Abstract
Accelerated atherosclerosis is the leading cause of death in type 1 diabetes, but the mechanism of type 1 diabetes-accelerated atherosclerosis is not well understood, in part due to the lack of a good animal model for the long-term studies required. In an attempt to create a model for studying diabetic macrovascular disease, we have generated type 1 diabetic Akita mice lacking the low density lipoprotein receptor (Ins2AkitaLdlr−/−). Ins2AkitaLdlr−/− mice were severely hyperglycemic with impaired glucose tolerance. Compared with Ldlr−/− mice, 20-week-old Ins2AkitaLdlr−/− mice fed a 0.02% cholesterol AIN76a diet showed increased plasma triglyceride and cholesterol levels, and increased aortic root cross-sectional atherosclerotic lesion area [224% (P < 0.001) in males and 30% (P < 0.05) in females]. Microarray and quantitative PCR analyses of livers from Ins2AkitaLdlr−/− mice revealed altered expression of lipid homeostatic genes, including sterol-regulatory element binding protein (Srebp)1, liver X receptor (Lxr)α, Abca1, Cyp7b1, Cyp27a1, and Lpl, along with increased expression of pro-inflammatory cytokine genes, including interleukin (Il)1α, Il1β, Il2, tumor necrosis factor (Tnf)α, and Mcp1. Immunofluorescence staining showed that the expression levels of Mcp1, Tnfα, and Il1β were also increased in the atherosclerotic lesions and artery walls of Ins2AkitaLdlr−/− mice. Thus, the Ins2AkitaLdlr−/− mouse appears to be a promising model for mechanistic studies of type 1 diabetes-accelerated atherosclerosis.
Keywords: type 1 diabetes, hyperglycemia, hyperlipidemia, Akita mouse, Ldlr knockout mouse
Accelerated atherosclerosis is the critical manifestation of macrovascular disease in both type 1 and 2 diabetics and the major etiology of morbidity and mortality in these individuals (1, 2). In 2000, the global excess mortality attributable to diabetes mellitus was estimated at 2.9 million deaths (3), with 80% the result of major cardiovascular events (4). Clinical studies of diabetic cardiovascular disease have mainly focused on type 2 diabetes. However, despite its younger age of onset, type 1 diabetes is also associated with a significantly increased risk of cardiovascular diseases, with an age-adjusted risk greater than 10 times the general population, which even exceeds that of type 2 diabetes (2, 5–8). The mechanism of type 1 diabetes-accelerated atherosclerosis is not well studied, and a major reason has been the lack of a good animal model (9). The most commonly used animal model of type 1 diabetes is the streptozotocin (STZ)-treated mouse. In this model, injection of STZ ablates pancreatic β-cells. Deficiencies of this model include considerable mouse-to-mouse variation; age dependency of pancreatic β-cell destruction; toxicity to other cells and tissues, including DNA damage and induction of carcinogenesis; instability of the diabetes phenotype due to pancreatic islet β-cell regeneration; and the difficulty of inducing diabetes in females (10–12). The variability and instability of STZ-treated mouse model make it especially difficult to perform the long-term experiments required for atherosclerosis studies.
The Akita mouse (Ins2Akita) carries a single copy of a dominant mutation in the Ins2 gene (Cys96Tyr). This mutation disrupts intramolecular disulfide bond formation causing improper folding of proinsulin. Proinsulin accumulates intracellularly and, by engorging the endoplasmic reticulum (ER) and triggering the ER stress response, leads to apoptosis of pancreatic β-cells (13). Despite the co-expression of a normal insulin gene allele, by 3 to 4 weeks of age, Ins2Akita mice exhibit hypoinsulinemia, hyperglycemia, polydipsia, and polyuria in the absence of obesity (14–16). On the C57BL/6J background on a chow diet, Ins2Akita mice have persistent hyperglycemia with fasting blood glucose levels of greater than 400 mg/dl (14–16). The Ins2Akita model has been used to study diabetic microvascular complications, such as retinopathy, neuropathy, and nephropathy (9). However, macrovascular diabetic complications, such as atherosclerotic cardiovascular disease, have not been examined.
In our initial studies we found Ins2Akita mice on the C57BL/6J background fed the 0.02% cholesterol AIN76a diet (low cholesterol, low fat) from weaning to 20 weeks of age averaged total cholesterol levels of ∼112 mg/dl and triglycerides of 52 mg/dl and had no signs of atherosclerotic lesions at the aortic root (data not shown). The latter was not surprising since mice are normally atherosclerosis-resistant and much higher lipid levels are required to foster lesion development (17). Therefore, to enable studies of the effect of hyperglycemia on atherosclerosis, we bred the Ins2Akita trait onto the atherosclerosis-susceptible Ldlr−/− background and compared Ins2AkitaLdlr−/− to Ldlr−/− controls. We chose the Ldlr−/− background over the apoE−/− background because its plasma lipid profile more closely resembles that of most atherosclerosis-prone humans. We also chose the 0.02% cholesterol AIN76a diet to avoid the additional stresses of obesity and insulin resistance apart from hyperglycemia present in other models (18). On the 0.02% cholesterol AIN76a diet at 20 weeks of age, Ins2AkitaLdlr−/− mice had higher levels of total, VLDL, and LDL cholesterol and triglycerides, as well as increased aortic root cross-sectional lesion areas. Liver gene expression revealed alteration in lipid homeostasis genes and increased expression of pro-inflammatory cytokine genes. Immunofluorescence staining showed that the expression levels of several pro-inflammatory cytokines were also increased in the atherosclerotic lesions and artery walls of Ins2AkitaLdlr−/− mice. These data suggest the Ins2AkitaLdlr−/− mouse is a promising model for mechanistic studies of accelerated macrovascular disease associated with type 1 diabetes.
MATERIAL AND METHODS
Animals
Ldlr−/− mice (B6.129S7-Ldlrtm1Her/J, stock no. 002207) and heterozygous Ins2Akita mice (C57BL/6-Ins2Akita/J, stock no. 003548) both on the C57BL/6J background were obtained from the Jackson Laboratory and subsequently crossed to generate Ldlr−/− and Ins2AkitaLdlr−/− mice. All animals were housed in the Rockefeller University Laboratory Animal Research Center under a protocol approved by the Institutional Animal Care and Use Committee in a specific pathogen-free environment in rooms with a light-dark cycle. Experimental mice were weaned and separated by gender at 28 days of age and fed a semisynthetic, modified AIN76 diet containing 0.02% cholesterol (Research Diets, New Brunswick, NJ) for 16 weeks until euthanasia at 20 weeks of age.
Intraperitoneal glucose tolerance test (IPGTT)
Three days prior to sacrifice mice were fasted for 6 h following the dark (feeding) cycle with free access to water. After fasting, mice were injected intraperitoneally with 2 mg glucose/g body weight. Plasma glucose values were obtained from venous blood from a small tail clip at 0, 15, 30, 60, and 120 min using the Ascensia Elite XL (Bayer HealthCare, Tarrytown, NY) handheld blood glucometer.
Tissue preparation
On the day of euthanasia, mice were fasted for 6 h following the dark (feeding) cycle. Immediately prior to euthanasia, the fasting plasma glucose was measured, and then mice were anesthetized by intraperitoneal injection with sodium pentobarbital (Henry Schein, Melville, NY). Mice were exsanguinated by left-ventricular puncture, and blood was collected into EDTA-containing syringes. Plasma was prepared by spinning at 16,000 g for 10 min. The circulation was flushed with PBS, and the heart was removed and stored frozen in Tissue-Tek OCT compound as we described before (19). Liver and other tissues were collected and stored in RNAlater solution (Life Technologies, Carlsbad, CA).
Blood analysis
Total cholesterol concentrations were determined enzymatically by a colorimetric method (Roche, Indianapolis, IN). Lipoproteins fractions were isolated by spinning 60 μl of plasma in a TL-100 ultracentrifuge (Beckman Coulter, Brea, CA) at its own density (1.006 g/ml) at 70,000 RPM for 3 h to harvest the supernatant and after adjusting the infranatant with solid KBr to a density of 1.063 g/ml, then spinning it for 70,000 RPM for 18 h to harvest the supernatant. The cholesterol content of each supernatant and the final infranatant were measured and taken to be VLDL (<1.006 g/ml), LDL (1.006 ≤ d ≤ 1.063 g/ml), and HDL (d >1.063 g/ml) cholesterol. Cholesterol concentrations in all three fractions were then determined enzymatically by a colorimetric method (Roche, Indianapolis, IN). Plasma triglyceride levels were determined enzymatically in the original plasma sample. Plasma insulin was measured using a rat/mouse insulin ELISA kit (EZRMI-13K, Linco Research, St. Charles, MO).
Quantification of atherosclerosis
To quantify atherosclerosis at the aortic root, OCT-embedded hearts were sectioned and stained with oil red O as described (19). The heart was oriented so that the three valves of the aortic root were in the same plane, and 12 μm sections were saved onto glass slides. Sections were stained with oil red O. The lesion area was quantified in every fourth section, and the average was reported for five measurements.
RNA isolation and quantitative real-time PCR analysis
Total RNA was isolated from the livers of male Ins2AkitaLdlr−/− and Ldlr−/− mice using the RNeasy mini kit (Qiagen USA, Valencia, CA) according to the manufacturer-supplied protocol. Quantitative real-time PCR (QPCR) was performed using gene-specific primers and the SYBR green PCR kit (Life Technologies, Carlsbad, CA) in an ABI 7900 system (Life Technologies) as described before (20). All samples were quantified using the comparative Ct method for relative quantification of gene expression, normalized to GAPDH (21). The primer sets utilized in this study are shown in supplementary Table I.
Microarray and gene set analyses
Total RNA samples were prepared from the livers of Ins2AkitaLdlr−/− and Ldlr−/− mice using the RNeasy mini kit (Qiagen USA, Valencia, CA). cRNA was prepared and hybridized to the MouseRef-8 v2.0 Illumina Genome-Wide Expression BeadChip Array (Illumina, San Diego, CA). The hybridized BeadChip was washed and labeled with streptavidin-Cy3 and scanned with Illumina BeadScan by the Rockefeller University Genomics Resource Center according to the manufacturer's protocol (Illumina). BeadStudio 3.2 software was used for background correction of the imported scanned image. Gene expression profiles were analyzed using the GeneSpring GX 10.0 software (Agilent Technologies, Santa Clara, CA).
In addition to microarray analysis, gene set enrichment analysis (GSEA) was conducted using the GSEA desktop application (http://www.broad.mit.edu/GSEA) (Broad Institute, Cambridge, MA) (22, 23). This application ranks the expression of members of a gene set in Ins2AkitaLdlr−/− versus Ldlr−/−. To determine whether a gene set is significantly enriched, GSEA identifies significant changes in gene sets by assigning each a calculated enrichment score (ES). For each gene set, ES was calculated by using weighted Kolmogorov-Smirnov statistics to measure the proximity of the gene set to the top of the Ins2AkitaLdlr−/−-effect ranked list. A highly positive ES indicated that the gene set or pathway was collectively upregulated by the Akita mutation intervention, while a highly negative ES indicated downregulation. To account for differences in numbers of genes in each set, the normalized ES (NES) was used to compare analysis results across gene sets. Gene set analysis was conducted using the canonical pathway (C2) and the transcription factor target (C3) databases from the molecular signature database (MSigDb) available at the Broad Institute, MIT, website. Positively or negatively enriched gene sets were considered significant with a P value ≤ 0.05 and a false discovery rate (FDR) ≤ 25%.
On the basis of the difference in the expression of each gene between Ins2AkitaLdlr−/− and Ldlr−/− mice, global functions, networks, and canonical pathways were also analyzed by Ingenuity Pathway Analysis (IPA) (http://www.ingenuity.com) (Ingenuity Systems, Redwood City, CA). IPA is web-based software designed to organize biological information in a way that allows one to gain a high-level overview of the general biology associated with microarray data (24–26). In this study, the gene list from GeneSpring GX 10.0 was generated with gene identifiers, and the difference in gene expression was entered into IPA. The IPA Canonical Pathway Analysis tool was used to identify the signaling pathways associated with the database. The functional analysis identified the molecular and cellular function that was most significant to the data set as a whole and generated functional interpretation of microarray data (26).
Immunohistochemistry
Immunohistochemistry were performed on 12 μm sections of aortic roots freshly embedded in OCT. Sections were first fixed in 100% ice-cold acetone for 15 min and then washed with PBS for 20 min. Sections were permeabilized with PBS + 0.1% Triton ×100 (PBST) for 10 min. Nonspecific binding was reduced by incubating slides in 10% rabbit sera diluted in PBST for 20 min at room temperature. Sections were then incubated with rabbit antibodies against mouse Mcp1 (1:50; Abcam, Cambridge, MA), tumor necrosis factor (Tnf)α (1:100; Abcam,) or interleukin (Il)1β (1:50; Abcam) at 4°C for 12-15 h. Sections were rinsed with PBS and incubated with Alexa 594-labeled goat anti-rabbit secondary antibodies (1:500; Life Technologies). The nuclei were stained by mounting the slides with DAPI medium (Vector Laboratories, Burlingame, CA). Images were acquired with a Nikon fluorescence microscopy (Nikon, Melville, NY).
Statistical analysis
All data are expressed as mean ± SD unless indicated otherwise. Statistically significant differences between two groups were analyzed by t-test for data normally distributed and by the Mann-Whitney test for data not normally distributed using Prism version 4.0 (GraphPad Prism Software, San Diego, CA). Grubbs's test was also performed to detect significant outliers (P < 0.05).
RESULTS
Ins2AkitaLdlr−/− mice demonstrate an insulin-deficient diabetic phenotype
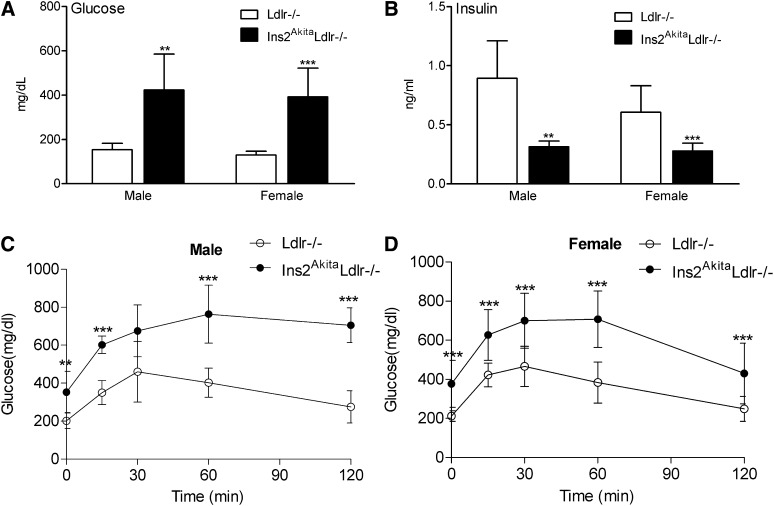
At 20 weeks of age, Ins2AkitaLdlr−/− mice had significantly decreased body weight in males, but female body weight remained the same (supplementary Fig. IA). The Akita mutation also increased mortality of Ldlr−/− mice in both males and females (supplementary Fig. IB). Ins2AkitaLdlr−/− mice were severely hyperglycemic with fasting blood glucose levels of 423 ± 162 mg/dl in males and 392 ± 130 mg/dl in females, compared with Ldlr−/− mice with glucose levels of 153 ± 29 mg/dl in males and 129 ± 17 mg/dl in females (Fig. 1A). In addition, Ins2AkitaLdlr−/− mice had low fasting plasma insulin levels of 0.31 ± 0.05 ng/ml in males and 0.28 ± 0.07 ng/ml in females, compared with Ldlr−/− mice with insulin levels of 0.89 ± 0.32 ng/ml in males and 0.61 ± 0.22 ng/ml in females (Fig. 1B). Finally, Ins2AkitaLdlr−/− mice of both genders showed impaired glucose tolerance compared with Ldlr−/− mice as measured by the IPGTT (Fig. 1C, D).
Fig.1.
Ins2AkitaLdlr−/− mice demonstrate insulin-deficient diabetic phenotypes. Ins2AkitaLdlr−/− mice were bred to Ldlr−/− mice to generate Ins2AkitaLdlr−/− and Ldlr−/− littermates. Four-week-old Ins2AkitaLdlr−/− and Ldlr−/− mice were fed a semisynthetic low-cholesterol AIN76a diet (0.02% cholesterol) for 16 weeks. (A) Plasma insulin and (B) glucose levels were measured by standard methods. (C, D) Intraperitoneal glucose tolerance tests (IPGTT) were performed after a 6 h fast on male (C) and female (D) Ins2AkitaLdlr−/− and Ldlr−/− mice (n = 8-14 per group, **P < 0.01 and ***P < 0.001).
Ins2AkitaLdlr−/− mice show increased cholesterol and triglyceride levels due to increased VLDL and LDL
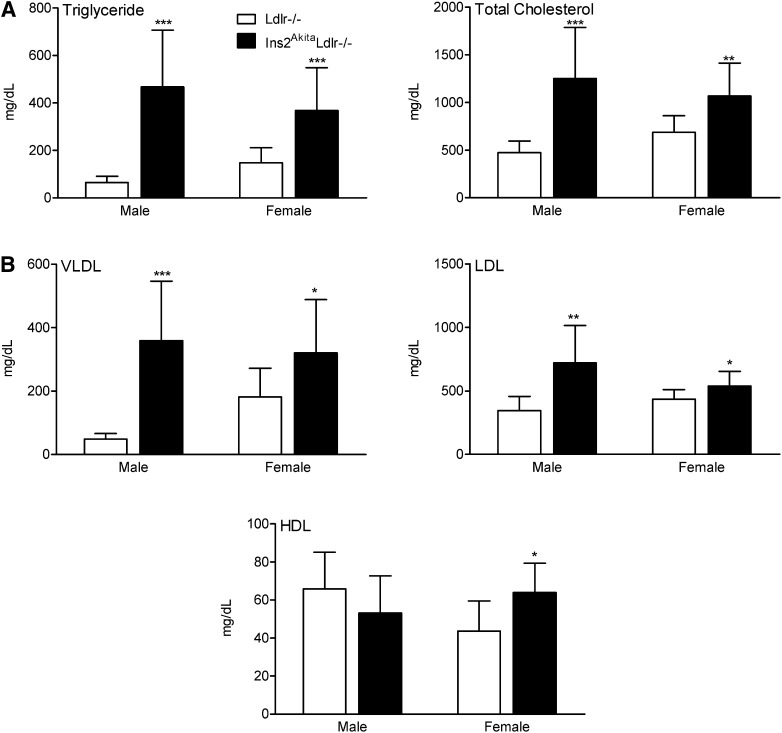
Also at 20 weeks of age, Ins2AkitaLdlr−/− mice had increased fasting cholesterol levels of 1,252 ± 536 mg/dl in males and 1,067 ± 347 mg/dl in females, compared with Ldlr−/− mice with cholesterol levels of 473 ± 124 mg/dl in males and 688 ± 173 mg/dl in females. Similarly, Ins2AkitaLdlr−/− mice had increased fasting triglyceride levels of 467 ± 239 mg/dl in males and 368 ± 181 mg/dl in females, compared with Ldlr−/− mice with triglyceride levels of 65 ± 26 mg/dl in males and 148 ± 64 mg/dl in females (Fig. 2A). Comparison of lipoprotein cholesterol levels between Ins2AkitaLdlr−/− and Ldlr−/− mice revealed VLDL cholesterol levels increased 7-fold in males and 1.8-fold in females; LDL cholesterol levels increased 2-fold in males but only 24% in females; and HDL cholesterol levels were unchanged in males but slightly and significantly increased in females (Fig. 2B).
Fig.2.
Akita mutation induces hyperlipidemia in Ldlr−/− mice. Four-week-old male and female Ldlr−/− and Ins2AkitaLdlr−/− mice were fed a semisynthetic 0.02% cholesterol-AIN76a diet for 16 weeks. The plasma levels of (A) triglyceride and total cholesterol and (B) lipoprotein fraction cholesterols (VLDL, LDL, and HDL) were measured in male and female Ldlr−/− and Ins2AkitaLdlr−/− mice (n = 8-14 per group, *P < 0.05, **P < 0.01, and ***P < 0.001).
Atherosclerosis is accelerated in diabetic hyperlipidemic Ins2AkitaLdlr−/− mice
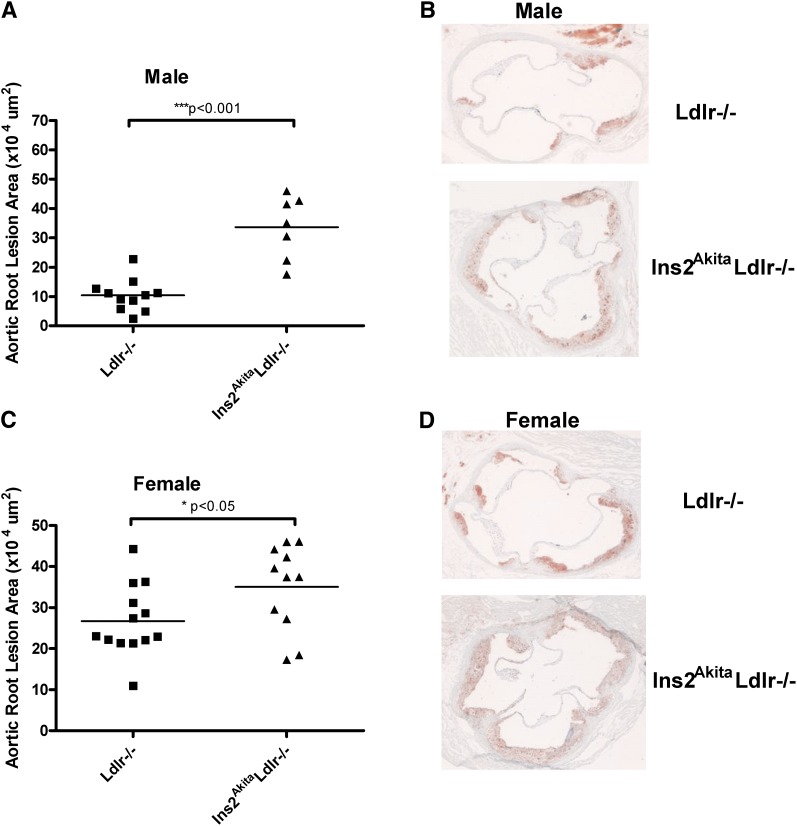
The Ins2AkitaLdlr−/− and Ldlr−/− mice were sacrificed at 20 weeks of age, and aortic root cross-sectional lesion areas were determined (Fig. 3). Compared with Ldlr−/− mice, Ins2AkitaLdlr−/− mice had increased cross-sectional lesion areas of 224% in males and 30% in females (P < 0.001and P < 0.05, respectively).
Fig.3.
Atherosclerosis is accelerated in diabetic Ins2AkitaLdlr−/− mice. Four-week-old male and female Ldlr−/− and Ins2AkitaLdlr−/− mice were fed a semisynthetic 0.02% cholesterol AIN76a diet for 16 weeks. (A, C) Quantitative analysis of the lesion size in the aortic root of male and female Ldlr−/− and Ins2AkitaLdlr−/− mice (n = 7-13 per group, **P < 0.001 and *P < 0.05). (B, D) Representative oil red O-stained sections of atherosclerotic lesion area in the aortic root of male and female Ldlr−/− and Ins2AkitaLdlr−/− mice.
Akita mutation affects hepatic genes involved in lipid metabolism and inflammation
To elucidate possible molecular mechanisms through which the Akita mutation might augment hyperlipidemia and atherosclerosis in Ldlr−/− mice, gene expression microarray studies were performed comparing livers from Ins2AkitaLdlr−/− and Ldlr−/− mice. Genes with more than 1.5-fold expression changes are listed in supplementary Table II.
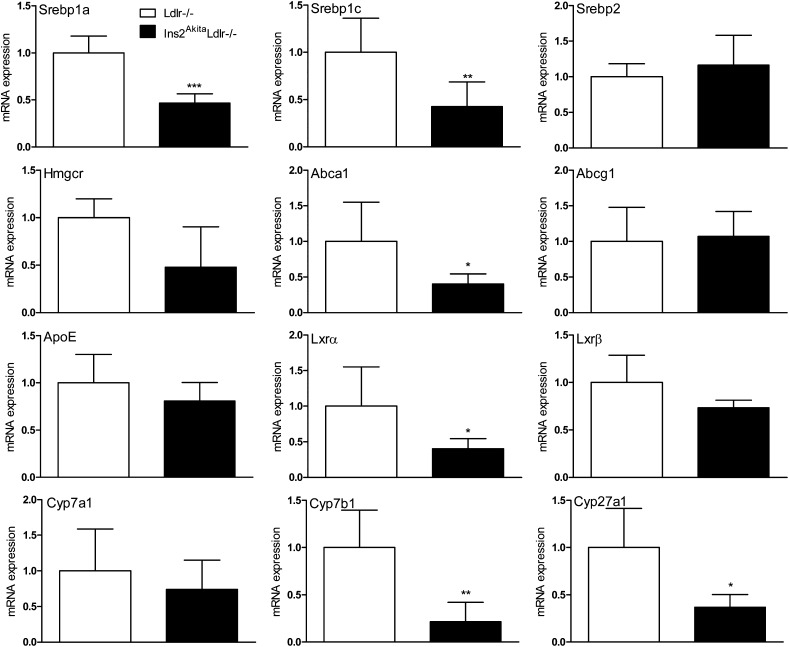
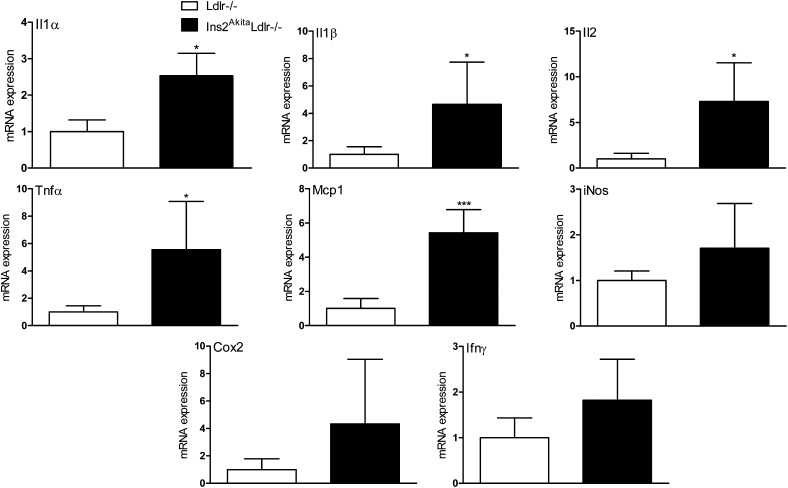
To confirm the results of the microarray analysis and to investigate other genes involved in lipid homeostasis that were not revealed by the microarray analysis, hepatic gene expression in Ins2AkitaLdlr−/− and Ldlr−/− mice were evaluated by QPCR (Fig. 4). Expression of several key genes involved in lipid homeostasis, including sterol-regulatory element binding protein (Srebp)1c, liver X receptor (Lxr)α, Abca1, Lpl, Cyp7b1, and Cyp27a1, were significantly altered in Ins2AkitaLdlr−/−. Furthermore, the expression of pro-inflammatory cytokine genes, including Il1α, Il1β, Il2, Tnfα, and Mcp1, were significantly increased in Ins2AkitaLdlr−/− mice (Fig. 5).
Fig.4.
Hepatic genes involved in lipid homeostasis are significantly altered in Ins2AkitaLdlr−/− mice. Total RNA was isolated from the livers of 20-week-old male Ldlr−/− and Ins2AkitaLdlr−/− mice. The expression levels of hepatic genes involved in lipid homeostasis and other atherosclerosis-related genes were measured by quantitative PCR (n = 5 per group, *P < 0.05, **P < 0.01, and ***P < 0.001).
Fig.5.
Akita mutation increases mRNA levels of pro-inflammatory cytokine genes in the livers of Ins2AkitaLdlr−/− mice. Total RNA was isolated from the livers of 20-week-old male Ldlr−/− and Ins2AkitaLdlr−/− mice. The expression levels of pro-inflammatory cytokine genes were measured by quantitative PCR (n = 5 per group, *P < 0.05 and ***P < 0.001).
GSEA and IPA of genes differentially expressed between Ins2AkitaLdlr−/− and Ldlr−/− mice
Microarray results were further analyzed by GSEA and IPA to identify gene sets and pathways affected by breeding the Ins2Akita mutation onto the Ldlr−/− background. GSEA is a computational method that determines whether an a priori defined set of genes shows statistically significance between two biological states (22). Comparing Ins2AkitaLdlr−/− to Ldlr−/− mice, GSEA analysis identified seven downregulated and seven upregulated gene sets (Table 1). The downregulated gene sets in Ins2AkitaLdlr−/− mice include pathways involved in complement and coagulation cascades, soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) interactions in vesicular transport, biosynthesis of steroids, ether lipid metabolism, 1 and 2 methylnaphthalene degradation, linoleic acid metabolism, and bile acid biosynthesis. The upregulated gene sets include pathways involved in carbon fixation, glutathione metabolism, oxidative phosphorylation, pentose phosphate pathway, metabolism of xenobiotics by cytochrome p450, citrate cycle, and hematopoietic cell lineage.
TABLE 1.
GSEA Analysis of Altered Hepatic Gene Networks in Ins2 LDLR Mice
| Rank | Gene Set (Pathway) | Genes Within Gene Set | NES | P | FDR |
| Negatively Enriched Gene Sets | |||||
| 1 | HSA04610_COMPLEMET_AND_COAGULATION CASCADES | Cfh, Serping1, Cfi, Plg, C4b, Serpina5, C3ar1, Tfpi, Cpb2, Serpind1, Proc, F2r, F12, Masp1, C1s, Masp2, C9, C6, C8a, F8, F7, F2,Cfd, C8b, Klkb1, F11 | −2.32 | <0.001 | 0.005 |
| 2 | HSA04130_SNARE_INTERACTIONS_IN_VESICULAR_TRANSPORT | Snap23, Bnip1, Sec22b, Stx3, Gosr2, Stx18, Vamp2, Ykt6, Vamp5, Stx7, Bet1 | −2.14 | <0.001 | 0.003 |
| 3 | HSA00100_BIOSYNTHESIS_OF_STEROIDS | Dhcr7,Vkorc1, Mvd, Hsd17b7, Pmvk, Sqle, Idi1, Sc4mol,Nsdhl,Fdps, Ggcx, Ebp | −2.13 | <0.001 | 0.003 |
| 4 | HSA00565_ETHER_LIPID_METABOLISM | Agps, Enpp6, Pla2g10, Pla2g12a, Chpt1, Ppap2c, Pafah2, Ppap2a,Enpp2, Agpat4, Pla2g6, Agpat2, Ppap2b, Pla2g12b | −2.04 | 0.005 | 0.004 |
| 5 | HSA00624_1_AND_2_METHYLNAPHTHALENE DEGRADATION | Dhrs2, Nat6, Dhrs2, Acad9, Acad8, Dhrs1, Adh4 | −1.57 | 0.027 | 0.117 |
| 6 | HSA00591_LINOLEIC_ACID_METABOLISM | Pla2g10, Pla2g12a, Rdh11, Rdh14, Cyp1a2, Hsd3b7, Pla2g6, Pla2g12b | −1.53 | 0.020 | 0.131 |
| 7 | HSA00120_BILE_ACID_BIOSYNTHESIS | Acaa2, Cyp7a1, Acad9, Baat, Aldh2, Rdh11, Rdh14, Srd5a2. Soat2, Cyp27a1, Hsdeb7, Srd5a1, Adh4 | −1.53 | 0.010 | 0.115 |
| Positively Enriched Gene Sets | |||||
| 1 | HSA00710_CARBON_FIXATION | Mdh2, Tktl1, Fbp2, Aldoa, Tpi1, Got2, Aldob | 1.88 | <0.001 | 0.015 |
| 2 | HSA00480_GLUTATHIONE_METABOLISM | Gsta1, Idh2, Gpx3, Gsta2, Mgst3, Gpx4, Gstm3, Gstm1, Gstm2, Gpx7, Gstk1, Gstt2 | 1.74 | 0.002 | 0.060 |
| 3 | HSA00190_OXIDATIVE_PHOSPHORYLATION | Atp6v1b2, Sdhb, Ndufa5, Ndufv2, Ndufb4, Atp5a1, Ndufa7, Ndufs4, Ndufs8, Atp6v0b, Atp5 h, Uqcrc1, Cox10, Ppa1, Ndufb5, Ndufb7, Ndufv1, Cox7b, Ndufa10, Cox5b, Cox7a2, Cox6a1, Cox8a, Ndufb9, Cox4i1, Atp5e, Cox7c, Atp5j2, Ndub10, Ndufs6, Atp5g1, Ndufs7, Ndufb8, Cox6c, Ndufa2 | 1.70 | <0.001 | 0.074 |
| 4 | HSA00030_PENTOSE_PHOSPHATE_PATHWAY | Tktl1, Prps1, H6pd, Fbp2, Aldoa, Aldob, Pgls | 1.62 | 0.008 | 0.149 |
| 5 | HSA00980_METABOLISM_OF_XENOBIOTICS_BY_CYTOCHROME_P450 | Gsta1, Aldh3b1, Gsta2, Mgst3, Ephx1, Gstm3, Adh7, Gstm1, Gstm2, Aldh1a3, Gstk1, Gstt2, Cyp1b1 | 1.60 | 0.007 | 0.140 |
| 6 | HSA00020_CITRATE_CYCLE | Idh2, Mdh2, Sdh8, Idh3b, Aco2, Ogdh | 1.60 | 0.015 | 0.120 |
| 7 | HSA04640_HEMATOPOIETIC_CELL_LINEAGE | Il3ra, Cd8b, Epo, Cd44, Tnf, Cd36, Cd22, Ilr5a, Il7, Il7r, Il2ra, Gp1ba, Itgam, Itga2b, Cd34, Csf3r, Cd3e, Csf3, Cd5, Il1r2, Cd7, Epor, Gp1bb | 1.54 | 0.007 | 0.215 |
In addition to GSEA, the microarray results were analyzed by IPA Canonical Pathway Analysis. The genes from microarray results were overlaid onto the global signaling pathways developed from information contained in the Ingenuity Pathways Knowledge Base (26). The IPA revealed differential expression of genes in lipid and carbohydrate metabolism and cell death (Table 2). Both analyses indentified significantly changed gene sets or pathways related to lipid homeostasis in the livers of Ins2AkitaLdlr−/− mice.
TABLE 2.
Ingenuity Pathway Analysis of Altered Hepatic Gene Expression in Ins2 LDLR Mice
| Network | P | Fold Change | Gene | UniGene Number | Description |
| Lipid metabolism | 7.07E-04 - 2.79E02 | 2.02 | Abcc3 | Mm.23942 | ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| 3.15 | Lpl | Mm.1514 | Lipoprotein lipase | ||
| 5.12 | Por | Mm.3863 | P450 (cytochrome) oxidoreductase | ||
| −2.05 | Gck | Mm.220358 | Glucokinase | ||
| −2.43 | Gnas | Mm.125770 | Guanine nucleotide binding protein, α stimulating complex locus | ||
| −2.03 | Idh1 | Mm.9925 | Isocitrate dehydrogenase 1 (NADP+), soluble | ||
| −2.49 | Insig1 | Mm.30221 | Insulin induced gene 1 | ||
| −2.62 | Insig2 | Mm.27136 | Insulin induced gene 2 | ||
| −2.21 | Scp2 | Mm.379011 | Sterol carrier protein 2 | ||
| −2.03 | Srebf1 | Mm.278701 | Sterol regulatory element binding transcription factor 1 | ||
| Carbohydrate metabolism | 1.79E-04 - 2.67E-02 | 2.18 | H6pd | Mm.22183 | Hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| 2.29 | Irs2 | Mm.407207 | Insulin receptor substrate 2 | ||
| 3.15 | Lpl | Mm.1514 | Lipoprotein lipase | ||
| 2.52 | Stat3 | Mm.249934 | Signal transducer and activator of transcription 3 (acute-phase response factor) | ||
| −2.24 | Avpr1a | Mm.4351 | Arginine vasopressin receptor 1A | ||
| −2.05 | Gck | Mm.220358 | Glucokinase | ||
| −2.43 | Gnas | Mm.125770 | Guanine nucleotide binding protein, α stimulating complex locus | ||
| −2.61 | Ppp1r3b | Mm.247126 | Protein phosphatase 1, regulatory (inhibitor) subunit 3B | ||
| −2.30 | Ppp1r3c | Mm.24724 | Protein phosphatase 1, regulatory (inhibitor) subunit 3C | ||
| −2.21 | Scp2 | Mm.379011 | Sterol carrier protein 2 | ||
| −4.81 | Slc2a2 | Mm.18443 | Solute carrier family 2 (facilitated glucose transporter), member 2 | ||
| −2.03 | Srebf1 | Mm.278701 | Sterol regulatory element binding transcription factor 1 | ||
| Cell death | 2.91E-03 - 2.91E-03 | 2.85 | Mt1e | Mm.147226 | Metallothionein 1E |
| 2.12 | Mt1f | Mm.192991 | Metallothionein 1F | ||
| −2.54 | Cpb2 | Mm.24242 | Carboxypeptidase B2 (plasma) | ||
| −2.84 | Hspd1 | Mm.476770 | Heat shock 60 kDa protein 1 (chaperonin) |
Akita mutation increases inflammatory cytokine expression in the atherosclerotic lesions
The significantly increased mRNA levels of pro-inflammatory cytokines in the livers of Ins2AkitaLdlr−/− mice and accelerated atherosclerosis in these mice promoted us to investigate the protein content of these cytokines in the atherosclerotic lesions. Immunofluorescence staining showed that the expression levels of several key inflammatory cytokines, including Mcp1, Tnfα, and Il1β, were increased in the atherosclerotic lesions and artery walls of Ins2AkitaLdlr−/− mice (Fig. 6). Therefore, the enhanced inflammation in Ins2AkitaLdlr−/− mice may contribute, at least in part, to the increased atherosclerosis development in these mice.
Fig.6.
Akita mutation increases the pro-inflammatory cytokine levels in the atherosclerotic lesions and artery walls of Ins2AkitaLdlr−/− mice. Sections of atherosclerotic lesion areas in the aortic root of male Ldlr−/− and Ins2AkitaLdlr−/− mice were stained with rabbit antibodies against mouse Mcp1, Tnfα, or Il1β followed by fluorescein-labeled goat anti-rabbit secondary antibody (red). The nuclei were stained with DAPI (blue).
DISCUSSION
There is a major need for a good animal model to study the mechanism of type 1 diabetes-accelerated atherosclerosis. To address this need, we bred the Ins2Akita mice onto the atherosclerosis-prone Ldlr−/− background, and we showed augmented levels of VLDL and LDL cholesterol as well as increased aortic root cross-sectional lesion areas. Since the liver plays a central role in lipid metabolism and controls aspects of inflammation relevant to atherosclerosis, we also assessed the alteration of hepatic gene expression by microarray and QPCR determinations. We found the Ins2Akita mutation altered expression of hepatic lipid metabolism genes, including Srebp1, Lxrα, Abca1, Lpl, Cyp7b1, and Cyp27a1, and increased expression of pro-inflammatory cytokine genes. In further analysis, both GSEA and IPA indicated alterations in lipid and carbohydrate metabolism genes, whereas IPA alone indicated altered expression of cell-death genes.
The Srebp and Lxr transcription factor genes are known to be particularly important for regulation of lipid metabolism. In the liver of Ins2AkitaLdlr−/− mice, Srebp1a and Srebp1c mRNA levels were significantly downregulated, but Srebp2 mRNA levels remained the same. Insulin is a known direct stimulator of Srebp1 transcription (27), so a likely cause of decreased Srebp1 mRNA levels in the liver is the hypoinsulinemia of the Ins2AkitaLdlr−/− mice. However, since Srebp1 stimulates fatty acid and triglyceride synthesis, it is unlikely that decreased Srebp1 mRNA levels are the primary explanation for the Akita mutation-mediated hyperlipidemia.
Lxrα is a nuclear hormone receptor that acts as a cholesterol sensor. It responds to excess cellular cholesterol by upregulating genes involved in promoting reverse cholesterol transport, increasing cellular cholesterol export, and promoting decreased intestinal cholesterol absorption and increased hepatic cholesterol excretion (28). Breeding the Lxrα knockout trait to the apoE−/− background resulted in accelerated atherosclerosis (29, 30). Thus, the decreased Lxrα mRNA level in the livers of Ins2AkitaLdlr−/− mice likely contributed to the increase in atherosclerosis observed. It has also been reported that Ins2Akita mice have reduced levels of Lxrα mRNA in their kidneys (31). Thus the Akita mutation might result in reduced Lxrα mRNA in multiple tissues, but in no case is the mechanism known.
Cyp7b1 and Cyp27a1 are two enzymes involved in cholesterol catabolism and bile acid biosynthesis, and both were significantly decreased in Ins2AkitaLdlr−/− mice. Cyp7b1 catalyzes the 7α-hydroxylation of oxysterols, an early step in the alternative bile acid biosynthesis pathway, and Cyp7b1-deficient mice had elevated plasma and tissue levels of 25- and 27-hydroxycholesterol (32). Cyp27a1 catalyzes the oxidative cleavage of the sterol side chain in the bile acid biosynthesis pathway and 27-hydroxylation of cholesterol (33). Cyp27a1 plays an important role in cholesterol catabolism and atherosclerosis protection, and mutations in the human Cyp27a1 gene have been linked to the accumulation of cholesterol and premature atherosclerosis in cerebrotendinous xanthomatosis patients (33, 34). Therefore, the decreased expression of both of these cholesterol catabolism genes might contribute to the increased lipid levels and accelerated atherosclerosis of Ins2AkitaLdlr−/− mice.
Lpl mRNA levels were increased more than 4-fold in the liver of Ins2Akita Ldlr−/− mice. LPL is the rate-controlling enzyme involved in plasma lipoprotein triglyceride hydrolysis and is responsible for the generation of fatty acids from circulating triglyceride-rich lipoproteins (35). The Lpl gene is normally expressed mainly in adipose tissue and muscle, not in liver. We have previously generated Lpl-deficient mice (36) as well as transgenic mice expressing Lpl exclusively in muscle (37), adipose tissue (38), or liver (39). Lpl-deficient mice develop lethal hypertriglyceridemia within the first day of life, at which point they have markedly reduced intracellular lipid stores (36, 38). Interestingly, mice with liver-specific overexpression of Lpl have increased liver lipid droplets, VLDL production, and plasma ketone body levels (39), and they are also insulin resistant (40). Liver Lpl expression may increase at times of metabolic stress and shunt circulating triglycerides to the liver. The increased liver Lpl expression in Ins2AkitaLdlr−/− mice might be due to the defect in liver insulin signaling and impaired ability of insulin to suppress endogenous glucose production in these mice. The increased hepatic Lpl can increase VLDL production and might contribute to the Akita mutation-induced hyperlipidemia in Ins2AkitaLdlr−/− mice.
The precise mechanisms through which the Akita mutation modulates lipid metabolism and cholesterol levels in Ldlr−/− mice remain to be determined. Nevertheless, the altered expression levels of genes suggest that the Akita mutation can affect multiple genes involved in lipid homeostasis in Ldlr−/− mice and might provide us some insight into how type 1 diabetes induces abnormal lipid metabolism in patients.
In addition to hyperlipidemia, inflammation plays a critical role in the initiation and progression of atherosclerosis. We found that gene sets related to inflammation were also significantly altered in the livers of Ins2AkitaLdlr−/− mice, with many pro-inflammatory cytokine genes upregulated (Fig. 5). Furthermore, the protein levels of several pro-inflammatory cytokines, including Mcp1, Tnfα, and Il1β, were increased in the atherosclerotic lesions and artery walls of Ins2AkitaLdlr−/− mice. These cytokines play an important role in atherosclerosis initiation and progression. For example, monocytes are attracted by Mcp1 to lesion-prone areas; this is considered a critical step in atherosclerosis initiation (41, 42). Deficiency of Mcp1 in mice significantly reduces atherosclerotic lesions in Ldlr−/− mice (43). Tnfα and Il1β can be produced by immune cells (e.g., macrophages, T cells) within the lesion and contribute to atherosclerosis progression (44). Lxr might be important for regulation of inflammatory cytokines in Ins2AkitaLdlr−/− mice, since Lxr can inhibit immune and inflammatory responses by repressing Nf-κb-dependent induction of inflammatory gene expression, including Il-1β, Tnfα, and Mcp1 (28). Furthermore, one of the Lxr target genes, Abca1, can also modulate inflammatory response by promoting cellular cholesterol efflux and inhibiting Toll-like receptor (TLR) signaling (45). Abca1-deficient macrophages have increased lipid rafts and enhanced inflammatory responses (46). Taken together, reduced Lxr and Abca1 expression levels might, at least in part, account for the increased inflammatory cytokine gene expression in livers of Ins2AkitaLldr−/− mice.
GSEA and IPA have also identified significantly altered genes or gene networks related to other pathways, including cell death, complement and coagulation cascades, SNARE interactions in vesicular transport, methylnaphthalene degradation, and linoleic acid metabolism. It is not clear whether the identified gene networks have any effect on type 1 diabetic cardiovascular complications. It would be interesting to further investigate the effects of the Akita mutation on these gene networks and explore the link between the altered expression levels of those genes and the pathophysiological phenotype of Ins2AkitaLdlr−/− mice.
In summary, we generated a genetically induced type 1 diabetic mouse model (Ins2AkitaLdlr−/−) to study the mechanism of diabetes-accelerated atherosclerosis. Ins2AkitaLdlr−/− mice retain type 1 diabetic phenotypes, but they have increased plasma lipid levels and accelerated atherosclerosis. Multiple genes involved in lipid homeostasis and inflammation have been significantly altered in Ins2AkitaLdlr−/− mice. Our findings suggest a critical role for the diabetic milieu in the pathophysiology of dyslipidemia and atherosclerosis. The Ins2AkitaLdlr−/− mouse may represent an important model for studying the effect of type 1 diabetes on lipid homeostasis and atherosclerosis, and it may contribute to our understanding of the mechanism of diabetes-accelerated atherosclerosis.
Supplementary Material
Acknowledgments
The authors thank Katie Tsang and Helen Yu for technical assistance; Dr. Connie Zhao and the Genomics Resource Center for microarray experiments; and Ms. Natasha Levenkova for data analysis.
Footnotes
Abbreviations:
- ER
- endoplasmic reticulum
- ES
- enrichment score
- FDR
- false discovery rate
- GSEA
- gene set enrichment analysis
- IPA
- Ingenuity Pathway Analysis (software)
- IPGTT
- intraperitoneal glucose tolerance test
- Il
- interleukin
- Lxr
- liver X receptor
- NES
- normalized ES
- QPCR
- quantitative real-time PCR
- SNARE
- soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- Srebp
- sterol-regulatory element binding protein
- STZ
- streptozotocin
- Tnf
- tumor necrosis factor
This work was supported by National Institutes of Health Grants U01 HL-070524 (J.L.B.) and P30 HL-101300 (C.Z.); American Heart Association Grant 09SDG2150176 (C.Z.); and the Medical Student Program of the Rockefeller University Center for Clinical and Translational Science (funded in part by National Institutes of Health Grant UL1 RR-024143) (N.K.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figures and two tables.
REFERENCES
- 1.Calles-Escandon J., Cipolla M. 2001. Diabetes and endothelial dysfunction: a clinical perspective. Endocr. Rev. 22: 36–52. [DOI] [PubMed] [Google Scholar]
- 2.Retnakaran R., Zinman B. 2008. Type 1 diabetes, hyperglycaemia, and the heart. Lancet. 371: 1790–1799. [DOI] [PubMed] [Google Scholar]
- 3.Roglic G., Unwin N., Bennett P. H., Mathers C., Tuomilehto J., Nag S., Connolly V., King H. 2005. The burden of mortality attributable to diabetes: realistic estimates for the year 2000. Diabetes Care. 28: 2130–2135. [DOI] [PubMed] [Google Scholar]
- 4.Coccheri S. 2007. Approaches to prevention of cardiovascular complications and events in diabetes mellitus. Drugs. 67: 997–1026. [DOI] [PubMed] [Google Scholar]
- 5.Libby P., Nathan D. M., Abraham K., Brunzell J. D., Fradkin J. E., Haffner S. M., Hsueh W., Rewers M., Roberts B. T., Savage P. J., et al. 2005. Report of the National Heart, Lung, and Blood Institute-National Institute of Diabetes and Digestive and Kidney Diseases Working Group on cardiovascular complications of type 1 diabetes mellitus. Circulation. 111: 3489–3493. [DOI] [PubMed] [Google Scholar]
- 6.Krolewski A. S., Kosinski E. J., Warram J. H., Leland O. S., Busick E. J., Asmal A. C., Rand L. I., Christlieb A. R., Bradley R. F., Kahn C. R. 1987. Magnitude and determinants of coronary artery disease in juvenile-onset, insulin-dependent diabetes mellitus. Am. J. Cardiol. 59: 750–755. [DOI] [PubMed] [Google Scholar]
- 7.Orchard T. J., Costacou T., Kretowski A., Nesto R. W. 2006. Type 1 diabetes and coronary artery disease. Diabetes Care. 29: 2528–2538. [DOI] [PubMed] [Google Scholar]
- 8.Laing S. P., Swerdlow A. J., Slater S. D., Burden A. C., Morris A., Waugh N. R., Gatling W., Bingley P. J., Patterson C. C. 2003. Mortality from heart disease in a cohort of 23,000 patients with insulin-treated diabetes. Diabetologia. 46: 760–765. [DOI] [PubMed] [Google Scholar]
- 9.Hsueh W., Abel E. D., Breslow J. L., Maeda N., Davis R. C., Fisher E. A., Dansky H., McClain D. A., McIndoe R., Wassef M. K., et al. 2007. Recipes for creating animal models of diabetic cardiovascular disease. Circ. Res. 100: 1415–1427. [DOI] [PubMed] [Google Scholar]
- 10.Bugger H., Boudina S., Hu X. X., Tuinei J., Zaha V. G., Theobald H. A., Yun U. J., McQueen A. P., Wayment B., Litwin S. E., et al. 2008. Type 1 diabetic akita mouse hearts are insulin sensitive but manifest structurally abnormal mitochondria that remain coupled despite increased uncoupling protein 3. Diabetes. 57: 2924–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldberg I. J., Dansky H. M. 2006. Diabetic vascular disease: an experimental objective. Arterioscler. Thromb. Vasc. Biol. 26: 1693–1701. [DOI] [PubMed] [Google Scholar]
- 12.Bolzan A. D., Bianchi M. S. 2002. Genotoxicity of streptozotocin. Mutat. Res. 512: 121–134. [DOI] [PubMed] [Google Scholar]
- 13.Yoshinaga T., Nakatome K., Nozaki J., Naitoh M., Hoseki J., Kubota H., Nagata K., Koizumi A. 2005. Proinsulin lacking the A7-B7 disulfide bond, Ins2Akita, tends to aggregate due to the exposed hydrophobic surface. Biol. Chem. 386: 1077–1085. [DOI] [PubMed] [Google Scholar]
- 14.Wang J., Takeuchi T., Tanaka S., Kubo S. K., Kayo T., Lu D., Takata K., Koizumi A., Izumi T. 1999. A mutation in the insulin 2 gene induces diabetes with severe pancreatic beta-cell dysfunction in the Mody mouse. J. Clin. Invest. 103: 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kayo T., Koizumi A. 1998. Mapping of murine diabetogenic gene mody on chromosome 7 at D7Mit258 and its involvement in pancreatic islet and beta cell development during the perinatal period. J. Clin. Invest. 101: 2112–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izumi T., Yokota-Hashimoto H., Zhao S., Wang J., Halban P. A., Takeuchi T. 2003. Dominant negative pathogenesis by mutant proinsulin in the Akita diabetic mouse. Diabetes. 52: 409–416. [DOI] [PubMed] [Google Scholar]
- 17.Breslow J. L. 1996. Mouse models of atherosclerosis. Science. 272: 685–688. [DOI] [PubMed] [Google Scholar]
- 18.Mulvihill E. E., Assini J. M., Sutherland B. G., DiMattia A. S., Khami M., Koppes J. B., Sawyez C. G., Whitman S. C., Huff M. W. 2010. Naringenin decreases progression of atherosclerosis by improving dyslipidemia in high-fat-fed low-density lipoprotein receptor-null mice. Arterioscler. Thromb. Vasc. Biol. 30: 742–748. [DOI] [PubMed] [Google Scholar]
- 19.Teupser D., Persky A. D., Breslow J. L. 2003. Induction of atherosclerosis by low-fat, semisynthetic diets in LDL receptor-deficient C57BL/6J and FVB/NJ mice: comparison of lesions of the aortic root, brachiocephalic artery, and whole aorta (en face measurement). Arterioscler. Thromb. Vasc. Biol. 23: 1907–1913. [DOI] [PubMed] [Google Scholar]
- 20.Zhou C., King N., Chen K. Y., Breslow J. L. 2009. Activation of PXR induces hypercholesterolemia in wild-type and accelerates atherosclerosis in apoE deficient mice. J. Lipid Res. 50: 2004–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian A., Kuehn H., Gould J., Tamayo P., Mesirov J. P. 2007. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics. 23: 3251–3253. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvano S. E., Xiao W., Richards D. R., Felciano R. M., Baker H. V., Cho R. J., Chen R. O., Brownstein B. H., Cobb J. P., Tschoeke S. K., et al. 2005. A network-based analysis of systemic inflammation in humans. Nature. 437: 1032–1037. [DOI] [PubMed] [Google Scholar]
- 25.Li C. J., Li R. W., Wang Y. H., Elsasser T. H. 2007. Pathway analysis identifies perturbation of genetic networks induced by butyrate in a bovine kidney epithelial cell line. Funct. Integr. Genomics. 7: 193–205. [DOI] [PubMed] [Google Scholar]
- 26.Liu X., Lu R., Xia Y., Sun J. 2010. Global analysis of the eukaryotic pathways and networks regulated by Salmonella typhimurium in mouse intestinal infection in vivo. BMC Genomics. 11: 722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldstein J. L., DeBose-Boyd R. A., Brown M. S. 2006. Protein sensors for membrane sterols. Cell. 124: 35–46. [DOI] [PubMed] [Google Scholar]
- 28.Zelcer N., Tontonoz P. 2006. Liver X receptors as integrators of metabolic and inflammatory signaling. J. Clin. Invest. 116: 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradley M. N., Hong C., Chen M., Joseph S. B., Wilpitz D. C., Wang X., Lusis A. J., Collins A., Hseuh W. A., Collins J. L., et al. 2007. Ligand activation of LXR beta reverses atherosclerosis and cellular cholesterol overload in mice lacking LXR alpha and apoE. J. Clin. Invest. 117: 2337–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calkin A. C., Tontonoz P. 2010. Liver X receptor signaling pathways and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 30: 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proctor G., Jiang T., Iwahashi M., Wang Z., Li J., Levi M. 2006. Regulation of renal fatty acid and cholesterol metabolism, inflammation, and fibrosis in Akita and OVE26 mice with type 1 diabetes. Diabetes. 55: 2502–2509. [DOI] [PubMed] [Google Scholar]
- 32.Li-Hawkins J., Lund E. G., Turley S. D., Russell D. W. 2000. Disruption of the oxysterol 7alpha-hydroxylase gene in mice. J. Biol. Chem. 275: 16536–16542. [DOI] [PubMed] [Google Scholar]
- 33.Li T., Chen W., Chiang J. Y. 2007. PXR induces CYP27A1 and regulates cholesterol metabolism in the intestine. J. Lipid Res. 48: 373–384. [DOI] [PubMed] [Google Scholar]
- 34.Cali J. J., Hsieh C. L., Francke U., Russell D. W. 1991. Mutations in the bile acid biosynthetic enzyme sterol 27-hydroxylase underlie cerebrotendinous xanthomatosis. J. Biol. Chem. 266: 7779–7783. [PMC free article] [PubMed] [Google Scholar]
- 35.Hotamisligil G. S., Shargill N. S., Spiegelman B. M. 1993. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 259: 87–91. [DOI] [PubMed] [Google Scholar]
- 36.Weinstock P. H., Bisgaier C. L., Aalto-Setala K., Radner H., Ramakrishnan R., Levak-Frank S., Essenburg A. D., Zechner R., Breslow J. L. 1995. Severe hypertriglyceridemia, reduced high density lipoprotein, and neonatal death in lipoprotein lipase knockout mice. Mild hypertriglyceridemia with impaired very low density lipoprotein clearance in heterozygotes. J. Clin. Invest. 96: 2555–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levak-Frank S., Radner H., Walsh A., Stollberger R., Knipping G., Hoefler G., Sattler W., Weinstock P. H., Breslow J. L., Zechner R. 1995. Muscle-specific overexpression of lipoprotein lipase causes a severe myopathy characterized by proliferation of mitochondria and peroxisomes in transgenic mice. J. Clin. Invest. 96: 976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinstock P. H., Levak-Frank S., Hudgins L. C., Radner H., Friedman J. M., Zechner R., Breslow J. L. 1997. Lipoprotein lipase controls fatty acid entry into adipose tissue, but fat mass is preserved by endogenous synthesis in mice deficient in adipose tissue lipoprotein lipase. Proc. Natl. Acad. Sci. USA. 94: 10261–10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merkel M., Weinstock P. H., Chajek-Shaul T., Radner H., Yin B., Breslow J. L., Goldberg I. J. 1998. Lipoprotein lipase expression exclusively in liver. A mouse model for metabolism in the neonatal period and during cachexia. J. Clin. Invest. 102: 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J. K., Fillmore J. J., Chen Y., Yu C., Moore I. K., Pypaert M., Lutz E. P., Kako Y., Velez-Carrasco W., Goldberg I. J., et al. 2001. Tissue-specific overexpression of lipoprotein lipase causes tissue-specific insulin resistance. Proc. Natl. Acad. Sci. USA. 98: 7522–7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lusis A. J. 2000. Atherosclerosis. Nature. 407: 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Libby P. 2002. Inflammation in atherosclerosis. Nature. 420: 868–874. [DOI] [PubMed] [Google Scholar]
- 43.Gu L., Okada Y., Clinton S. K., Gerard C., Sukhova G. K., Libby P., Rollins B. J. 1998. Absence of monocyte chemoattractant protein-1 reduces atherosclerosis in low density lipoprotein receptor-deficient mice. Mol. Cell. 2: 275–281. [DOI] [PubMed] [Google Scholar]
- 44.Hansson G. K., Libby P. 2006. The immune response in atherosclerosis: a double-edged sword. Nat. Rev. Immunol. 6: 508–519. [DOI] [PubMed] [Google Scholar]
- 45.Yvan-Charvet L., Wang N., Tall A. R. 2010. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler. Thromb. Vasc. Biol. 30: 139–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koseki M., Hirano K., Masuda D., Ikegami C., Tanaka M., Ota A., Sandoval J. C., Nakagawa-Toyama Y., Sato S. B., Kobayashi T., et al. 2007. Increased lipid rafts and accelerated lipopolysaccharide-induced tumor necrosis factor-alpha secretion in Abca1-deficient macrophages. J. Lipid Res. 48: 299–306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.