 75.971053 | C/T | 0.19 | 51.6 | 5′ UTR | T-Ag | No T-Ag |
| rs36972615 | 76.049338 | G/C | 0.19 | 51.6 | CN | Asp 927 | Glu 927 |
| rs36331951 | 76.050775 | A/G | 0.21 | 53.2 | 3′ UTR | A | G |
| rs29630739 | 76.051098 | A/G | 0.28 | 45.0 | 3′ UTR | A | G |
| rs33755426 | 76.051115 | G/A | 0.28 | 45.0 | 3′ UTR | G | A |
| rs13469910 | 76.051946 | C/T | 0.28 | 45.0 | 3′ UTR | C | T |
| rs36266306 | 76.052269 | A/G | 0.19 | 51.6 | 3′ UTR | G | A |
| rs36248044 | 76.052312 | A/G | 0.21 | 53.2 | 3′ L | G | A |
| rs37033324 | 76.052937 | A/G | 0.19 | 51.6 | 3′ L | A | G |
Notes: SNP = reference single-nucleotide polymorphism identification number; Location (Mbp) = location on mouse chromosome 5 in megabase pairs (build 37); Observed SNP = base pair substitution with minor (allele 1) listed first; Allele 1 Frequency = the ratio of mice in the total population (n = 213) that have allele 1; Percentage of Phenotype = a comparison of the mean static compliance (Cl) of mice with allele 1 to that of allele 2 divided by the maximum difference between the polar strains (NZW/LacJ vs. CZECH/EiJ) × 100, based on alleles assigned by the Mouse Phenome Database (http://www.jax.org/phenome); Region = location near (L) or within (CN = nonsynonymous) the gene, including the untranslated region (UTR); Allele 1 = the possible consequence of the minor allele as determined by the Transcription Element Search System (30) for the 5′ region, in the coding sequence, or in the 3′ UTR. Allele 2 = major allele. The predicted transcriptional binding site use the TRANSFAC abbreviation.
Figure 2.
Phenotype of mice with genetic variants in the Kit promoter. (A) Static compliance (Cl) was significantly lower in mice that have the C allele of a promoter single-nucleotide polymorphism (SNP), rs37967821, than in mice with the T allele. The C allele had a frequency of 0.21 (n = 46 mice) among the 19 strains (n = 213 mice) genotyped and could explain 53.2% of the Cl difference observed in the extreme mouse strains CZECHII/EiJ and NZW/LacJ. (B) Cl was significantly lower in mice that have the C allele of a promoter SNP, rs37998535, than in mice with the G allele. The C allele had a frequency of 0.19 (n = 40 mice) among the 19 mouse strains phenotyped and could explain 51.6% of the Cl difference observed between the extreme strains. Values represent means ± standard errors and differences between alleles were assessed by analysis of variance followed by an all-pairwise multiple-comparison procedure (Holm-Sidak method) (*P < 0.001)
Mice Deficient in c-Kit Develop Spontaneous Airspace Enlargement
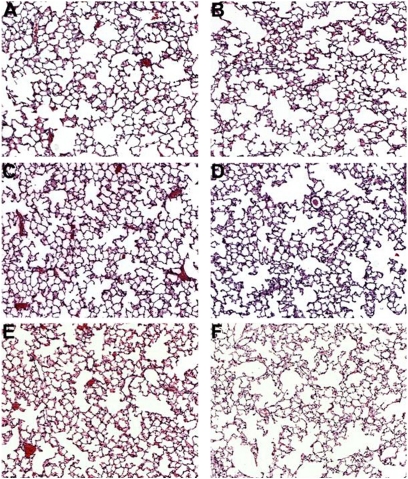
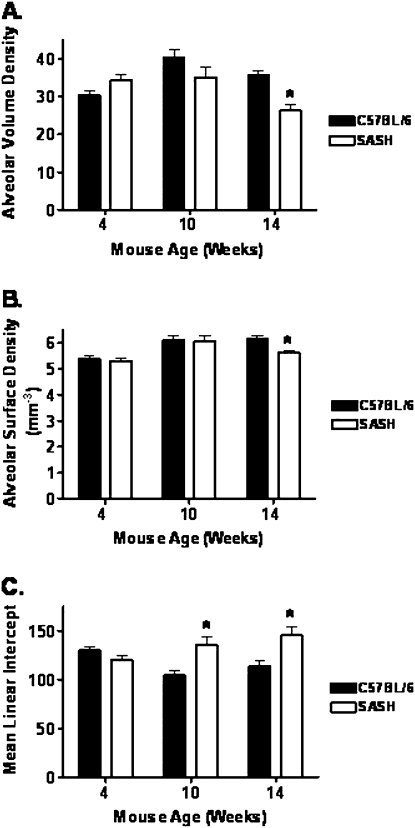
To investigate the role of c-Kit in lung development and changes in lung function over time, we compared the lung histology of B6.Cg-KitW-sh/HNihrJaeBsmJ mice (referred to here as SASH for convenience) with that of C57BL/6J (control) mice at 4, 10, and 14 weeks of age. At 4 and 10 weeks of age there was no discernible difference between histological sections based on genotype (Figure 3, compare panel A with panel B, and panel C with panel D). By 14 weeks of age we observed prominent alveolar enlargement in the lungs of SASH mice (Figure 3F) when compared with C57BL/6J control lungs (Figure 3E). We then measured alveolar volume density, alveolar surface density, and mean linear intercept to quantify the histological changes we observed (Figure 4). At 14 weeks of age the SASH mice had significant decreases in both alveolar volume density (Figure 4A) and alveolar surface density (Figure 4B). At both 10 and 14 weeks of age the SASH mice had significantly increased mean linear intercept (Figure 4C) when compared with C57BL/6J control mice.
Figure 3.
SASH (B6.Cg-KitW-sh/HNihrJaeBsmJ) mice develop spontaneous airspace enlargement when compared with C57BL/6J mice. SASH mice have a sequence inversion in the 5′ regulatory region of Kit, and lack KIT mRNA expression in the lung. Hematoxylin and eosin images were captured at ×10 magnification for (A) 4-week C57BL/6J, (B) 4-week SASH, (C) 10-week C57BL/6J, (D) 10-week SASH, (E) 14-week C57BL/6J, and (F) 14-week SASH mice. No difference was noted between lung histological sections obtained at 4 weeks between (A) C57BL/6J and (B) SASH mice. By 10 weeks, (C) C57BL/6J mouse lung developed normally whereas (D) SASH mouse lung began to show signs of airspace enlargement. Histology at 14 weeks revealed that (E) C57BL/6J lung had intact parenchyma and well-preserved alveoli whereas (F) SASH lung showed enlarged (emphysema-like) airspaces.
Figure 4.
SASH (B6.Cg-KitW-sh/HNihrJaeBsmJ) mice develop delayed abnormal morphology when compared with C57BL/6J mice. Analysis of (A) alveolar volume density, (B) alveolar surface area, and (C) mean linear intercept by MetaMorph software reflects observed histological changes in SASH lung histology. Data represent means ± standard errors. *Significant difference (P < 0.05) as determined by Student t test (n = 10–13).
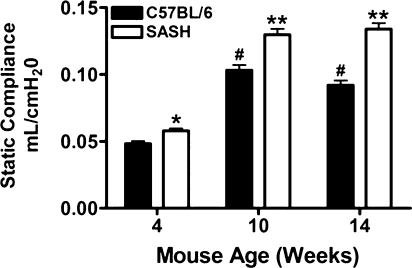
Static Lung Compliance Increases Significantly over Time in Mice Deficient in c-Kit
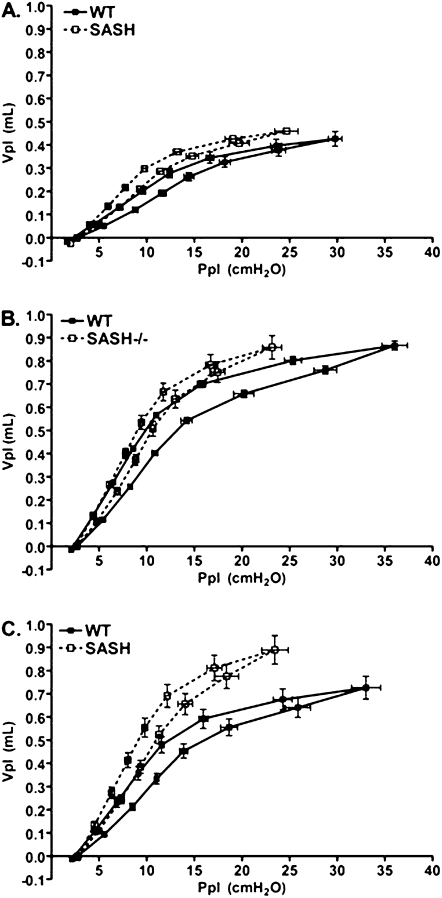
To assess whether the altered histology was accompanied by changes in lung mechanics at baseline, we measured Cl in both C57BL/6J and SASH mice at 4, 10, and 14 weeks of age (Figure 5). As early as 4 weeks of age, Cl in SASH mice was significantly increased compared with Cl of C57BL/6J mice (Figure 5). The observed differences persisted and increased through 14 weeks. We also observed that the pressure–volume curves in SASH mice were shifted upward and to the left during inflation and deflation when compared with C57BL/6J mice, indicating a more dynamically compliant lung (Figure 6). Lung volumes as determined by saline fill volume in SASH mice were also significantly greater than in C57BL/6J mice (see Figure E1 in the online supplement).
Figure 5.
SASH (B6.Cg-KitW-sh/HNihrJaeBsmJ) mice develop delayed increased static lung compliance when compared with C57BL/6J mice. Static lung compliance was determined from tracheal airway pressure and volume, which was generated by applying a 2-second sine wave volume perturbation to the tracheal cannula with an amplitude of 0.2 ml and a frequency of 2.5 Hz. Data represent means ± standard errors. *P < 0.05 versus C57BL/6J, #P < 0.05 versus C57BL/6J at 4 weeks, **P < 0.05 versus SASH at 4 weeks and C57BL/6J at 10 and 14 weeks. Statistical differences were determined by Student t test (n = 5–8).
Figure 6.
SASH (B6.Cg-KitW-sh/HNihrJaeBsmJ) mice develop delayed decreased airway pressure at higher volumes when compared with C57BL/6J mice. Pressure–volume loops were generated by stepwise (seven equal steps) inflation and deflation of the lungs of C57BL/6J mice and SASH mice at baseline. (A) Four weeks (n = 12); (B) 10 weeks (n = 10–14); (C) 14 weeks (n = 11). Ppl = airway tracheal pressure measured at each indicated volume (Vpl) on inflation and deflation limbs. WT = wild type.
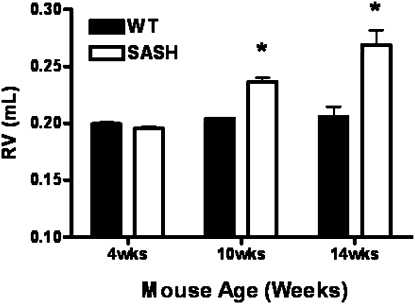
Residual Volume and Ex Vivo Lung Compliance Are Significantly Increased in Mice Deficient in c-Kit
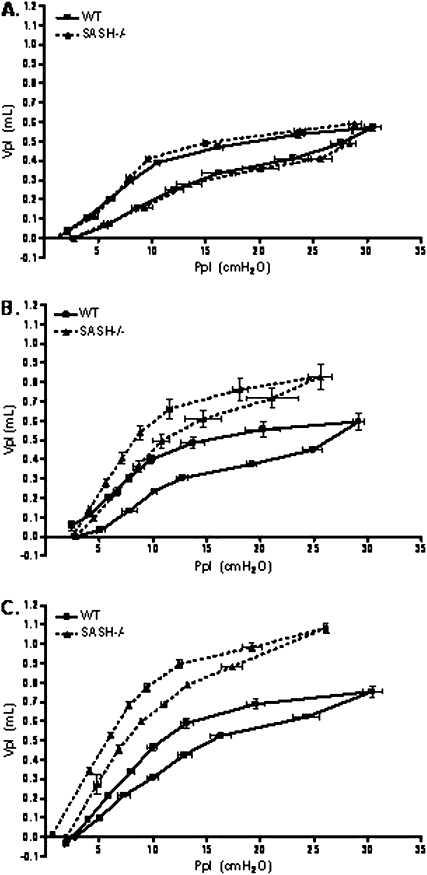
Emphysema is characterized by increased lung residual volume and reduced lung compliance. Therefore, we measured lung residual volumes directly from degassed lungs in both C57BL/6J and SASH mice. We did not observe differences in either residual volume or ex vivo lung compliance at 4 weeks of age. However, at both 10 and 14 weeks SASH mice had significantly increased residual volume when compared with age-matched C57BL/6J mice (Figure 7). Total excised lung weight was not significantly different between the strains (data not shown). Consistent with our in vivo pressure–volume loops (Figure 6), we observed that ex vivo pressure–volume loops in 10- or 14-week-old SASH mice were shifted upward and to the left, indicating a more dynamically compliant lung when compared with C57BL/6J mice (Figure 8).
Figure 7.
SASH (B6.Cg-KitW-sh/HNihrJaeBsmJ) mice develop delayed increase in residual volume when compared with C57BL/6J mice. Direct measurement of residual volume from degassed lungs in age-matched C57BL/6J and SASH mice was performed. Data represent means + SEM. *P < 0.01 (n = 4 or 5). RV = residual volume; WT = wild type.
Figure 8.
SASH (B6.Cg-KitW-sh/HNihrJaeBsmJ) mice develop delayed decreased airway pressure at higher volumes when compared with C57BL/6J mice. Ex vivo pressure–volume loops were generated by stepwise (seven equal steps) inflation and deflation of surgically excised lungs from C57BL/6J mice and SASH mice. (A) Four weeks (n = 4); (B) 10 weeks (n = 5); (C) 14 weeks (n = 4). Ppl = airway tracheal pressure measured at each indicated volume (Vpl) on inflation and deflation limbs. WT = wild type.
c-Kit Regulates Epithelial Progenitor Cell Expansion
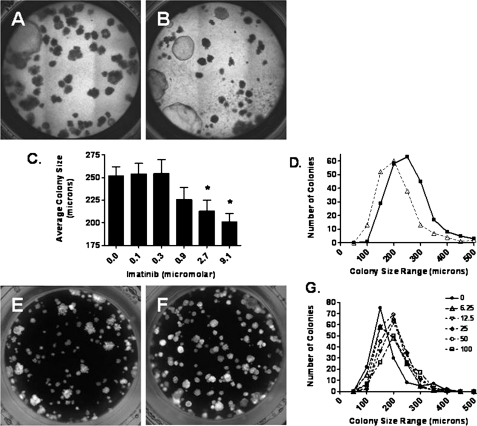
We did not observe differences in the number or properties of EpCAM+ lung epithelial cells by flow cytometric analysis of total cells isolated from lungs of SASH mice compared with C57BL/6J mice (Figures E2 and E3). However, treatment of cultured lung epithelial progenitor cells in the presence of imatinib, an inhibitor of c-Kit tyrosine kinase, resulted in a dose-dependent inhibition of epithelial growth as shown by significant decreases in the mean size and size distribution of colonies (Figures 9A–9D). Treatment of epithelial progenitor cells with stem cell factor (SCF), the ligand for c-Kit tyrosine kinase, increased the size distribution of epithelial colonies (Figures 9E–9G).
Figure 9.
Primary lung epithelial cell growth is impaired by imatinib. Cells expressing epithelial cell adhesion molecule (EpCAM+ cells) were isolated from the lung by flow cytometric sorting and grown in culture with (A) control or (B) imatinib at 9.1 μM. The average radius of all colonies for each condition was determined. (C) The mean radius of colonies is reduced by imatinib (2.7 and 9.1 μM) when compared with saline treatment (*P < 0.01). (D) Number of colonies was characterized by radius, and the number of large colonies was greater with saline treatment when compared with imatinib (9.1 μM). EpCAM+ cells were isolated from the lung by flow cytometric sorting and grown in culture with (E) control or (F) stem cell factor at 50 ng/ml. (G) Number of colonies was characterized by radius, and the number of large colonies was greater with stem cell factor treatment (6.25–100 ng/ml) when compared with control.
DISCUSSION
An emerging global health problem, COPD is rapidly becoming the third most common cause of death and is the only major disease whose contribution to morbidity and mortality continues to increase (31). Emphysema, a key component of COPD, is characterized structurally by progressive loss of alveolar architecture and increased alveolar size, and functionally by a concomitant loss of gas exchange capacity in the lung (32). Thus, airspace enlargement and reduced lung compliance constitute a major cause of declining lung function, dyspnea, hypoxemia, and decreased quality of life in patients. Because no effective therapy exists to attenuate loss of lung function in patients with emphysema, insights into the cell biology of tissue loss could contribute to novel pharmacological interventions that could improve the prognosis of patients with COPD.
In our previous genetic analysis of lung function of age-matched offspring derived from the JF1/Ms mouse strain crossed with the C3H/HeJ mouse strain, we observed a QTL on mouse chromosome 5 that was associated with lung compliance. This analysis was based on an initial assessment of nine mouse strains, of which JF1/Ms and C3H/HeJ demonstrated the greatest phenotypic difference. In the present study, we found that CZECH/EiJ and NZW/LacJ had a slightly greater difference in lung compliance, but were quite similar to JF1/Ms and C3H/HeJ, respectively. On the basis of the likely candidate genes contained in the QTL region on chromosome 5, we selected Kit for additional analysis. By expanding the phenotyping of mouse strains by an additional 10 strains, we developed additional information useful in associating genotype with phenotypes.
On the basis of the results with these 19 mouse strains, an analysis of Kit genetic variants revealed several SNPs with possible functional consequences. Two promoter SNPs (rs37967821 and rs3799853) were of special interest. The first, rs37967821, alters a conserved sequence of an HOXB4-binding site such that it is converted to an MYB1 site. In the mouse lung, HOXB4 is expressed in mesenchyme and epithelial cells throughout development (33). In HSCs, HOXB4 treatment can produce HSC expansion (34), whereas HOXB4 expression in HSCs can reduce long-term engraftment (35). The other promoter SNP, rs3799853, could eliminate a putative C/EBPα-binding site. Lung-specific gene-targeted Cepba mice have mildly altered fetal lung development, but at 90 days these mice have abnormal alveolar histology (diminished alveolar septa, increased mean linear intercept), mucous cell hyperplasia, and inflammation, features consistent with COPD (36). Previously, Didon and colleagues (37) reported that C/EBP-binding activity is increased in the airway epithelial cells of healthy smokers compared with never-smokers, whereas C/EBP-binding activity was not increased in the epithelium of smokers with COPD. Thus, these investigators proposed that inappropriate C/EBP activity could render the epithelium incompetent for efficient regeneration.
Both of the promoter SNPs have relatively high allelic frequency (∼20%), which could explain much (∼50%) of the phenotypic difference noted between the polar mouse strains. It is noteworthy that the CZECHII/EiJ and JF1/Ms mouse strains share the same genotype for these SNPs whereas the NZW/LacJ and C3H/HeJ mouse strains share the opposing genotype for these SNPs. Thus, the genotypes are consistent and serve to link our previous genome-wide analysis with this candidate gene.
In addition to the promoter differences, SNPs associated with Cl were identified in other regions of the Kit gene. A nonsynonymous SNP (rs36972615) could produce an aspartic acid-to-glutamic acid conversion at amino acid position 927. This amino acid substitution is likely to have minor consequence to protein structure because these amino acids share identical side chain polarity (polar), side chain charge (negative), and hydropathy index (−3.5). Three of the 3′ UTR SNPs (rs33755426, rs36266306, and rs36248044) involved G-from-A conversions that could disrupt segments of the polyadenosine mRNA region, a region that often alters mRNA stability. Together, our SNP interrogation suggests that regulation of Kit gene expression (transcription or message stabilization) may be more important than amino acid substitutions in the mouse. It should be noted that as with any study of genetic variants, the association may be not be due to the exact SNP that we examined but rather to another SNP in linkage disequilibrium with the SNPs identified in Kit. Furthermore, we do not know the functional consequences of each individual SNP identified to be associated with changes in Cl. Thus, more direct functional evidence that Kit has a role in determining lung compliance was needed.
Further supporting the possible role of this candidate gene, lung function was examined in SASH mice that harbor an insertion in the 5′ UTR of Kit. Importantly, the KitW-sh mutation diminishes KIT mRNA expression in mast cells and mesenchyme cells of the lung on E13 and in adult lung, whereas expression in other tissues is normal (25). We observed altered lung histology in the SASH mouse that was accompanied by alterations in lung volumes and both static and dynamic compliance, which are indicative of an emphysema-like phenotype. These observations suggest that signaling through the KIT receptor tyrosine kinase is essential for maintenance of normal lung parenchymal homeostasis in mice. The specific molecular mechanism by which the presence of KIT spares the lung from developing spontaneous airspace enlargement during normal growth is worthy of additional investigation. Nonetheless, the SASH mouse provides an additional model of emphysema in which delayed-onset alveolar enlargement occurs in the absence of differences in inflammation. This suggests that dysregulation of Kit expression may be a more proximal cause of emphysema and possibly an event elicited by inflammation or inflammatory mediators.
The pathogenesis of COPD remains an area of considerable interest (38,39). Two hypotheses have been proposed for the etiology of emphysema in COPD, namely the proteinase–antiproteinase imbalance hypothesis (40, 41) and the oxidant–antioxidant hypothesis (42). Accordingly, mouse models of spontaneous emphysema have features consistent with one of these two hypotheses (43, 44). In the pallid mouse, for example, spontaneous emphysema is associated with an antiproteinase deficiency (45) that is presumed to increase the burden of active neutrophil elastase in the lung (46). Mice deficient in Toll-like receptor-4, the receptor for bacterial LPS, develop emphysema spontaneously that is associated with an imbalance in antioxidant capacity (44). It remains controversial whether spontaneous airspace enlargement observed in murine models is homologous to human emphysematous lung disease (47).
Human studies of COPD have focused mainly on chromosomes 2 and 12 and on a distal region of chromosome 4 (∼145 cM). Human KIT is located on chromosome 4 (55 Mbp) and near/in a region (∼55–70 Mbp) found to have suggestive linkage to both moderate airflow obstruction (48) and postbronchodilator flow limitation (49, 50). In addition, KIT is near a region (48–56 cM) associated with the ratio of FEV1 to FVC in the Framingham Heart Study (51). To our knowledge, there are no human genome-wide association studies of lung compliance. Our studies using divergent strains of mice provide a candidate gene on human chromosome 4, which could contribute to lung compliance. Studies in animal models could additionally provide insight into the molecular and cellular mechanisms that regulate lung homeostasis.
Here, we have reported that many pathognomonic features of emphysema are evident in the SASH mouse. c-KIT protein is a recognized distinguishing marker for HSCs and is a receptor for c-KIT ligand, also known as stem cell factor. Given the association with stem cell function, we consider the possibility that Kit dysregulation may limit normal stem cell function and thereby contribute to the pathogenesis of airspace enlargement due to an inappropriate maintenance of alveolar structure, rather than loss of lung tissue due to dysregulation of response to injury. The role of stem cells in the maintenance of lung architecture remains an area of substantial controversy (52), with conflicting reports regarding whether cells derived from bone marrow can function as lung epithelial progenitor cells (17, 53). However, previous work suggests a role for bone marrow–derived cells in lung repair after injury with combined radiation and elastase (19, 21), and bone marrow transplantation can salvage spontaneous emphysema observed in the tight skin (Tsk) mouse that harbors a spontaneous fibrillin 1 mutation (54). Furthermore, in a murine model of elastase-induced emphysema, intranasal hepatocyte growth factor causes a reduction in airspace enlargement, a return of static lung compliance to control levels, and repair of alveolar wall destruction that was associated with an increase in the expression of bone marrow–derived c-KIT–positive cells (55). Overall, previous studies suggest a potential role for bone marrow–derived cells in protection against the development of airspace enlargement in mice in the context of lung injury. However, we did not use a model of lung injury in our studies. Our findings suggest that c-KIT derived from either epithelia or fibroblasts contributes to normal lung homeostasis.
To determine whether KIT protein expression varied in mouse lung epithelial cells, we performed flow cytometric analysis (56) on dissociated lung cells to quantify alveolar and airway epithelial cell populations. No detectible difference was noted between the SASH and C57BL/6J mouse strains among either EpCAMpos/Sca-1neg (alveolar epithelial cells) or EpCAMpos/Sca-1low (conducting airway epithelial cells) populations within total lung cell preparations (Figures E2 and E3). However, we did observe differences in the capacity of epithelial progenitor cells to undergo in vitro clonal expansion in a manner dependent on c-Kit. Because of the limitations of our assay, it remains unclear whether these effects are either a direct or indirect effect of fibroblast-derived c-KIT on epithelia homeostasis. Evidence supports that bone marrow stromal cells can both attenuate parenchymal injury in a paracrine manner (57) and prevent arrested alveolar growth through paracrine activity (58). Furthermore, lung stromal cells can regulate the growth of lung epithelial cells (59). Interestingly, KIT mRNA is highly expressed in the developing lungs of mice (E14.5) in stromal cells that are in close proximity to epithelia (Figure E4). In humans, KIT-immunoreactive interstitial cells have been observed in the alveolar septa of 12 normal newborn lungs, but were absent in lungs from 2 cases of congenital alveolar capillary dysplasia (18). On the basis of these observations, we hypothesize that Kit expression in interstitial mesenchyme or bone marrow stromal cells facilitates an appropriate alveolar septa microenvironment, which helps maintain alveolar structure possibly through local paracrine activity.
Our study has a limitation in that at this time we cannot identify or assign the specific cell types that are required to express Kit to maintain adult lung structure, but rather highlights the temporal consequences of Kit deficiency to the development of lung structure and function. Although the specific role of Kit in bone marrow–derived stem cells in this model remains unknown, our study supports a role for Kit expression in lung development and maintenance of normal lung architecture in a monopodial branched lung, as exists in rodent models, and suggests that a better understanding of strategies to control Kit-dependent signaling in lung injury and repair may be of value to restore normal lung homeostasis.
Supplementary Material
Acknowledgments
The authors appreciate insightful comments from Caroline L. Hollingsworth, M.D., M.P.H. Infrastructure support was generously provided by Mary Sunday, M.D., Ph.D., and Richard L. Auten, M.D.
Supported by the NIH (ES015675, ES016126, ES016659, ES016347, HL077763, HL090146, and HL085655) and the German Ministry of Education and Research (BMBF)/National Genome Research Network (NGFN) NGFN2:1GR0430.
Contributions: Conception and design: J.Y.L., K.G., Z.L., S.N.A., A.B., B.S., W.M.F., G.L., H.S., J.W.H.; analysis and interpretation: J.Y.L., K.G., Z.L., E.N.P., S.D., H.C., B.B., A.B., B.S., W.M.F., G.L., H.S., J.W.H.; drafting the manuscript for important intellectual content: J.Y.L., D.M.B., B.S., W.M.F., G.L., H.S., J.W.H.
This article has an online supplement, which is available from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201007-1157OC on March 11, 2011
Author Disclosure: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Minoo P. Transcriptional regulation of lung development: emergence of specificity. Respir Res 2000;1:109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kimura J, Deutsch GH. Key mechanisms of early lung development. Pediatr Dev Pathol 2007;10:335–347. [DOI] [PubMed] [Google Scholar]
- 3.Shi W, Bellusci S, Warburton D. Lung development and adult lung diseases. Chest 2007;132:651–656. [DOI] [PubMed] [Google Scholar]
- 4.Warburton D, Bellusci S, De Langhe S, Del Moral PM, Fleury V, Mailleux A, Tefft D, Unbekandt M, Wang K, Shi W. Molecular mechanisms of early lung specification and branching morphogenesis. Pediatr Res 2005;57:26R–37R. [DOI] [PubMed] [Google Scholar]
- 5.Burrows B, Knudson RJ, Lebowitz MD. The relationship of childhood respiratory illness to adult obstructive airway disease. Am Rev Respir Dis 1977;115:751–760. [DOI] [PubMed] [Google Scholar]
- 6.Bridevaux PO, Gerbase MW, Probst-Hensch NM, Schindler C, Gaspoz JM, Rochat T. Long-term decline in lung function, utilisation of care and quality of life in modified GOLD stage 1 COPD. Thorax 2008;63:768–774. [DOI] [PubMed] [Google Scholar]
- 7.Engstrom G, Hedblad B, Janzon L. Reduced lung function predicts increased fatality in future cardiac events: a population-based study. J Intern Med 2006;260:560–567. [DOI] [PubMed] [Google Scholar]
- 8.Friedman GD, Klatsky AL, Siegelaub AB. Lung function and outcome of myocardial infarction. N Engl J Med 1976;295:1323. [DOI] [PubMed] [Google Scholar]
- 9.Strachan DP. Ventilatory function, height, and mortality among lifelong non-smokers. J Epidemiol Community Health 1992;46:66–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss ST. Lung function and airway diseases. Nat Genet 2010;42:14–16. [DOI] [PubMed] [Google Scholar]
- 11.Hirschhorn JN. Genomewide association studies—illuminating biologic pathways. N Engl J Med 2009;360:1699–1701. [DOI] [PubMed] [Google Scholar]
- 12.Reinhard C, Meyer B, Fuchs H, Stoeger T, Eder G, Ruschendorf F, Heyder J, Nurnberg P, de Angelis MH, Schulz H. Genomewide linkage analysis identifies novel genetic loci for lung function in mice. Am J Respir Crit Care Med 2005;171:880–888. [DOI] [PubMed] [Google Scholar]
- 13.Roskoski R Jr. Structure and regulation of Kit protein-tyrosine kinase—the stem cell factor receptor. Biochem Biophys Res Commun 2005;338:1307–1315. [DOI] [PubMed] [Google Scholar]
- 14.Roskoski R Jr. Signaling by Kit protein-tyrosine kinase—the stem cell factor receptor. Biochem Biophys Res Commun 2005;337:1–13. [DOI] [PubMed] [Google Scholar]
- 15.Duttlinger R, Manova K, Chu TY, Gyssler C, Zelenetz AD, Bachvarova RF, Besmer P. W-sash affects positive and negative elements controlling c-kit expression: ectopic c-kit expression at sites of kit-ligand expression affects melanogenesis. Development 1993;118:705–717. [DOI] [PubMed] [Google Scholar]
- 16.Giangreco A, Arwert EN, Rosewell IR, Snyder J, Watt FM, Stripp BR. Stem cells are dispensable for lung homeostasis but restore airways after injury. Proc Natl Acad Sci USA 2009;106:9286–9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotton DN, Fabian AJ, Mulligan RC. Failure of bone marrow to reconstitute lung epithelium. Am J Respir Cell Mol Biol 2005;33:328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang KT, Rajadurai VS, Walford NQ, Hwang WS. Alveolar capillary dysplasia: absence of CD117 immunoreactivity of putative hemangioblast precursor cells. Fetal Pediatr Pathol 2008;27:127–140. [DOI] [PubMed] [Google Scholar]
- 19.Yamada M, Kubo H, Kobayashi S, Ishizawa K, Numasaki M, Ueda S, Suzuki T, Sasaki H. Bone marrow–derived progenitor cells are important for lung repair after lipopolysaccharide-induced lung injury. J Immunol 2004;172:1266–1272. [DOI] [PubMed] [Google Scholar]
- 20.Ishizawa K, Kubo H, Yamada M, Kobayashi S, Numasaki M, Ueda S, Suzuki T, Sasaki H. Bone marrow–derived cells contribute to lung regeneration after elastase-induced pulmonary emphysema. FEBS Lett 2004;556:249–252. [DOI] [PubMed] [Google Scholar]
- 21.Abe S, Boyer C, Liu X, Wen FQ, Kobayashi S, Suzuki T, Mizuno S, Nakamura T, Sasaki H. Cells derived from the circulation contribute to the repair of lung injury. Am J Respir Crit Care Med 2004;170:1158–1163. [DOI] [PubMed] [Google Scholar]
- 22.Ishizawa K, Kubo H, Yamada M, Kobayashi S, Suzuki T, Mizuno S, Nakamura T, Sasaki H. Hepatocyte growth factor induces angiogenesis in injured lungs through mobilizing endothelial progenitor cells. Biochem Biophys Res Commun 2004;324:276–280. [DOI] [PubMed] [Google Scholar]
- 23.Ganguly K, Stoeger T, Wesselkamper SC, Reinhard C, Sartor MA, Medvedovic M, Tomlinson CR, Bolle I, Mason JM, Leikauf GD, Candidate genes controlling pulmonary function in mice: transcript profiling and predicted protein structure. Physiol Genomics 2007;31:410–421. [DOI] [PubMed] [Google Scholar]
- 24.Reinhard C, Eder G, Fuchs H, Ziesenis A, Heyder J, Schulz H. Inbred strain variation in lung function. Mamm Genome 2002;13:429–437. [DOI] [PubMed] [Google Scholar]
- 25.Lyon MF, Glenister PH. A new allele sash (Wsh) at the W-locus and a spontaneous recessive lethal in mice. Genet Res 1982;39:315–322. [DOI] [PubMed] [Google Scholar]
- 26.Nagle DL, Kozak CA, Mano H, Chapman VM, Bucan M. Physical mapping of the Tec and Gabrb1 loci reveals that the Wsh mutation on mouse chromosome 5 is associated with an inversion. Hum Mol Genet 1995;4:2073–2079. [DOI] [PubMed] [Google Scholar]
- 27.Schulz H, Johner C, Eder G, Ziesenis A, Reitmeier P, Heyder J, Balling R. Respiratory mechanics in mice: strain and sex specific differences. Acta Physiol Scand 2002;174:367–375. [DOI] [PubMed] [Google Scholar]
- 28.Auten RL Jr, Mason SN, Tanaka DT, Welty-Wolf K, Whorton MH. Anti-neutrophil chemokine preserves alveolar development in hyperoxia-exposed newborn rats. Am J Physiol Lung Cell Mol Physiol 2001;281:L336–L344. [DOI] [PubMed] [Google Scholar]
- 29.Dunnill MS. Quantitative methods in the study of pulmonary pathology. Thorax 1962;17:320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. Curr Protoc Bioinformatics 2008;Chapter 2:Unit 2.6. [DOI] [PubMed]
- 31.Murray CJ, Lopez AD. Alternative projections of mortality and disability by cause 1990–2020: Global Burden of Disease Study. Lancet 1997;349:1498–1504. [DOI] [PubMed] [Google Scholar]
- 32.Shapiro SD, Ingenito EP. The pathogenesis of chronic obstructive pulmonary disease: advances in the past 100 years. Am J Respir Cell Mol Biol 2005;32:367–372. [DOI] [PubMed] [Google Scholar]
- 33.Volpe MV, Wang KT, Nielsen HC, Chinoy MR. Unique spatial and cellular expression patterns of Hoxa5, Hoxb4, and Hoxb6 proteins in normal developing murine lung are modified in pulmonary hypoplasia. Birth Defects Res A Clin Mol Teratol 2008;82:571–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Y, Chen J, Young NS. Expansion of haematopoietic stem cells from normal donors and bone marrow failure patients by recombinant hoxb4. Br J Haematol 2009;144:603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabayoyong WB, Salas JG, Bonde S, Zavazava N. HOXB4-transduced embryonic stem cell–derived Lin-c-kit+ and Lin-Sca-1+ hematopoietic progenitors express H60 and are targeted by NK cells. J Immunol 2009;183:5449–5457. [DOI] [PubMed] [Google Scholar]
- 36.Didon L, Roos AB, Elmberger GP, Gonzalez FJ, Nord M. Lung-specific inactivation of CCAAT/enhancer binding protein α causes a pathological pattern characteristic of COPD. Eur Respir J 2010;35:186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Didon L, Qvarfordt I, Andersson O, Nord M, Riise GC. Decreased CCAAT/enhancer binding protein transcription factor activity in chronic bronchitis and COPD. Chest 2005;127:1341–1346. [DOI] [PubMed] [Google Scholar]
- 38.Sharafkhaneh A, Hanania NA, Kim V. Pathogenesis of emphysema: from the bench to the bedside. Proc Am Thorac Soc 2008;5:475–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taraseviciene-Stewart L, Voelkel NF. Molecular pathogenesis of emphysema. J Clin Invest 2008;118:394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gadek JE, Fells GA, Zimmerman RL, Crystal RG. Role of connective tissue proteases in the pathogenesis of chronic inflammatory lung disease. Environ Health Perspect 1984;55:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janoff A, Sloan B, Weinbaum G, Damiano V, Sandhaus RA, Elias J, Kimbel P. Experimental emphysema induced with purified human neutrophil elastase: tissue localization of the instilled protease. Am Rev Respir Dis 1977;115:461–478. [DOI] [PubMed] [Google Scholar]
- 42.Repine JE, Bast A, Lankhorst I; Oxidative Stress Study Group. Oxidative stress in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 1997;156:341–357. [DOI] [PubMed] [Google Scholar]
- 43.Keil M, Lungarella G, Cavarra E, van Even P, Martorana PA. A scanning electron microscopic investigation of genetic emphysema in tight-skin, pallid, and beige mice, three different C57 BL/6J mutants. Lab Invest 1996;74:353–362. [PubMed] [Google Scholar]
- 44.Zhang X, Shan P, Jiang G, Cohn L, Lee PJ. Toll-like receptor 4 deficiency causes pulmonary emphysema. J Clin Invest 2006;116:3050–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martorana PA, Brand T, Gardi C, van Even P, de Santi MM, Calzoni P, Marcolongo P, Lungarella G. The pallid mouse. A model of genetic α1-antitrypsin deficiency. Lab Invest 1993;68:233–241. [PubMed] [Google Scholar]
- 46.Cavarra E, Martorana PA, Gambelli F, de Santi M, van Even P, Lungarella G. Neutrophil recruitment into the lungs is associated with increased lung elastase burden, decreased lung elastin, and emphysema in α1 proteinase inhibitor-deficient mice. Lab Invest 1996;75:273–280. [PubMed] [Google Scholar]
- 47.Mouded M, Egea EE, Brown MJ, Hanlon SM, Houghton AM, Tsai LW, Ingenito EP, Shapiro SD. Epithelial cell apoptosis causes acute lung injury masquerading as emphysema. Am J Respir Cell Mol Biol 2009;41:407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silverman EK, Mosley JD, Palmer LJ, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ, et al. Genome-wide linkage analysis of severe, early-onset chronic obstructive pulmonary disease: airflow obstruction and chronic bronchitis phenotypes. Hum Mol Genet 2002;11:623–632. [DOI] [PubMed] [Google Scholar]
- 49.DeMeo DL, Celedon JC, Lange C, Reilly JJ, Chapman HA, Sylvia JS, Speizer FE, Weiss ST, Silverman EK. Genome-wide linkage of forced mid-expiratory flow in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2004;170:1294–1301. [DOI] [PubMed] [Google Scholar]
- 50.Hersh CP, Soto-Quiros ME, Avila L, Lake SL, Liang C, Fournier E, Spesny M, Sylvia JS, Lazarus R, Hudson T, et al. Genome-wide linkage analysis of pulmonary function in families of children with asthma in Costa Rica. Thorax 2007;62:224–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilk JB, Chen TH, Gottlieb DJ, Walter RE, Nagle MW, Brandler BJ, Myers RH, Borecki IB, Silverman EK, Weiss ST, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet 2009;5:e1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kotton DN, Fine A. Lung stem cells. Cell Tissue Res 2008;331:145–156. [DOI] [PubMed] [Google Scholar]
- 53.Wagers AJ, Sherwood RI, Christensen JL, Weissman IL. Little evidence for developmental plasticity of adult hematopoietic stem cells. Science 2002;297:2256–2259. [DOI] [PubMed] [Google Scholar]
- 54.Adachi Y, Oyaizu H, Taketani S, Minamino K, Yamaguchi K, Shultz LD, Iwasaki M, Tomita M, Suzuki Y, Nakano K, et al. Treatment and transfer of emphysema by a new bone marrow transplantation method from normal mice to Tsk mice and vice versa. Stem Cells 2006;24:2071–2077. [DOI] [PubMed] [Google Scholar]
- 55.Hegab AE, Kubo H, Yamaya M, Asada M, He M, Fujino N, Mizuno S, Nakamura T. Intranasal HGF administration ameliorates the physiologic and morphologic changes in lung emphysema. Mol Ther 2008;16:1417–1426. [DOI] [PubMed] [Google Scholar]
- 56.Teisanu RM, Lagasse E, Whitesides JF, Stripp BR. Prospective isolation of bronchiolar stem cells based on immunophenotypic and autofluorescence characteristics. Stem Cells 2009;27:612–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aslam M, Baveja R, Liang OD, Fernandez-Gonzalez A, Lee C, Mitsialis SA, Kourembanas S. Bone marrow stromal cells attenuate lung injury in a murine model of neonatal chronic lung disease. Am J Respir Crit Care Med 2009;180:1122–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Haaften T, Byrne R, Bonnet S, Rochefort GY, Akabutu J, Bouchentouf M, Rey-Parra GJ, Galipeau J, Haromy A, Eaton F, et al. Airway delivery of mesenchymal stem cells prevents arrested alveolar growth in neonatal lung injury in rats. Am J Respir Crit Care Med 2009;180:1131–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McQualter JL, Yuen K, Williams B, Bertoncello I. Evidence of an epithelial stem/progenitor cell hierarchy in the adult mouse lung. Proc Natl Acad Sci USA 2010;107:1414–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.