Abstract
Saturated free fatty acids (FFA) induce hepatocyte lipoapoptosis, a key mediator of liver injury in nonalcoholic fatty liver disease (NAFLD). Lipoapoptosis involves the upregulation of the BH3-only protein PUMA, a potent pro-apoptotic protein. Given that dysregulation of hepatic microRNA expression has been observed in NAFLD, we examined the role of miRNA in regulating PUMA expression during lipotoxicity. By in silico analysis, we identified two putative binding sites for miR-296-5p within the 3′ untranslated region (UTR) of PUMA mRNA. Enforced miR-296-5p levels efficiently reduced PUMA protein expression in Huh-7 cells, while antagonism of miR-296-5p function increased PUMA cellular levels. Reporter gene assays identified PUMA 3′UTR as a direct target of miR-296-5p. The saturated FFA, palmitate, repressed miR-296-5p expression; and Huh-7 cells were sensitized to palmitate-induced lipotoxicity by antagonism of miR-296-5p function using a targeted locked nucleic acid (LNA). Finally, miR-296-5p was reduced in liver samples from nonalcoholic steatohepatitis (NASH) patients compared with patients with simple steatosis (SS) or controls. Also miR-296-5p levels inversely varied with PUMA mRNA levels in human liver specimens. Our results implicate miR-296-5p in the regulation of PUMA expression during hepatic lipoapoptosis. We speculate that enhancement of miR-296-5p expression may represent a novel approach to minimize apoptotic damage in human fatty liver diseases.
Keywords: BH3-only proteins, free fatty acid, hepatic steatosis, microRNA, p53-upregulated mediator of apoptosis
Nonalcoholic fatty liver disease (NAFLD) is commonly associated with the metabolic syndrome and represents a growing epidemic afflicting up to 30% of the American population (1). NAFLD covers a wide spectrum of chronic liver diseases, ranging from simple steatosis (SS), to nonalcoholic steatohepatitis (NASH), to fibrosis, and ultimately, to cirrhosis (2). This syndrome is characterized by elevated levels of circulating free fatty acids (FFA) and hepatocyte apoptosis, which increase in relationship to NAFLD severity (3–5). This relationship has generated the concept that the pathogenesis of NAFLD is related to the lipotoxic effect of FFA on liver cells.
Saturated FFAs are directly hepatotoxic and induce lipoapoptosis (6–8). Lipoapoptosis in vitro requires upregulation of the pro-apoptotic BH3-only protein PUMA. PUMA knockdown in Huh-7 cells or genetic deficiency of Puma in mouse primary hepatocytes partially protects liver cells from lipotoxic insults (7). Hepatic PUMA mRNA and protein levels are increased in NAFLD patients (7), and overexpression of cellular PUMA promotes cell death (9). The increased cellular levels of PUMA protein directly promote mitochondrial dysfunction, resulting in caspase 3/7 activation and culminating in cellular demise (10). During the lipotoxic insult, the mechanisms resulting in upregulation of PUMA mRNA are complex and mediated by multiple p53-independent transcriptional processes, including those regulated by activator protein (AP)-1 and C/EBP homologous protein (CHOP) (7, 8). The cellular regulation of PUMA is, however, multifaceted and also includes posttranscriptional processes (11). For example, only approximately 50% of PUMA induction by FFA can be accounted for by transcriptional regulation (8). Given its cytotoxic potential, additional insights into the different levels of PUMA regulation during lipoapoptosis may help develop novel approaches to treating human fatty liver diseases.
A recent study has reported altered hepatic expression of several microRNAs in human NASH (12). MicroRNAs are short noncoding RNAs of approximately 22 nucleotides long. MicroRNAs negatively regulate protein expression via binding to complementary sequences within the 3′untranslated region (UTR) of target mRNA. The bound mRNA remains untranslated, leading to reduced levels of the corresponding protein, or it can be degraded, resulting in decreased levels of the corresponding mRNA (13, 14). Reports of microRNA-targeted genes in the context of human fatty liver disease are very limited, and a potential role for microRNAs in regulating PUMA expression during lipoapoptosis has not been explored.
In the current study, we identified microRNA miR-296-5p as a direct negative regulator of PUMA expression during hepatocyte lipoapoptosis, adding to our understanding of PUMA expression in this liver disease. In NASH patients, hepatic miR-296-5p levels were reduced, and its downregulation was significantly associated with increased PUMA expression. These findings have potential relevance for the development of new targeted therapies to minimize PUMA-mediated apoptotic damage in patients with fatty liver diseases.
MATERIALS AND METHODS
Cells
Huh-7 and KMCH cells, a human hepatoma cell line and a human cholangiocarcinoma cell line, respectively, were cultured in Dulbecco's modified Eagle's medium containing glucose (25 mM), 100,000 units/liter penicillin, 100 mg/liter streptomycin, and 10% fetal bovine serum. KMCH cells were used only as another cellular model to study the functional repression of PUMA 3′UTR by microRNA-296-5p in a luciferase reporter gene assay.
Fatty acid treatment
Palmitic acid and oleic acid (Sigma Aldrich, St. Louis, MO) were prepared as previously described (7). The concentrations of FFA used in the experiments were comparable to the fasting FFA plasma concentrations observed in human nonalcoholic steatohepatitis (15, 16). The concentration of the vehicle, isopropyl alcohol, was 0.5% in final incubations.
MicroRNA target prediction
To identify potential PUMA-binding microRNAs, we used two algorithms, TargetScan (Release 5.1) and MicroCosm (Version 5). Both programs indicated that miR-296-5p is a putative candidate targeting the 3′UTR of PUMA.
Plasmid constructs and cellular transfection
The precursor sequence of miR-296 (pre-miR-296) was amplified by PCR from genomic human DNA using the following primers: forward, 5′-CACTGAGATGGGACAGG-3′; reverse, 5′-CTCTTGTATCGACTGTC-3′. The amplified product was subcloned into the pCR2.1-TOPO vector, sequenced to ensure accuracy of the DNA sequence, and then cloned into the BamHI/NotI sites of the pcDNA3.1 expression vector (miR-296 expression vector). Stable clones of Huh-7 cells overexpressing miR-296-5p were generated by transfecting cells with 2 µg/ml of the miR-296 expression vector using FuGENE HD transfection reagent (Roche Applied Science, Indianapolis, IN). Stably transfected clones were selected in medium containing 3,000 mg/l G418, and selected clones were screened by real-time PCR analysis for mature miR-296-5p.
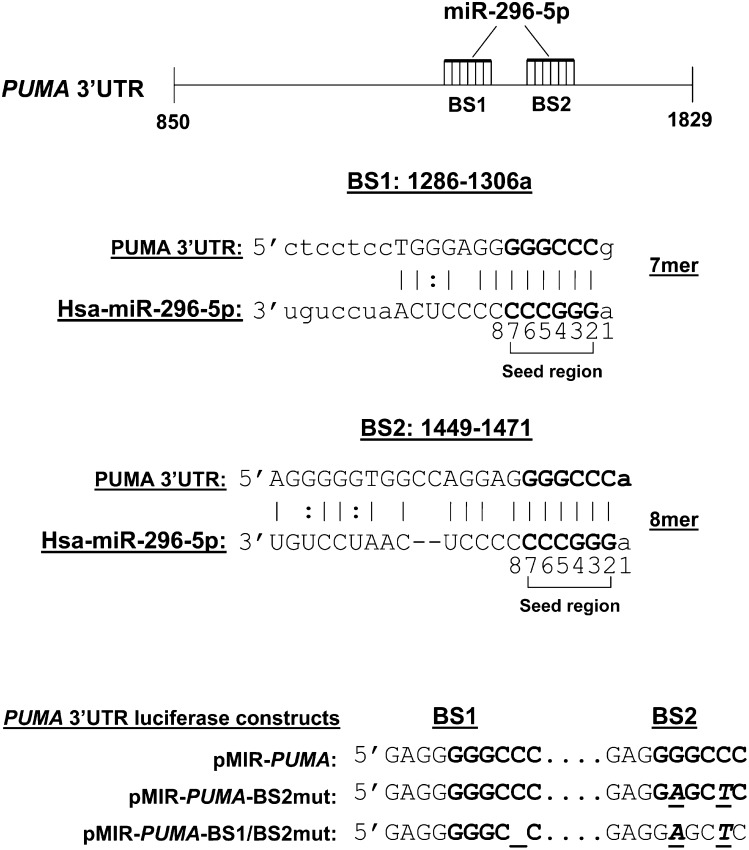
The human PUMA 3′UTR (AF354654, 980 base pair, position +850 to +1829), harboring two miR-296-5p target sequences (binding site 1, BS1, position 1299/1306; binding site 2, BS2, position 1465/1471), was amplified from genomic DNA using the following primers: forward, 5′-ACGGAGCTCGTGCCTGCACCCGCCCGGTG-3′; reverse, 5′-ACGAAGCTTCACTGTTCCAATCTGATTTT-3′. A SacI and HindIII restriction site were added, respectively, at the 5′ end of the forward and reverse primer (underlined sequence). PCR amplified human PUMA 3′UTR was then cloned in pMIR-REPORT luciferase vector (Ambion) downstream of the luciferase gene and designated as pMIR-PUMA; automated sequencing confirmed the identity and fidelity of the insert. Mutation of the miR-296-5p target sequence BS2 was generated by site-directed mutagenesis (QuickChange II site-directed mutagenesis kit, Stratagene, La Jolla, CA) using the following primers: BS2mutF 5′-GGGTGGCCAGGAGGAGCTCAGACTGTGAATCC-3′, BS2mutR 5′-GGATTCACAGTCTGAGCTCCTCCTGGCCACCC-3′ (altered nucleotides are underlined and bold) (pMIR-PUMA-BS2mut). Several attempts to mutate the miR-296-5p target sequence BS1 by site-directed mutagenesis were unsuccessful. To overcome this technical problem, we adopted a different strategy to mutate BS1 based on nucleotide deletion. Both miR-296-5p seed sequences in the PUMA 3′UTR contain a cleavage site for the restriction endonuclease ApaI (GGGCCC), one of which was lost in the mutated BS2 generated above. Based on this observation, a single nucleotide deletion of the miR-296-5p target sequence BS1 was generated by digestion of the pMIR-PUMA-BS2mut with ApaI, followed by treatment with Mung bean nuclease (New England BioLabs, Ipswich, MA) to cleave single-stranded overhangs and religation of the plasmid using T4 DNA ligase. This strategy resulted in deletion of a C nucleotide at position 1304 of BS1. The double mutant plasmid was designated as pMIR-PUMA-BS1/BS2mut (Fig. 1). The miR-296 expression vector and pMIR luciferase plasmids were transiently transfected using FuGENE HD transfection reagent (Roche Applied Science), and assays or treatments were carried out 24 h later.
Fig. 1.
Putative miR-296-5p binding sites in PUMA mRNA. Schematic representation of the PUMA 3′UTR (#AF354654) nucleotides 863-1843. Computational algorithms were used to predict microRNAs that bind to a target sequence in the 3′UTR of PUMA. Two putative miR-296-5p binding sites were identified at position 1299/1306 (BS1) and position 1465/1471 (BS2) of PUMA 3′UTR. The complementarity of the 5′ seed region of miR-296-5p with BS1 and BS2 is indicated in the lower panel. According to Grimson et al. (17), BS1 is a 7mer site which contains the seed match (6 nucleotide at position 2-7) augmented by a match to microRNA nucleotide 8; BS2 is a 8mer site which contains the seed match flanked by both the match at position 8 and an A at position 1. As described in Materials and Methods, deletion or mutation of critical residues of BS1 and BS2 were generated for luciferase reporter gene assay.
Downregulation of miR-296 levels was through transfection (lipofectamine 2000, Invitrogen, Carlsbad, CA) for 24 h with miRCURY LNA microRNA inhibitor (50 nM) for miR-296-5p (Exiqon, Woburn, MA). Scramble LNA (Exiqon) was used as a control.
Quantitative real-time PCR and determination of miR-296-5p half-life
Total RNA was isolated using Trizol reagent (Invitrogen). Real-time PCR for human PUMA was performed as previously described in detail (7) using SYBR green fluorescence technology. For microRNA RT-PCR, 50 ng of total RNA was converted into cDNA using specific primers for miR-296-5p, miR-221, miR-222, miR-483-3p, or the internal control Z30 (Applied Biosystems, Foster City, CA) and the TaqMan micro-RNA reverse transcription kit (Applied Biosystems). Next, TaqMan real-time PCR was performed using specific primers for the different microRNAs or Z30. MicroRNA expression levels were expressed relative to the internal control according to the comparative threshold cycle (Ct) method. All real-time PCR was performed on a LightCycler 480 instrument (Roche Applied Science). miR-296-5p's half-life was examined in vehicle-treated cells or palmitate-treated cells in the presence of the transcription inhibitor actinomycin D (Sigma Aldrich) over a 6 h time course. Half-life (t1/2) was calculated based on the decay constant.
Immunoblot analysis
Whole cell lysates were prepared as previously described (7). An equal amount of protein (50 µg) was resolved by SDS-PAGE on a 15% acrylamide gel, transferred to nitrocellulose membranes, and incubated with primary antibodies. Antibodies used were obtained from the following sources: anti-PUMA (Ab 54288, Abcam, Inc., Cambridge, MA), anti-Bim (BD Biosciences, San Jose, CA) and anti-β-actin (Santa Cruz Biotechnology, Santa Cruz, CA). Membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies (Biosource International, Camarillo, CA). Bound antibody was visualized using chemiluminescent substrate (ECL, Amersham, Arlington Heights, IL) and was exposed to Kodak X-OMAT film (Eastman Kodak, Rochester, NY).
Luciferase assay
Luciferase activity and protein concentration in supernatants were determined using the Luciferase Reporter Assay System (Promega, Madison, WI) and Bradford Reagent (Pierce, Rockford, IL), respectively. Luciferase activity was quantified in a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA) (7). The results are reported as relative light units per microgram total protein.
Quantitation of apoptosis
Cells were stained with 5 µg/ml 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI) for 30 min at 37°C and visualized under fluorescence microscopy (Nikon Eclipse TE200, Nikon Corporation, Japan). Apoptotic cells were quantified by counting 300 random cells per study. Cells with the characteristic nuclear changes of chromatin condensation and nuclear fragmentation were considered apoptotic (7). Apoptosis was expressed as a percentage of total cells counted. For caspase 3/7 activity, cells were plated in 96-well plates. The assay was performed using the commercially available Apo-ONE homogeneous caspase-3/7 assay (Promega) as previously described (7).
Patient population
The study was approved by the Mayo Institutional Review Board, and all patients gave written, informed consent for participation in medical research. Our cohort consisted of 56 patients who underwent liver biopsy at the time of bariatric surgery for medically complicated obesity at Mayo Clinic, Rochester, MN. The diagnosis of NAFLD was based on liver biopsy features. All patients had a body mass index (BMI) > 30kg/m2. Patients were divided into three histologic groups: obese normal (ON, normal liver biopsies, n = 18); simple steatosis (SS, n = 19); and NASH (steatosis, lobular and/or portal inflammation grade 1 and fibrosis stage 0-3, n = 19). Patients with NAFLD who had secondary causes of steatohepatitis (drugs or prior gastric surgery for obesity) and patients with other etiologies of chronic liver disease [excessive alcohol consumption, viral hepatitis (B, C), cholestatic liver disease, hemochromatosis, Wilson's disease, drug-induced liver disease, or α 1-antitrypsin deficiency], were excluded from this study. Total RNA was extracted from frozen tissue using RNeasy Mini Kit from Qiagen (Valencia, CA).
Statistical analysis
All data represent at least three independent experiments and are expressed as means ± SEM. of the mean. Differences between groups were compared using Student's t-test or one-way ANOVA with post hoc Dunnett test. The correlation between miR-296-5p levels and PUMA mRNA levels was analyzed by a two-tailed Pearson correlation test. Statistical significance was accepted at P < 0.05.
RESULTS
miR-296-5p negatively regulates PUMA expression
Among the different computationally predicted PUMA-binding miRNAs, we identified miR-296-5p as a putative candidate to regulate PUMA expression based on the guanine-cytosine (GC)-rich content of its seed region, allowing multiple sequence complementarity with the 3′UTR of PUMA. Indeed, two conserved miR-296-5p binding sites were identified at position 1299/1306 (BS1) and position 1465/1471 (BS2) of PUMA 3′UTR (Fig. 1). Both binding sites represented strong targets (7mer and 8mer) using the criteria of Grimson et al. (17).
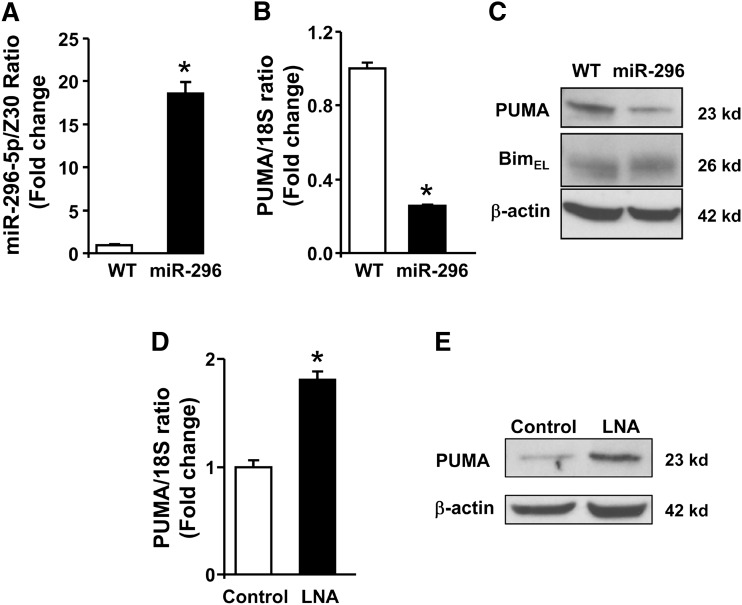
To test whether miR-296-5p regulates PUMA expression, Huh-7 cells were stably transfected with a pcDNA3.1 plasmid encoding the precursor sequence of miR-296. As assessed by real-time PCR, the level of mature miR-296-5p in cells stably transfected with this plasmid was efficiently increased compared with parental WT Huh-7 cells (Fig. 2A). Enhanced miR-296-5p expression resulted in a marked decrease of PUMA cellular mRNA and protein levels (Fig. 2B, C). This effect was specific as the protein levels of Bim, a BH3-only protein that lacks predicted miR-296-5p binding sites, remained unchanged in the Huh-7 cells overexpressing miR-296-5p (Fig. 2C). This forced overexpression of miR-296-5p cellular levels clearly identified that miR-296-5p regulates PUMA expression, albeit in an unphysiologic context. Therefore, in a converse experiment, miR-296-5p function was inhibited by transfecting WT Huh-7 cells with a locked nucleic acid (LNA) oligonucleotide that binds tightly to miR-296-5p, inhibiting its function. Antagonism of miR-296-5p was associated with a modest increase in PUMA mRNA levels (Fig. 2D) and a more substantial increase in protein levels compared with cells transfected with a control LNA (Fig. 2E). These observations suggest an effect of miR-296-5p on both PUMA transcript degradation and protein translation. Collectively, these results indicated that miR-296-5p negatively regulates PUMA cellular expression.
Fig. 2.
miR-296-5p negatively modulates PUMA expression. A: miR-296-5p levels were increased by stable transfection of Huh-7 cells (WT) with miR-296-5p expression vector (miR-296) and expressed in relation to the small housekeeping RNA Z30. B: PUMA mRNA was measured in WT and miR-296 stable cells by real-time PCR and expressed in relation to the housekeeping rRNA 18S. C: Immunoblot analysis was performed for PUMA and Bim protein expression in WT or miR-296 stable cells. β-actin was used as a control for protein loading. D: miR-296-5p function was inhibited by transient transfection for 24 h with a locked nucleic acid oligonucleotide specific to miR-296-5p (LNA). Scramble LNA were used as control. PUMA mRNA levels were increased in LNA-transfected cells. E: LNA-transfected as in panel D also resulted in significantly higher PUMA protein expression. Data represent the mean and standard error of three experiments; *P < 0.05.
miR-296-5p directly targets PUMA 3primeUTR
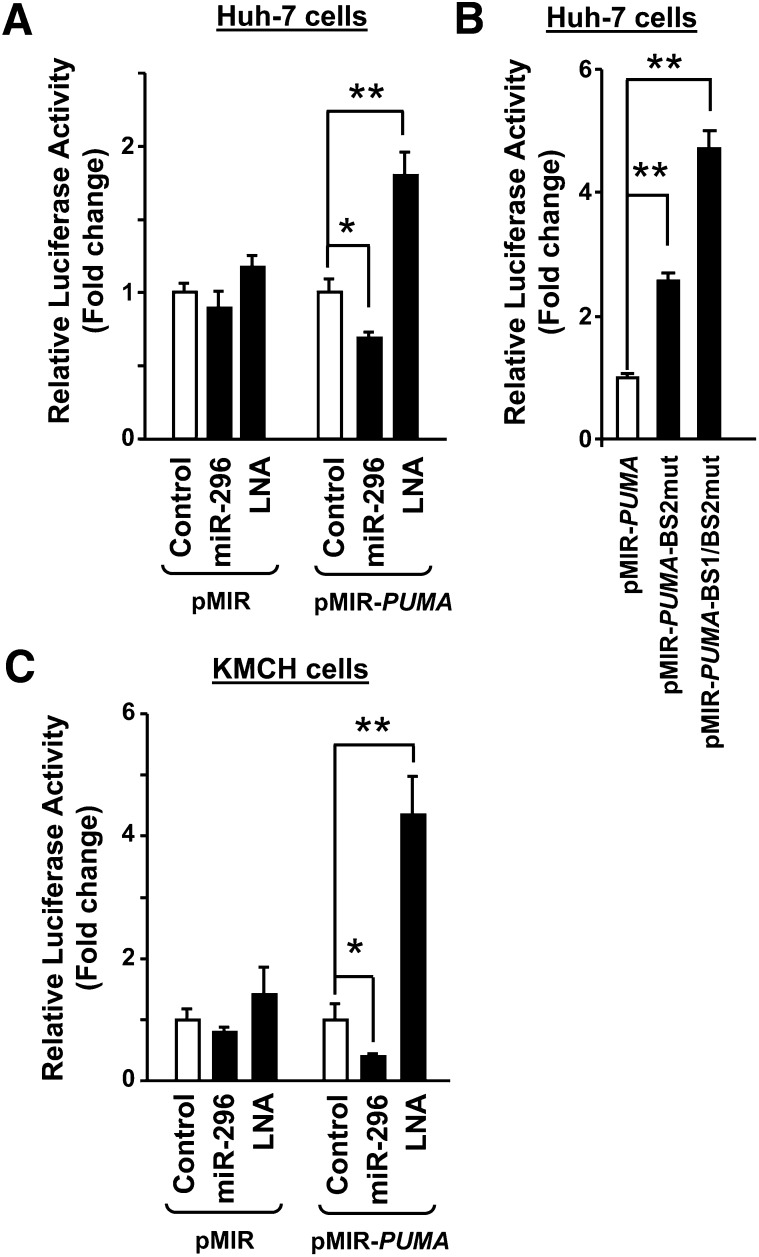
To demonstrate that PUMA mRNA is a direct target for miR-296-5p, the PUMA 3′UTR was cloned downstream of the luciferase coding sequence of the pMIR-report vector (pMIR-PUMA). Cotransfection of Huh-7 cells with the pMIR-PUMA plus the miR-296 expression vector significantly decreased luciferase activity (Fig. 3A). Conversely, antagonism of miR-296-5p by LNA transfection increased luciferase activity (Fig. 3A). As a control, modulation of miR-296 levels using the miR-296 expression vector or the LNA for miR-296-5p did not significantly modify luciferase activity when Huh-7 cells were transfected with pMIR empty vector (Fig. 3A). Functional derepression of luciferase activity was observed when critical residues of one or both miR-296-5p binding sites within PUMA 3′UTR (BS1 and BS2) were mutated (pMIR-PUMA-BS1/BS2mut) (Figs. 1 and 3B). These results indicate that miR-296-5p functionally binds to both PUMA 3′UTR binding sites.
Fig. 3.
PUMA mRNA is a direct target for miR-296-5p. PUMA 3′UTR was cloned downstream of the luciferase gene of the pMIR-report vector (pMIR-PUMA). Huh-7 (A and B) and KMCH cells (C) were transiently transfected with luciferase-containing plasmids as follows: pMIR as empty luciferase vector, pMIR-PUMA as luciferase plus 3′UTR from PUMA, pMIR PUMA-BS2mut as luciferase plus 3′UTR from PUMA with miR-296-5p binding site 2 mutated, and pMIR PUMA-BS1/BS2mut as luciferase plus 3′UTR from PUMA with both miR-296-5p binding sites mutated. When indicated, cells were cotransfected with the miR-296 expression vector (miR-296) or with a locked nucleic acid oligonucleotide specific to miR-296-5p (LNA). Relative luciferase activity was assessed 24 h after transfection as described in Materials and Methods. Data represent the mean and standard error of three experiments; *P < 0.05 and **P < 0.01.
These data were replicated in a different cellular model using the cholangiocarcinoma cell line, KMCH cells (Fig. 3C). These cells have slightly higher ratios of miR-296-5p/Z30 (0.51 ± 0.2) compared with Huh-7 cells (0.21 ± 0.02). Again luciferase activity was markedly decreased by enhanced expression of miR-296-5p, and increased after LNA-directed antagonism of miR-296-5p function (Fig. 3C). The changes in luciferase activity by miR-296-5p levels were quantitatively less than the changes in PUMA mRNA and proteins levels, indicating a possible inhibitory effect of miR-296-5p on PUMA protein translation in addition to an effect on PUMA mRNA stability. Thus, miR-296-5p directly downregulates PUMA cellular expression by binding to target sequences within the PUMA 3′UTR.
miR-296-5p expression protects Huh-7 cells from palmitate-mediated apoptosis
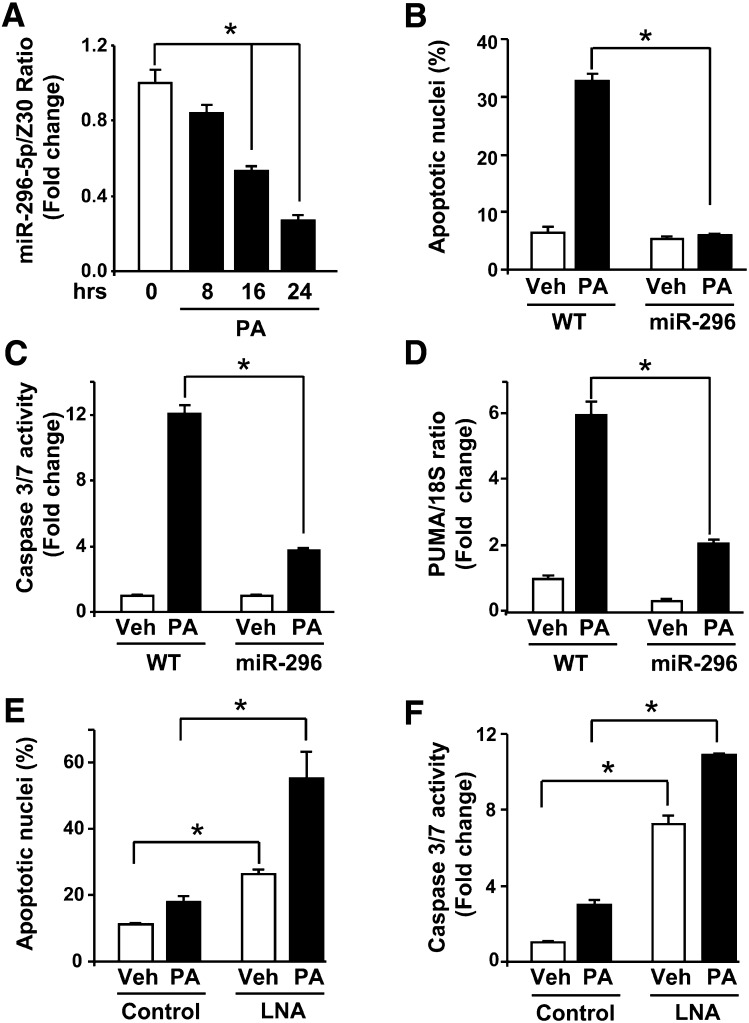
Increased PUMA expression contributes to saturated FFA-induced apoptosis (7, 8). Indeed, PUMA mRNA levels increases 4-fold at 8 h following treatment with palmitate (supplemental Fig. IA). By 16 h of treatment with palmitate, a further increase in PUMA mRNA expression (supplemental Fig. IA) was accompanied by an increase in PUMA cellular protein levels (supplemental Fig. IB). This late increase in PUMA mRNA and protein levels coincides with a 50% decrease in miR-296-5p levels at 16 h following treatment with the saturated FFA (Fig. 4A). As a control, treatment of Huh-7 cells with the nontoxic monounsaturated FFA oleate, which does not modulate PUMA mRNA expression (7), did not decrease miR-296-5p levels (supplemental Fig. II). To gain insight into the processes by which palmitate reduces miR-296-5p, we examined if palmitate reduces miR-296-5p's half-life (supplemental Fig. III). miR-296-5p's half-life was 5.9 h in vehicle-treated cells, and palmitate treatment did not significantly shortened the half-life of the mature microRNA. Taken together, saturated FFA-induced increase in PUMA mRNA and protein levels corresponds with a downregulation of miR-296-5p cellular levels prior to the onset of cell death. Palmitate-mediated reduction of miR-296-5p does not appear to be due to enhanced microRNA degradation but rather decreased microRNA generation.
Fig. 4.
Altered miR-296-5p levels modulate liver cell sensitivity to palmitate-induced apoptosis. A: WT Huh-7 cells were treated with vehicle (Veh) or palmitic acid (PA; 800 μM) for 8, 16, or 24 h, and then total RNA was analyzed for miR-296-5p expression. miR-296-5p is expressed in relation to the small housekeeping RNA Z30. B: Huh-7 (WT) or Huh-7 cells stably overexpressing miR-296-5p (miR-296) were treated with Veh or PA (800 μM), and 16 h after treatment, apoptotic nuclei were counted. C: Cells treated as in panel B were assayed for caspase 3/7 activity. D: WT and miR-296 stable Huh-7 cells were treated for 8 h with veh or PA (800 μM), and then total RNA was analyzed for PUMA expression by real-time PCR. E and F: WT Huh-7 cells were transfected with scrambled LNA (control) or miR-296 antagonist LNA (LNA) for 24 h. Cells were then treated with Veh or PA (200 μM) for 16 h and assayed for apoptotic nuclei (E) or caspase 3/7 activity (F). All data are expressed as mean and standard error for three experiments; *P < 0.01.
Given the above observations, we next sought to investigate whether changes in miR-296-5p levels might modulate saturated FFA-mediated apoptosis. In accord with previous observations (6–8), palmitate at the concentration of 800 μM induced significant apoptosis in Huh-7 cells. as assessed by both morphology (Fig. 4B) and caspase 3/7 activity (Fig. 4C). Enhanced expression of miR-296-5p in Huh-7 cells significantly blocked the palmitate-mediated increase of PUMA mRNA levels (Fig. 4D) and, consistent with this effect, markedly attenuated lipoapoptosis (Fig. 4B, C). Also, the combination of oleate (400 μM) with palmitate (800 μM), prevented the decrease in miR-296-5p levels (supplemental Fig. IVA) and subsequent increase in PUMA mRNA (supplemental Fig. IVB), a result consistent with the known cytoprotective functions of the unsaturated FFA in this model (18). Conversely, antagonism of miR-296-5p function by LNA transfection increased Huh-7 cell apoptosis compared with cells transfected with a control LNA (Fig. 4E, F), a result consistent with a previous report defining miR-296-5p as an anti-apoptotic microRNA (19). LNA-mediated inhibition of miR-296-5p function mimicked the downregulation of miR-296-5p levels by palmitate and further enhanced cell death following treatment with a low toxic concentration of palmitate (200 μM) (Fig. 4E, F). Three other microRNAs, miR-221/222 and miR-483-3p, also have been reported to negatively regulate PUMA expression (20) (21). Therefore we quantified the levels of miR-221, miR-222, and miR-483-3p during lipotoxicity. Contrary to the effect on miR-296-5p levels, incubation of Huh-7 cells with palmitate did not reduce the miR-221, miR-222, or miR-483-3p level (supplemental Fig. VA–C). Indeed, consistent with prior observations in NASH patients (12), miR-221, miR-222, and miR-483-3p levels were either increased or unchanged by palmitate, making these three microRNAs unlikely to be mechanistically related to increased PUMA expression in our model of lipotoxicity. Collectively, these data are consistent with saturated FFA-induced dysregulation of miR-296-5p, which enhanced cell sensitivity to palmitate-mediated apoptosis by increasing PUMA expression.
The level of miR-296-5p is decreased in NASH patients and inversely correlates with PUMA expression
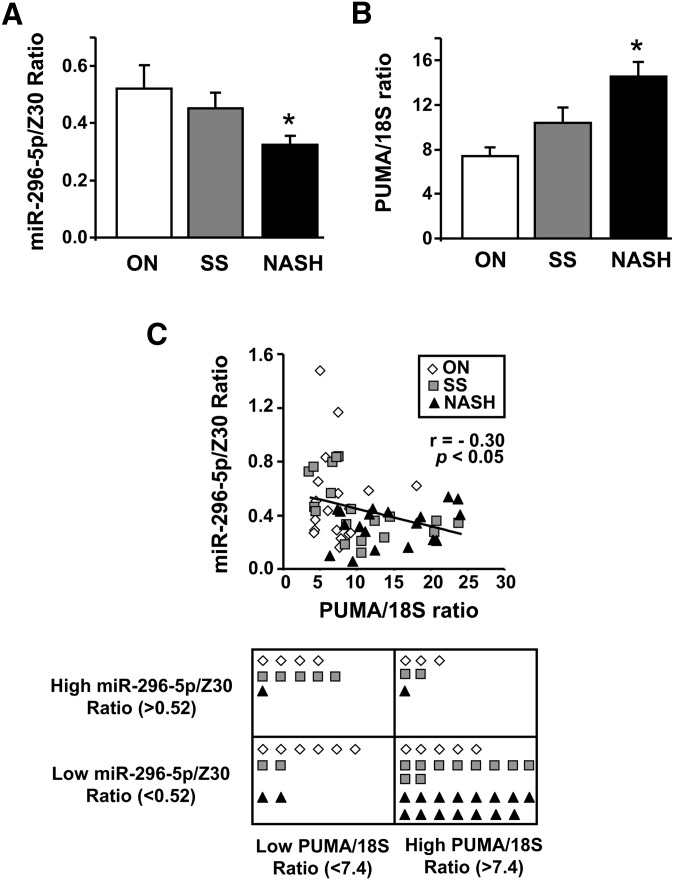
Because serum FFAs and hepatic PUMA expression are increased in NAFLD patients (3, 7), we sought to determine the levels of miR-296-5p in human liver specimens, comparing obese normal control liver specimens to those with simple steatosis, steatosis plus inflammation, and various degree of fibrosis (NASH). The level of miR-296-5p was decreased in liver tissue from patients with NASH compared with liver tissue from patients with simple steatosis and obese normal controls (P < 0.05) (Fig. 5A). This decrease in miR-296-5p levels was associated with an increase in PUMA mRNA (P < 0.05) (Fig. 5B) and proteins levels as previously reported (7). Cutoff levels of PUMA mRNA and miR-296-5p, corresponding to the mean value of the PUMA/18S ratio and miR-296-5p/Z30 ratio, respectively, in the obese normal group, were employed to classify patient results into four groups. On the basis of this classification, 27% of obese normal patients, 52% of simple steatosis patients, and 79% of NASH patients expressed high levels of PUMA mRNA and low levels of miR-296-5p (Fig. 5C). Thus, an inverse correlation between PUMA mRNA expression and miR-296-5p in liver tissue from obese patients with or without NAFLD was observed (Pearson correlation r = −0.30; P < 0.05) (Fig. 5C). These data suggest that repression of miR-296-5p expression in NAFLD may contribute to the pathogenesis of this fatty liver syndrome.
Fig. 5.
miR-296-5p levels are reduced in human NASH and inversely associate with increased PUMA expression. Total RNA were prepared from human liver biopsies of obese normal (ON, n = 18), simple steatosis (SS, n = 19), and NASH patients (n = 19). miR-296-5p levels (A) and PUMA mRNA expression (B) were assessed by real-time PCR. Data were normalized to their respective internal control Z30 or 18S. *P < 0.05, NASH versus ON or SS. An inverse correlation between miR-296-5p and PUMA expression was observed in human liver specimens (P < 0.05, Pearson's correlation test; r, correlation coefficient). Patients were divided into four groups based on high or low PUMA/18S ratio and high or low miR-296-5p/Z30 ratio. Cutoff values were based on the mean values obtained from the ON patient group (C).
DISCUSSION
The findings of the present study provide further mechanistic insights regarding the regulation of PUMA expression during hepatocyte apoptosis. Our results indicate the following: i) miR-296-5p, a microRNA predicted to target PUMA 3′UTR, specifically represses PUMA expression by degrading PUMA transcript and inhibiting PUMA protein translation; ii) suppression of PUMA expression by miR-296-5p is direct and sequence-specific; and iii) palmitate, the toxic saturated FFA, induces a decrease in miR-296-5p levels, which contributes to its associated lipotoxicity, in part, through the upregulation of PUMA expression. Consistent with our in vitro studies, our data also indicate that miR-296-5p levels are decreased in the liver of patients with NASH and that an inverse correlation exists between hepatic miR-296-5p and PUMA levels in subjects with or without NAFLD. These observations are more thoroughly discussed below.
The current study indicates that the saturated FFA palmitate decreases levels of miR-296-5p in liver cells. Mature microRNA levels are regulated by transcription, processing, nuclear export, and degradation. In our current study, palmitate treatment did not acutely modify miR-296-5p stability following transcriptional inhibition by actinomycin D, suggesting a possible effect of palmitate on transcription or processing of miR-296-5p. During apoptotic processes, Dicer, the cytosolic endoribonuclease that cleaves the microRNA precursor into a mature form, can be cleaved by caspase-3 with loss of function (22, 23). Palmitate-induced caspase-3 activation may alter Dicer function, resulting in decreased generation of the processed and functionally active miR-296-5p.
Our previous studies (8) have implicated both the transcription factors AP-1 and CHOP in the transcriptional regulation of PUMA expression during lipoapoptosis. However, we observed that inhibition of both AP-1 and CHOP decreased PUMA induction by palmitate by 50% (8), implicating other mechanisms that might regulate PUMA expression. Palmitate decreases miR-296-5p levels, which results in increase in PUMA expression with associated toxicity; and overexpression of miR-296-5p limits palmitate-induced PUMA expression also by approximately 50%. These data suggest that miR-296-5p represents an additional factor in the regulation of PUMA expression during lipotoxicity. Decrease in miR-296-5p levels in response to palmitate occurred temporally later compared with the activation of CHOP and AP-1 (7), but it coincided with a later increase in both PUMA mRNA and protein levels; these observations are consistent with an effect of miR-296-5p on both PUMA transcript degradation and protein translation. Therefore, it is plausible that CHOP- and AP-1-mediated transcriptional regulation of PUMA may account for early increases of PUMA mRNA by palmitate, whereas dysregulation of miR-296-5p during lipoapoptosis accounts for a later increase in PUMA mRNA and protein levels. Our studies indicate that PUMA expression is regulated, in part, by AP-1 and CHOP at the transcriptional level and by the microRNA miR-296-5p at the posttranscriptional level, and both of these processes may equally regulate PUMA expression, albeit differentially over time, during lipoapoptosis.
Insulin resistance, elevated serum FFA, increased PUMA expression, and hepatocyte apoptosis are important features of human NAFLD (1, 3, 4, 7). NAFLD progression not only correlates with an increase in hepatocyte apoptosis (4) but also with a dysregulation of the hepatic expression of several microRNAs (12), suggesting a possible role for microRNAs in regulating cell death in the context of human fatty liver disease. However, studies of microRNA-mRNA interaction in this disease context are very limited. For example, miR-122, which is decreased throughout the progression of NAFLD (12), regulates the expression of key hepatic lipogenic genes (24). Similarly, miR-34a, which is increased in NAFLD, was found to participate in disease development by targeting the nicotinamide adenine dinucleotide (NAD)-dependent deacetylase Sirtuin-1, implicated in the control of lipid and glucose metabolism (25) and the regulation of cellular apoptosis (26). We have previously demonstrated that PUMA mRNA is upregulated in human NASH (7). Herein, we report that miR-296-5p is aberrantly downregulated in liver biopsies from patients with NASH and associated with the increase in PUMA mRNA. Reduced miR-296-5p levels in obese patients with or without NAFLD significantly correlate with increased expression of PUMA, possibly linking miR-296-5p-regulated PUMA expression with increased hepatocyte apoptosis in human NAFLD. Consistent with these results, miR-296-5p levels are also decreased in the visceral adipose tissue of patients with NAFLD (27) and may contribute to the adipocyte apoptosis associated with human obesity (28). Finally, the involvement of miR-296-5p in fatty liver disease is also supported by the observation that a region on human chromosome 20, including the intergenic locus coding for miR-296-5p (20q13.3), is linked to human obesity (29).
The previous study by Cheung et al. (12) did not report a change in miR-296-5p expression in NASH patients, while our current study identified this microRNA as repressed by saturated FFA and decreased in NASH liver samples. This apparent discrepancy is most likely due to methodological differences. Specifically, microarray technology as used in the previous study involves assessing a large quantity of targets simultaneously, in essence, performing hundreds of parallel hybridization experiments. The ideal conditions for an individual reaction (e.g., miR-296-5p) are not identical due to sequence and context differences, and thus, the hybridization conditions for the experiment as a whole are a compromise. The miR-296-5p sequence contains 66.7% GC content (14 of 21 bases), and thus, the ideal hybridization conditions (to maximize specificity and sensitivity) for this microRNA are likely to differ from the conditions used for the microarray experiment. The use of targeted assessment by real-time PCR, as performed in the current study, bypasses the compromise in reaction conditions and is more sensitive and specific for each target analyzed.
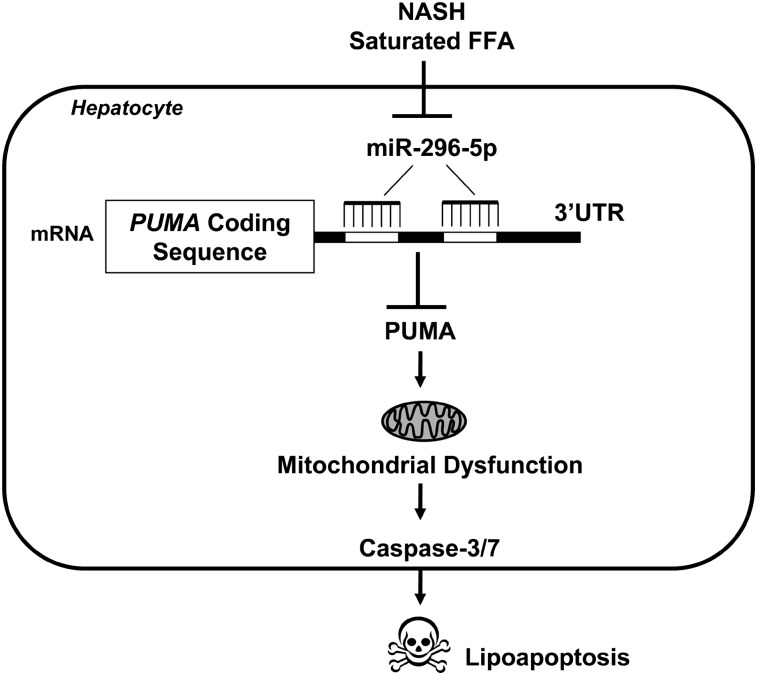
Although most of NASH patients expressed high levels of PUMA mRNA, approximately half of the patients with simple steatosis also had increased levels of PUMA mRNA, suggesting that PUMA may not be the sole driver of cytotoxicity in the context of NAFLD. Other BH3-only proteins, such as Bim, also contribute to lipoapoptosis (6) and cooperate with PUMA in cell death processes (30). Also, NASH patients display increased expression of death receptors, such as Fas and TRAIL death receptor (DR)5, which commonly mediate hepatocyte apoptosis (4, 5, 31). NAFLD progression is the result of a multifactorial process, and which combination of pro-death effectors is crucial in the pathogenesis of the disease remains to be clarified. Nonetheless, loss of miR-296-5p levels likely represents an additional mechanism resulting in PUMA upregulation in the context of human fatty liver disease (Fig. 6). Mechanisms to enhance miR-296-5p expression could prove useful to minimize liver damage in human NASH. Given the interest of the pharmaceutical industry in microRNA therapy (32), the concept of miR-296-5p-directed liver therapy warrants further study in preclinical NAFLD animal models.
Fig. 6.
Altered miR-296-5p regulates PUMA expression and hepatocyte lipoapoptosis. mir-296-5p directly binds to PUMA 3′UTR and negatively regulates its expression. In NASH patients with increased circulating FFA, hepatic miR-296 expression is reduced, resulting in increases in PUMA with associated lipotoxicity.
Supplementary Material
Acknowledgments
The authors thank Courtney Riddle for her excellent secretarial assistance.
Footnotes
Abbreviations:
- AP-1
- activator protein-1
- BH3
- Bcl-2 homology domain 3
- BS
- binding site
- CG
- guanine-cytosine
- CHOP
- C/EBP homologous protein; mut, mutant
- LNA
- locked nucleic acid
- NAFLD
- nonalcoholic fatty liver disease
- NASH
- nonalcoholic steatohepatitis
- ON
- obese normal
- PUMA
- p53-upregulated mediator of apoptosis
- SS
- simple steatosis
- UTR
- untranslated region
- WT
- wild-type
This work was supported by National Institute of Health Grants DK-41876 (G.J.G.), 5R01 DK-069757-05 (M.C.R.), K01 DK-079875 (J.L.M.), and P30 DK-084567 (Clinical Core of the Mayo Clinic Center for Cell Signaling in Gastroenterology); and by the Mayo Foundation. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of five figures.
REFERENCES
- 1.Browning J. D., Szczepaniak L. S., Dobbins R., Nuremberg P., Horton J. D., Cohen J. C., Grundy S. M., Hobbs H. H. 2004. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 40: 1387–1395. [DOI] [PubMed] [Google Scholar]
- 2.Adams L. A., Lymp J. F., St. Sauver J., Sanderson S. O., Lindor K. D., Feldstein A., Angulo P. 2005. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. 129: 113–121. [DOI] [PubMed] [Google Scholar]
- 3.Nehra V., Angulo P., Buchman A. L., Lindor K. D. 2001. Nutritional and metabolic considerations in the etiology of nonalcoholic steatohepatitis. Dig. Dis. Sci. 46: 2347–2352. [DOI] [PubMed] [Google Scholar]
- 4.Feldstein A. E., Canbay A., Angulo P., Taniai M., Burgart L. J., Lindor K. D., Gores G. J. 2003. Hepatocyte apoptosis and fas expression are prominent features of human nonalcoholic steatohepatitis. Gastroenterology. 125: 437–443. [DOI] [PubMed] [Google Scholar]
- 5.Kahraman A., Schlattjan M., Kocabayoglu P., Yildiz-Meziletoglu S., Schlensak M., Fingas C. D., Wedemeyer I., Marquitan G., Gieseler R. K., Baba H. A., et al. 2010. Major histocompatibility complex class I-related chains A and B (MIC A/B): a novel role in nonalcoholic steatohepatitis. Hepatology. 51: 92–102. [DOI] [PubMed] [Google Scholar]
- 6.Barreyro F. J., Kobayashi S., Bronk S. F., Werneburg N. W., Malhi H., Gores G. J. 2007. Transcriptional regulation of Bim by FoxO3A mediates hepatocyte lipoapoptosis. J. Biol. Chem. 282: 27141–27154. [DOI] [PubMed] [Google Scholar]
- 7.Cazanave S. C., Mott J. L., Elmi N. A., Bronk S. F., Werneburg N. W., Akazawa Y., Kahraman A., Garrison S. P., Zambetti G. P., Charlton M. R., et al. 2009. JNK1-dependent PUMA expression contributes to hepatocyte lipoapoptosis. J. Biol. Chem. 284: 26591–26602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cazanave S. C., Elmi N. A., Akazawa Y., Bronk S. F., Mott J. L., Gores G. J. 2010. CHOP and AP-1 cooperatively mediate PUMA expression during lipoapoptosis. Am. J. Physiol. Gastrointest. Liver Physiol. 299: G236–G243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jabbour A. M., Heraud J. E., Daunt C. P., Kaufmann T., Sandow J., O'Reilly L. A., Callus B. A., Lopez A., Strasser A., Vaux D. L., et al. 2009. Puma indirectly activates Bax to cause apoptosis in the absence of Bid or Bim. Cell Death Differ. 16: 555–563. [DOI] [PubMed] [Google Scholar]
- 10.Kim H., Tu H. C., Ren D., Takeuchi O., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. 2009. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell. 36: 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J., Zhang L. 2008. PUMA, a potent killer with or without p53. Oncogene. 27 (Suppl. 1): S71–S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung O., Puri P., Eicken C., Contos M. J., Mirshahi F., Maher J. W., Kellum J. M., Min H., Luketic V. A., Sanyal A. J. 2008. Nonalcoholic steatohepatitis is associated with altered hepatic microRNA expression. Hepatology. 48: 1810–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 14.Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 466: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belfort R., Harrison S. A., Brown K., Darland C., Finch J., Hardies J., Balas B., Gastaldelli A., Tio F., Pulcini J., et al. 2006. A placebo-controlled trial of pioglitazone in subjects with nonalcoholic steatohepatitis. N. Engl. J. Med. 355: 2297–2307. [DOI] [PubMed] [Google Scholar]
- 16.Sanyal A. J., Campbell-Sargent C., Mirshahi F., Rizzo W. B., Contos M. J., Sterling R. K., Luketic V. A., Shiffman M. L., Clore J. N. 2001. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology. 120: 1183–1192. [DOI] [PubMed] [Google Scholar]
- 17.Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 27: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei Y., Wang D., Gentile C. L., Pagliassotti M. J. 2009. Reduced endoplasmic reticulum luminal calcium links saturated fatty acid-mediated endoplasmic reticulum stress and cell death in liver cells. Mol. Cell. Biochem. 331: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng A. M., Byrom M. W., Shelton J., Ford L. P. 2005. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 33: 1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C. Z., Zhang J. X., Zhang A. L., Shi Z. D., Han L., Jia Z. F., Yang W. D., Wang G. X., Jiang T., You Y. P., et al. 2010. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol. Cancer. 9: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veronese A., Lupini L., Consiglio J., Visone R., Ferracin M., Fornari F., Zanesi N., Alder H., D'Elia G., Gramantieri L., et al. 2010. Oncogenic role of miR-483–3p at the IGF2/483 locus. Cancer Res. 70: 3140–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakagawa A., Shi Y., Kage-Nakadai E., Mitani S., Xue D. 2010. Caspase-dependent conversion of Dicer ribonuclease into a death-promoting deoxyribonuclease. Science. 328: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghodgaonkar M. M., Shah R. G., Kandan-Kulangara F., Affar E. B., Qi H. H., Wiemer E., Shah G. M. 2009. Abrogation of DNA vector-based RNAi during apoptosis in mammalian cells due to caspase-mediated cleavage and inactivation of Dicer-1. Cell Death Differ. 16: 858–868. [DOI] [PubMed] [Google Scholar]
- 24.Esau C., Davis S., Murray S. F., Yu X. X., Pandey S. K., Pear M., Watts L., Booten S. L., Graham M., McKay R., et al. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3: 87–98. [DOI] [PubMed] [Google Scholar]
- 25.Lee J., Kemper J. K. 2010. Controlling SIRT1 expression by microRNAs in health and metabolic disease. Aging. 2: 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamakuchi M., Ferlito M., Lowenstein C. J. 2008. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. USA. 105: 13421–13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Estep M., Armistead D., Hossain N., Elarainy H., Goodman Z., Baranova A., Chandhoke V., Younossi Z. M. 2010. Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment. Pharmacol. Ther. 32: 487–497. [DOI] [PubMed] [Google Scholar]
- 28.Alkhouri N., Gornicka A., Berk M. P., Thapaliya S., Dixon L. J., Kashyap S., Schauer P. R., Feldstein A. E. 2010. Adipocyte apoptosis, a link between obesity, insulin resistance, and hepatic steatosis. J. Biol. Chem. 285: 3428–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lembertas A. V., Perusse L., Chagnon Y. C., Fisler J. S., Warden C. H., Purcell-Huynh D. A., Dionne F. T., Gagnon J., Nadeau A., Lusis A. J., et al. 1997. Identification of an obesity quantitative trait locus on mouse chromosome 2 and evidence of linkage to body fat and insulin on the human homologous region 20q. J. Clin. Invest. 100: 1240–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erlacher M., Labi V., Manzl C., Bock G., Tzankov A., Hacker G., Michalak E., Strasser A., Villunger A. 2006. Puma cooperates with Bim, the rate-limiting BH3-only protein in cell death during lymphocyte development, in apoptosis induction. J. Exp. Med. 203: 2939–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malhi H., Barreyro F. J., Isomoto H., Bronk S. F., Gores G. J. 2007. Free fatty acids sensitise hepatocytes to TRAIL mediated cytotoxicity. Gut. 56: 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown B. D., Naldini L. 2009. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Natl. Rev. Genet. 10: 578–585. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.