Abstract
Rationale: Idiopathic pulmonary fibrosis is a progressive, uniformly fatal interstitial lung disease. An acute exacerbation of idiopathic pulmonary fibrosis is an episode of acute respiratory worsening without an identifiable etiology. Occult viral infection has been proposed as a possible cause of acute exacerbation.
Objectives: To use unbiased genomics-based discovery methods to define the role of viruses in acute exacerbation of idiopathic pulmonary fibrosis.
Methods: Bronchoalveolar lavage and serum from patients with acute exacerbation of idiopathic pulmonary fibrosis, stable disease, and acute lung injury were tested for viral nucleic acid using multiplex polymerase chain reaction, pan-viral microarray, and high-throughput cDNA sequencing.
Measurements and Main Results: Four of forty-three patients with acute exacerbation of idiopathic pulmonary fibrosis had evidence of common respiratory viral infection (parainfluenza [n = 1], rhinovirus [n = 2], coronavirus [n = 1]); no viruses were detected in the bronchoalveolar lavage from stable patients. Pan-viral microarrays revealed additional evidence of viral infection (herpes simplex virus [n = 1], Epstein-Barr virus [n = 2], and torque teno virus [TTV] [n = 12]) in patients with acute exacerbation. TTV infection was significantly more common in patients with acute exacerbation than stable controls (P = 0.0003), but present in a similar percentage of acute lung injury controls. Deep sequencing of a subset of acute exacerbation cases confirmed the presence of TTV but did not identify additional viruses.
Conclusions: Viral infection was not detected in most cases of acute exacerbation of idiopathic pulmonary fibrosis. TTV was present in a significant minority of cases, and cases of acute lung injury; the clinical significance of this finding remains to be determined.
Keywords: acute lung injury, virus, infection, pulmonary fibrosis, etiology
AT A GLANCE COMMENTARY.
Scientific Knowledge on the Subject
The etiology of acute exacerbation of idiopathic pulmonary fibrosis remains unknown. Occult viral infection has been proposed as one possible cause of acute exacerbation of idiopathic pulmonary fibrosis.
What This Study Adds to the Field
This study uses the most current genomics-based technologies to investigate the possible infectious etiology of acute exacerbations of idiopathic pulmonary fibrosis. Most cases demonstrate no evidence of viral infection. Torque teno virus was present in a significant minority of cases, and cases of acute lung injury.
Idiopathic pulmonary fibrosis (IPF) is a progressive fibrotic lung disease with no known cause or cure (1, 2). Although some patients experience a steady progression of disease over time, many have periods of relative stability punctuated by episodes of acute respiratory worsening that can be fatal (3, 4). When no identifiable cause for this acute worsening is found, it is termed an “acute exacerbation” of IPF (5).
Acute exacerbation of IPF is defined as an idiopathic acute worsening of dyspnea characterized radiologically by the presence of bilateral ground glass abnormality on high-resolution computed tomography scan of the chest (5). It is estimated that between 5% and 10% of patients with IPF experience an acute exacerbation annually, with more physiologically advanced disease at higher risk (4–6). It remains unclear whether acute exacerbation of IPF represents a primary acceleration of the underlying fibroproliferative process in IPF or is a clinically occult secondary complication (e.g., infection) (5, 7).
Acute exacerbation of IPF is often accompanied by fever, increased cough, and myalgia, suggesting an infectious etiology. Respiratory viruses have been considered a particularly likely cause, based on the similarities in clinical and radiologic presentation between acute exacerbation of IPF and viral pneumonitis and the poor sensitivity of standard methods of viral detection (5). Also, there may be an increased incidence of acute exacerbation of IPF in East Asia, a finding that could be explained by an environmental factor, such as an endemic virus (8–10). Preliminary evaluations of the role of infection in acute exacerbation have yielded mixed results (11, 12).
In this study, we tested the hypothesis that acute exacerbation of IPF is caused by occult viral infection. We prospectively collected bronchoalveolar lavage (BAL) from patients experiencing acute exacerbation of IPF and controls (stable IPF and acute lung injury [ALI]) and used multiplex polymerase chain reaction (PCR) and a pan-viral microarray discovery platform to test for the presence of known and novel viruses. Next-generation parallel sequencing (deep sequencing) was applied to a subset of acute exacerbation samples to increase the sensitivity of viral detection.
METHODS
Study Population
Patients with acute exacerbation of IPF were identified prospectively from two centers (University of Ulsan, Korea, and Tosei General Hospital, Japan). Sequential subjects with acute exacerbation of IPF were enrolled if they underwent bronchoscopy and were consented. Diagnostic criteria for acute exacerbation of IPF were prespecified according to established criteria (5). All patients with acute exacerbation had negative clinical evaluation for infectious causes including routine bacterial and viral BAL antibody titers and cultures for respiratory syncytial virus, influenza A and B, human parainfluenza viruses, adenovirus, human cytomegalovirus (CMV), herpes simplex virus (HSV), and varicella-zoster virus. Control patients with stable IPF (defined by the absence of acute exacerbation) and ALI were identified from an existing cohort at a single center (University of Ulsan, Korea) and underwent bronchoscopy at the time of diagnosis. IPF and ALI were defined by consensus criteria (1, 13, 14). BAL from a case of IPF that was positive for rhinovirus and CMV was included as a blinded positive control. All centers received approval from their institutional review board or equivalent, and all patients provided informed consent.
Sample Collection and Processing
In all cases, bronchoscopy was performed as part of patients' clinical evaluations. In most cases, BAL was collected within the first 48 hours of admission to the hospital. In general, BAL was performed in a single subsegment of the right middle lobe or lingula, with at least 100 ml of sterile saline instilled. Blood was not collected as part of the initial study protocol. A subset of acute exacerbation samples and stable IPF controls underwent phlebotomy at the time of bronchoscopy as part of a separate ongoing repository study. These samples were available to us for analysis. All samples were stored at −80°C until ready for processing. Total RNA was extracted from 200 μl of each sample using the RNeasy mini kit (Qiagen, Inc., Valencia, CA) according to the manufacturer's instructions.
PCR Analysis
A blinded, nested respiratory multiplex PCR was run on the BAL samples from acute exacerbation of IPF and stable IPF controls for prespecified respiratory viruses (influenza virus, human parainfluenza virus, respiratory syncytial virus, human rhinovirus, human enterovirus, human coronavirus, human metapneumovirus, and human adenovirus) (15). PCR was also performed to confirm viral signatures present on pan-viral microarray analysis (HSV), Epstein-Barr virus, and torque teno virus (TTV) using published nested primer sets (16–18). Additional PCR for TTV was performed on BAL samples from ALI controls and on serum samples from a subset of acute exacerbation of IPF and stable IPF controls to define better the epidemiology of TTV.
Pan-Viral Microarray Analysis
Acute exacerbation of IPF samples and stable IPF controls were randomly amplified to generate cDNA, which was hybridized blindly to a pan-viral microarray as previously described (19). Arrays were scanned using the Axon 4000B scanner (Molecular Devices, Sunnyvale, CA) and intensities were calculated using GenePix 6.0 (Molecular Devices). The presence of a viral signature was determined using Cluster 3.0 (20) and E-predict (21). All microarray data were deposited to the Gene Expression Omnibus (GEO) under the accession GSE27578.
Deep Sequencing Analysis
BALs from a subgroup of study patients with acute exacerbation of IPF were selected for deep sequencing on the Illumina Genome Analyzer IIx platform (Illumina, San Diego, CA) based on the presence of both fever and myalgia, symptoms suggestive of a viral-like illness. Deep sequencing libraries were prepared and analyzed as described in the online supplement. The deep sequencing reads were submitted to the Short Read Archive (SRA) under the accession SRX042016.
Statistical Methods
Clinical data are expressed as means or percentages, unless otherwise stated. The primary comparison was between BAL samples from acute exacerbation of IPF and stable IPF controls. Additional comparisons were made between BAL samples from acute exacerbation of IPF and ALI controls, and serum samples from acute exacerbation of IPF and stable IPF controls. In all cases, intergroup comparisons were performed conservatively using nonparametric methods (Wilcoxon signed-rank test) and chi-square/Fisher exact analyses as appropriate. Regression analysis was performed to determine the relationship between clinical factors, PCR positivity, and survival. Clinical data analysis was performed using SAS 9.1 (SAS Institute, Cary, NC). Statistical significance was defined as a P value less than 0.05.
RESULTS
Patient Characteristics
Sixty patients with acute exacerbation of IPF (52 Korean, 8 Japanese) were identified between 2006 and 2009. Forty-three of these underwent bronchoscopy and were enrolled in the study. The median time from diagnosis to acute exacerbation was 85 days. Their clinical characteristics are summarized in Table 1. Twelve (28%) of the acute exacerbation of patients with IPF presented with both fever and myalgia, suggestive of a viral-like illness. Forty patients with stable IPF and twenty-nine patients with ALI were included as controls. Patients with ALI predominantly had lower respiratory tract infection (90%) as the underlying cause for their lung injury. These patients were all mechanically ventilated and had a mean PaO2/FiO2 ratio of 228. Serum was available from 22 acute exacerbation of patients with IPF and 31 stable IPF controls.
TABLE 1.
CLINICAL CHARACTERISTICS
| Variable | Acute Exacerbation (n = 43) | Stable (n = 40) | ALI (n = 29) |
|---|---|---|---|
| Age, yr | 65 | 66 | 60 |
| Male sex, % | 88 | 75 | 66 |
| Surgical lung biopsy, % | 21 | 28 | NA |
| Smoking, % | 84 | 75 | 48 |
| Baseline FVC, % | 73 | 79 | NA |
| Baseline DLCO, % | 60 | 70 | NA |
| Mechanical ventilation, % | 37 | NA | 100 |
| Immunosuppressive therapy, % | 60 | NA | NA |
Definition of abbreviations: ALI = acute lung injury; DLCO % = diffusing capacity for carbon monoxide percent predicted.
Immunosuppressive therapy includes corticosteroids with or without immunomodulator therapy.
Viral Detection by PCR and Pan-Viral Microarray
Four acute exacerbation of IPF BAL samples (9%) were positive for common respiratory viruses by initial multiplex PCR (two for rhinovirus, one for human coronavirus-OC43, and one for parainfluenza virus-1). All stable IPF samples were negative for common respiratory viruses (P = 0.12 compared with acute exacerbation of IPF samples) (Table 2). Array analysis of acute exacerbation of IPF BAL samples revealed the presence of TTV and several human herpesviruses, and accurately identified rhinovirus and CMV in the known positive control. To pursue these findings further, we performed sensitive genome-specific PCR reactions for HSV, Epstein-Barr virus, and TTV. This yielded 15 additional BAL positives in acute exacerbation of IPF samples (Table 3). Of these additional viruses, only TTV was significantly more common in acute exacerbation of IPF compared with stable controls (28% vs. 0%; P = 0.0003). Four BAL samples revealed double infections: two with TTV and rhinovirus, one with TTV and parainfluenza virus-1, and one with TTV and HSV. One BAL sample revealed a triple infection of TTV, Epstein-Barr virus, and coronavirus. Overall, 14 (33%) of acute exacerbation of IPF samples were positive for virus compared with no positives in the stable IPF samples (P <0.0001). There was no difference in the frequency of fever and myalgia between virus-positive and virus-negative cases, and there was no significant difference in the use of corticosteroid treatment.
TABLE 2.
RESPIRATORY VIRAL DETECTION IN ACUTE EXACERBATION AND STABLE IDIOPATHIC PULMONARY FIBROSIS
| Virus | Acute Exacerbation (n = 43) | Stable (n = 40) | P Value |
|---|---|---|---|
| Any respiratory virus (%) | 4 (9) | 0 (0) | 0.12 |
| Rhinovirus (%) | 2 (5) | 0 (0) | 0.49 |
| Coronavirus (%) | 1 (2) | 0 (0) | 1 |
| Parainfluenza (%) | 1 (2) | 0 (0) | 1 |
| Adenovirus (%) | 0 (0) | 0 (0) | – |
| Enterovirus (%) | 0 (0) | 0 (0) | – |
| Influenza (%) | 0 (0) | 0 (0) | – |
| Metapneumovirus (%) | 0 (0) | 0 (0) | – |
| Respiratory syncytial virus (%) | 0 (0) | 0 (0) | – |
TABLE 3.
ARRAY-BASED VIRAL DETECTION IN ACUTE EXACERBATION AND STABLE IDIOPATHIC PULMONARY FIBROSIS
| Virus | Acute Exacerbation (n = 43) | Stable (n = 40) | Acute lung injury (n = 29) | P Value* |
|---|---|---|---|---|
| Torque teno virus (%) | 12 (28) | 0 (0) | 7 (24) | 0.0003 |
| Epstein-Barr virus (%) | 2 (5) | 0 (0) | NA | 0.49 |
| Herpes simplex virus (%) | 1 (2) | 0 (0) | NA | 1 |
| Cytomegalovirus (%) | 0 (0) | 0 (0) | NA | – |
P value is for comparison of acute exacerbation with stable control.
Viral Detection by Deep Sequencing
BALs from 12 of the study patients with acute exacerbation of IPF were selected for deep sequencing to investigate the possibility of viruses being present but undetected by PCR and microarray. Of these samples, two were PCR-positive for tested viruses, one for TTV and one for TTV and HSV. After initial quality filtering, approximately 26 million pairs, or 52 million total reads, comprised the primary dataset. Each of the 12 barcoded acute exacerbation of IPF samples was represented with at least 3 million high-quality reads. Over 98% of the reads were derived from human origin, and of the remaining reads, approximately 0.1% were recognizably bacterial in origin. Only a few hundred were potentially attributable to known nonhuman eukaryotes and viruses. Aside from bacteriophages, only three viruses (two TTVs and one HSV) were found, consistent with the PCR results. After all stages of mapping to a sequence database were finished, approximately 0.6% of the original dataset remained without attribution.
Comparison of TTV-positive and TTV-negative Acute Exacerbation of IPF Samples
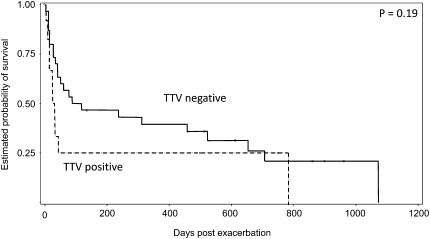
There were no significant differences in age, sex, baseline pulmonary function, or time to bronchoscopy in TTV-positive acute exacerbation of patients with IPF compared with TTV-negative acute exacerbation of IPF patients (data not shown). Patients who were TTV positive appeared sicker, with 58% requiring mechanical ventilation (vs. 29% in patients who were TTV negative; P = 0.09) and 75% dying at 60 days (vs. 42% in patients who were TTV negative; P = 0.06). Overall median survival time in the patients who were TTV positive was 29 days (vs. 88 d in patients who were TTV negative; P = 0.19) (Figure 1). Bivariate regression analysis of potential independent predictors of survival time (presence or absence of prednisone treatment at the time of BAL, mechanical ventilation and the time of BAL, and TTV positivity on BAL) revealed only mechanical ventilation as statistically significant (hazard ratio, 2.30; P = 0.03). TTV positivity was not an independent predictor of survival time (hazard ratio, 1.65; P = 0.20).
Figure 1.
Survival time in torque teno virus (TTV)–positive acute exacerbation of patients with idiopathic pulmonary fibrosis compared with TTV-negative acute exacerbation of patients with idiopathic pulmonary fibrosis. Mean survival time was 29 days versus 88 days, respectively. P = 0.19.
TTV Positivity in Acute Exacerbation of IPF and Stable IPF Serum
A total of 6 (27%) of 22 patients with acute exacerbation of IPF were PCR positive for TTV in serum, compared with 5 (16%) of 31 patients with stable IPF (P = 0.34). There was no relationship between serum and BAL positivity. Three (50%) of the six patients with acute exacerbation who were PCR positive in serum were also PCR positive in BAL. Four (25%) of the 16 patients with acute exacerbation who were PCR negative in serum were PCR positive in BAL.
TTV Positivity in ALI BAL
The statistically significant link between BAL-associated TTV and acute exacerbation of IPF prompted us to also examine BAL samples from 29 patients with ALI. TTV was detected in 7 (24%) of 29 BAL samples from ALI patients; this was not significantly different from the prevalence of TTV in BAL samples from acute exacerbation of IPF (28%; P = 0.73).
DISCUSSION
Using highly sensitive PCR, pan-viral microarrays, and deep sequencing technologies in a large, well-described cohort of patients with acute exacerbation of IPF and controls, we found that most cases of acute exacerbation of IPF had no evidence of an underlying viral infection. This suggests that viral infection is not a common cause of acute exacerbation of IPF.
Overall, we found viral nucleic acid in the BAL of 33% of patients with acute exacerbation of IPF; no viruses were found in samples from stable IPF controls. There were two rhinovirus-positive samples, one coronavirus-positive sample, and one parainfluenza virus–positive sample, suggesting that a small minority (9%) of acute exacerbations of IPF may be caused by occult infection with common respiratory viruses. Surprisingly, the most common virus detected in the BAL of acute exacerbation of IPF patients was TTV, which was present in 28% of acute exacerbation BAL samples. This finding was not unique to acute exacerbation of IPF because 24% of BAL samples from ALI controls were also TTV positive.
Two recent studies have commented indirectly on the possible role of occult viral infection in acute exacerbation of IPF. The first study performed gene expression microarrays on whole lung tissue from 8 patients who died of acute exacerbation of IPF, 23 patients with stable IPF, and 15 healthy controls (12). The authors concluded that acute exacerbation of IPF was characterized by a pattern of enhanced epithelial injury and proliferation, but found no gene expression profiles indicative of a response to viral or bacterial infection. In a second study of 27 patients presenting with acute decline in fibrotic lung disease (13 of whom had confirmed acute exacerbation of IPF), 5 had antigenic or PCR evidence of viral infection (one parainfluenza virus, two HSV, and two CMV infections), three of which were missed on standard viral culture (11).
Our study expands significantly on previously published reports. First, we take an unbiased approach to viral discovery using cutting-edge genomic methodology. It is the first study to do this in acute exacerbation of IPF. Our use of sequencing to confirm all suspected viruses rules out the possibility of spurious PCR results, a common pitfall of the technique. Second, our large cohort of well-defined patients with acute exacerbation with adequate controls allows for greater certainty regarding our conclusions. Third, we have identified an unexpected virus (TTV) that was associated with 33% of acute exacerbations, and that was absent in stable IPF.
The pathogenetic significance of TTV in acute exacerbation of IPF BAL is unclear. TTV is a nonenveloped single-stranded circular DNA virus that exists in a genetically diverse clade (22, 23). The virus seems to have broad tissue tropism because it has been detected in peripheral blood mononuclear cells (PBMCs) and bone marrow, spleen, liver, and lung (22). Infection with TTV in the human population is worldwide, with prevalences of viremia ranging from 8%–80% depending on the population studied and detection methodology used. When only considering the hemi-nested PCR of the N22 region used in this study, rates of TTV DNA found in healthy blood donors range from 8.4%–12% (24, 25) and do not seem to correlate with the geographic location of the patients. Most infected subjects are asymptomatic, and to date efforts to link TTV viremia with any acute or chronic pathologic state have been unsuccessful (22). Although there have been reports of TTV in the upper respiratory tract (nasopharynx and oral cavity) (26), TTV has not been identified in BAL fluids. TTV has previously been detected in the serum of 12 (36%) of 33 Japanese patients with IPF. In this study, TTV appeared more frequently in cases that progressed to acute exacerbations, and TTV positivity was suggested to correlate with worse survival (27). Our findings do not show a correlation between the presence of TTV in the serum and the presence of TTV in the BAL, or any correlation between serum TTV positivity and a diagnosis of acute exacerbation.
It is possible that de novo TTV infection in the lung causes direct alveolar epithelial cell injury and acute respiratory worsening. If so, this process does not seem to be unique to acute exacerbation of IPF because we detected TTV at a similar frequency in BAL from patients with ALI. Although this does not exclude a potential role for TTV in the pathogenesis of acute exacerbation of IPF, it is also compatible with the idea that inflammation or injury in the lung may nonspecifically trigger local TTV replication, or may result in increased vascular permeability in the lung allowing circulating virus to enter the alveolar compartment. In the latter two cases, the presence of TTV would represent a consequence of lung inflammation rather than its cause. The idea that local TTV replication might be enhanced by underlying inflammatory signaling is supported by in vitro studies of PBMCs from donors who are TTV negative (28). These PBMCs were infected in vitro with TTV, cultured with and without the presence of phytohemagglutinin, lipopolysaccharide, and interleukin-2, and then examined for evidence of TTV replication. In this experiment, TTV mRNA and replicative intermediates were only found in the stimulated PBMCs, consistent with an infection-amplifying role for inflammatory signaling.
The methodologies used in this study have unparalleled sensitivity for viral detection. The multiplex nested PCR is several fold more sensitive than virus culture and direct immunofluorescent tests, with the ability to amplify less than 10 copies of target nucleic acid (15). For viral discovery, however, PCR is of limited use because it identifies only a priori viral targets. Pan-viral microarray precludes the need for a preconceived list of targets, although even with its proved sensitivity, its benefit is dependent on the signal-to-noise ratio of the nucleic acid (19). The use of deep sequencing to look further for evidence of viral infection in a high-risk subpopulation of patients with acute exacerbation therefore adds confidence to our results, because it produces an unbiased, high-resolution description of the microbial landscape of the sample tested. This technology has been used to identify novel viruses in human diarrhea, and to describe the microbiome of the distal gut (31, 32), but never in BAL (29, 30). In the current study, two samples subjected to deep sequencing were positive for known viruses by PCR and pan-viral array screening. Using an efficient and sensitive pipeline for sorting reads, these positive PCR findings were confirmed, and no additional viruses were detected in these or the other samples tested. Interpretation of these findings must be tempered by the fact that existing computational methods for recognizing potential viral genomes are imperfect, and may fail to identify novel agents with only limited homology to known viral genera. The same is true of array-based viral detection methods.
One important limitation of this study is the potential for false-negative results because of the timing of sample collection. BAL was performed early in the course of hospitalization, most commonly in the first 48 hours after admission, and the median time from symptom onset to sampling was 7 days. Importantly, no difference in the time from symptom onset to sample collection was found between virus-positive and virus-negative cases. The duration of replicating virus in BAL is largely unknown, and it is possible that a virus could have stopped shedding during this time. In this study, we have maximized our likelihood of detecting virus by obtaining BAL samples early after admission and using highly sensitive viral detection techniques.
In summary, this study used unbiased, highly sensitive genomics-based discovery methods to investigate the role of viral infection in a large, well-characterized cohort of patients with acute exacerbation of IPF. The results of this study suggest that most cases of acute exacerbation of IPF are not caused by viral infection. Future research into the etiology of acute exacerbation of IPF should confirm these findings, further investigate the role of TTV, and consider other possible occult complications (e.g., aspiration) that may cause acute respiratory worsening in these patients.
Supplementary Material
Acknowledgments
Acknowledgment: The authors thank Amy Kistler for her effort with the initial sample processing and for offering her review. The authors also thank Drs. Naftali Kaminski and Talmadge E. King Jr. for their thoughtful input and guidance in the early stages of this project, and James Graham Ruby for his contribution to the sequencing library preparation.
Author contributions: Involvement in conception, hypothesis, and design of the study: S.C.W., D.S.K., Y.K., H.B., L.L., D.G., H.C.; selection of patients and acquisition of samples: D.S.K., Y.K., J.W.S., J.W.H., H.T.; acquisition of the data: S.C.W., D.S.K., Y.K., E.C., J.L., C.C., H.C.; analysis and interpretation of the data: S.C.W., D.S.K., Y.K., H.T., H.B., L.L., P.W., J.D., D.G., H.C.; substantial involvement in the writing and/or revision of the article: S.C.W., D.S.K., Y.K., H.T., P.W., J.D., D.G., H.C.
Supported by NHLBI HL 086516, HHMI, Packard Foundation, and Doris Duke Charitable Research Foundation.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201010-1752OC on February 25, 2011
Author Disclosure: S.C.W. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.S.K. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. Y.K. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. E.C. received funding for the Chui Lab from Abbott Diagnostics. J.S.L. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.W.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.W.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. H.T. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. C.C. received research support from Abbott Diagnostics. H.B. is a Board Member of the Health Effects Institute and was a consultant for Pharmaxis Ltd., KALOBIOS Pharmaceuticals, Inc., Merck Sharp & Dohme, and Genentech. He is employed by the University of California, San Francisco, and received grant support from GlaxoSmithKline. He receives royalties from McGraw-Hill Co., Blackwell Publishing Ltd, and Taylor & Francis. L.H.L. was on the Advisory Board for Intermune and Actelion. She received grant support from Intermune, Actelion, Johnson & Johnson, and Gilead. P.J.W. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.D. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. H.R.C.'s institution received consultancy fees from Boehringer-Ingelheim, FibroGen, Genentech, Gilead, Actelion, and Arresto. He received institutional grant support from the CHEST Foundation/ASP and received payment for the development of educational presentations from the France Foundation.
References
- 1.American Thoracic Society. Idiopathic pulmonary fibrosis: diagnosis and treatment. International consensus statement. American Thoracic Society (ATS), and the European Respiratory Society (ERS). Am J Respir Crit Care Med 2000;161:646–664. [DOI] [PubMed] [Google Scholar]
- 2.American Thoracic Society, European Respiratory Society. American Thoracic Society/European Respiratory Society International Multidisciplinary Consensus Classification of the Idiopathic Interstitial Pneumonias. Am J Respir Crit Care Med 2002;165:277–304. [DOI] [PubMed] [Google Scholar]
- 3.Kim DS, Collard HR, King TE, Jr. Classification and natural history of the idiopathic interstitial pneumonias. Proc Am Thorac Soc 2006;3:285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song JW, Hong SB, Lim CM, Koh Y, Kim DS. Acute exacerbation of idiopathic pulmonary fibrosis: incidence, risk factors, and outcome. Eur Respir J 2011;37:356–363. [DOI] [PubMed] [Google Scholar]
- 5.Collard HR, Moore BB, Flaherty KR, Brown KK, Kaner RJ, King TE, Jr Lasky JA, Loyd JE, Noth I, Olman MA, et al., Idiopathic Pulmonary Fibrosis Clinical Research Network Investigators. Acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2007;176:636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim DS, Park JH, Park BK, Lee JS, Nicholson AG, Colby T,. Acute exacerbation of idiopathic pulmonary fibrosis: frequency and clinical features. Eur Respir J 2006;27:143–150. [DOI] [PubMed] [Google Scholar]
- 7.Collard HR, Calfee CS, Wolters PJ, Song JW, Hong SB, Brady S, Ishizaka A, Jones KD, King TE, Jr Matthay MA, et al. Plasma biomarker profiles in acute exacerbation of idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 2010;299:L3–L7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azuma A, Nukiwa T, Tsuboi E, Suga M, Abe S, Nakata K, Taguchi Y, Nagai S, Itoh H, Ohi M, et al. Double-blind, placebo-controlled trial of pirfenidone in patients with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2005;171:1040–1047. [DOI] [PubMed] [Google Scholar]
- 9.Kubo H, Nakayama K, Yanai M, Suzuki T, Yamaya M, Watanabe M, Sasaki H. Anticoagulant therapy for idiopathic pulmonary fibrosis. Chest 2005;128:1475–1482. [DOI] [PubMed] [Google Scholar]
- 10.Taniguchi H, Ebina M, Kondoh Y, Ogura T, Azuma A, Suga M, Taguchi Y, Takahashi H, Nakata K, Sato A, et al., Pirfenidone Clinical Study Group in Japan. Pirfenidone in idiopathic pulmonary fibrosis. Eur Respir J 2010;35:821–829. [DOI] [PubMed] [Google Scholar]
- 11.Huie TJ, Olson AL, Cosgrove GP, Janssen WJ, Lara AR, Lynch DA, Groshong SD, Moss M, Schwarz MI, Brown KK, et al. A detailed evaluation of acute respiratory decline in patients with fibrotic lung disease: aetiology and outcomes. Respirology 2010;15:873–875. [DOI] [PubMed] [Google Scholar]
- 12.Konishi K, Gibson KF, Lindell KO, Richards TJ, Zhang Y, Dhir R, Bisceglia M, Gilbert S, Yousem SA, Song JW, et al. Gene expression profiles of acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2009;180:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med 2000;342:1334–1349. [DOI] [PubMed] [Google Scholar]
- 14.Bernard GR. Acute respiratory distress syndrome: a historical perspective. Am J Respir Crit Care Med 2005;172:798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam WY, Yeung AC, Tang JW, Ip M, Chan EW, Hui M, Chan PK. Rapid multiplex nested PCR for detection of respiratory viruses. J Clin Microbiol 2007;45:3631–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aurelius E, Johansson B, Skoldenberg B, Staland A, Forsgren M. Rapid diagnosis of herpes simplex encephalitis by nested polymerase chain reaction assay of cerebrospinal fluid. Lancet 1991;337:189–192. [DOI] [PubMed] [Google Scholar]
- 17.Ikuta K, Saiga K, Deguchi M, Sairenji T. Epstein-Barr virus DNA is detected in peripheral blood mononuclear cells of EBV-seronegative infants with infectious mononucleosis-like symptoms. Virus Genes 2003;26:165–173. [DOI] [PubMed] [Google Scholar]
- 18.Okamura A, Yoshioka M, Kubota M, Kikuta H, Ishiko H, Kobayashi K. Detection of a novel DNA virus (TTV) sequence in peripheral blood mononuclear cells. J Med Virol 1999;58:174–177. [DOI] [PubMed] [Google Scholar]
- 19.Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey HA, Ganem D, DeRisi JL. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci USA 2002;99:15687–15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998;95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urisman A, Fischer KF, Chiu CY, Kistler AL, Beck S, Wang D, DeRisi JL. E-Predict: a computational strategy for species identification based on observed DNA microarray hybridization patterns. Genome Biol 2005;6:R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hino S, Miyata H. Torque teno virus (TTV): current status. Rev Med Virol 2007;17:45–57. [DOI] [PubMed] [Google Scholar]
- 23.Mushahwar IK, Erker JC, Muerhoff AS, Leary TP, Simons JN, Birkenmeyer LG, Chalmers ML, Pilot-Matias TJ, Dexai SM. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc Natl Acad Sci USA 1999;96:3177–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Handa A, Dickstein B, Young NS, Brown KE. Prevalence of the newly described human circovirus, TTV, in United States blood donors. Transfusion 2000;40:245–251. [DOI] [PubMed] [Google Scholar]
- 25.Okamoto H, Takahashi M, Nishizawa T, Ukita M, Fukuda M, Tsuda F, Miyakawa Y, Mayumi M. Marked genomic heterogeneity and frequent mixed infection of TT virus demonstrated by PCR with primers from coding and noncoding regions. Virology 1999;259:428–436. [DOI] [PubMed] [Google Scholar]
- 26.Maggi F, Pifferi M, Fornai C, Andreoli E, Tempestini E, Vatteroni M, Presciuttini S, Marchi S, Pietrobelli A, Boner A, et al. TT virus in the nasal secretions of children with acute respiratory diseases: relations to viremia and disease severity. J Virol 2003;77:2418–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bando M, Ohno S, Oshikawa K, Takahashi M, Okamoto H, Sugiyama Y. Infection of TT virus in patients with idiopathic pulmonary fibrosis. Respir Med 2001;95:935–942. [DOI] [PubMed] [Google Scholar]
- 28.Mariscal LF, Lopez-Alcorocho JM, Rodriguez-Inigo E, Ortiz-Movilla N, de Lucas S, Bartolome J, Carreno V. TT virus replicates in stimulated but not in nonstimulated peripheral blood mononuclear cells. Virology 2002;301:121–129. [DOI] [PubMed] [Google Scholar]
- 29.Friaza V, la Horra C, Rodriguez-Dominguez MJ, Martin-Juan J, Canton R, Calderon EJ, Del Campo R. Metagenomic analysis of bronchoalveolar lavage samples from patients with idiopathic interstitial pneumonia and its antagonic relation with Pneumocystis jirovecii colonization. J Microbiol Methods 2010;82:98–101. [DOI] [PubMed] [Google Scholar]
- 30.Vannella KM, Moore BB. Viruses as co-factors for the initiation or exacerbation of lung fibrosis. Fibrogenesis Tissue Repair 2008;1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog 2008;4:e1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science 2006;312:1355–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.