Abstract
Lipid mediators are important in lung biochemistry and are derived from the enzymatic oxidation of arachidonic and docosahexaenoic acids, which are PUFAs that are present in phospholipids in cell membranes. In this study, MALDI imaging MS was used to determine the localization of arachidonate- and docosahexaenoate-containing phospholipids in mouse lung. These PUFA-containing phospholipids were determined to be uniquely abundant at the lining of small and large airways, which were unequivocally identified by immunohistochemistry. In addition, it was found that the blood vessels present in the lung were characterized by sphingomyelin molecular species, and lung surfactant phospholipids appeared evenly distributed throughout the lung parenchyma, indicating alveolar localization. This technique revealed unexpected high concentrations of arachidonate- and docosahexaenoate-containing phospholipids lining the airways in pulmonary tissue, which could serve as precursors of lipid mediators affecting airways biology.
Keywords: arachidonic acid, lipid localization, cryoembedding agent, lipid mediators, pulmonary tissue, mass spectrometry
Lipids play very important roles in lung biology as signaling molecules for inflammation (1), antiviral protectants (2), and as a structural component to prevent alveolar collapse (3). Leukotrienes and prostaglandins are lipid mediators that are involved in lung inflammation and are responsible for impairment of mucociliary clearance of particles, promotion of bronchoconstriction and mucous secretion, chemotactic recruitment of neutrophils to the airways, and facilitation of pulmonary edema (4, 5). In fact, drugs that are receptor antagonists of cysteinyl-leukotrienes or that interfere with cysteinyl-leukotriene production are frequently used in treating asthma (6). These lipid mediators are derived from the enzymatic hydrolysis of arachidonate-containing phospholipids, probably a phospholipase A2, and subsequent metabolism of free arachidonic acid to the oxygenated lipid mediator. Metabolites of docosahexaenoic acid are also emerging as important mediators of inflammation resolution in the lung (7), and likewise, the docosahexaenoate is stored in membrane phospholipids that must be hydrolyzed to generate free docosahexaenoic acid. Because these mediators act in an autocrine and paracrine manner, the specific localization of arachidonate- and docosahexaenoate-containing phospholipid precursors plays a critical role in driving the production of docosanoids, leukotrienes, and prostaglandins at local sites in the lung. In addition to phospholipids containing polyunsaturated fatty acyl groups, abundant phospholipids in the lung are present in lung surfactant (8, 9).
Chemical imaging techniques, such as immunohistochemistry and dye staining, are typically used to enable the detection and determination of the localization of biological compounds. However, the ability to image the location of specific lipids in tissues has been limited. Staining with Nile Red (10), Oil Red O (11), or BODIPY 493/503 (12) provides the localization of only the neutral lipids in a tissue. Antibodies for different phospholipid headgroups are available for studying their localization in tissues by immunohistochemical methods (13), but specific antibodies for a particular lipid molecular species of interest are not readily available. Neither of these methods provides spatial distribution information for specific lipid molecular species in a tissue, and consequently, there is a need for an effective method that indicates the localization and acyl chain composition of lipids in tissue. Localization of lipid molecular species in some tissues has been examined by microdissection of specific anatomical features followed by solvent extraction and LC/MS or LC/MS/MS analysis (14); however, the nonrigid nature of the lung, with interlaced architecture of airways and blood vessels of varying size and shape, makes this organ poorly suited for this approach.
MALDI imaging MS (IMS) has been developed as a powerful method for investigating the distribution of biomolecules within tissues (15). Analyte ions are produced directly from a tissue slice coated with MALDI matrix, and sequential mass spectra are acquired by rastering the laser beam across the tissue surface. This method is unique, compared with other imaging methods, because the distribution as well as the composition of many compounds of interest can be determined in a single experiment. Images can be obtained by plotting the intensity of the analytes of interest as a function of the x-y coordinates. Although much research in MALDI IMS has focused on the spatial distribution of proteins and peptides (15), the majority of ion current generated by MALDI IMS is related to desorption of phospholipids from the tissue (16). Recent studies have also revealed that the abundance of the phosphatidylcholine (PC)-related ions in MALDI IMS images is remarkably correlated with local concentrations of that PC lipid in tissue regions (17). Thus, MALDI IMS offers unique information about the molecular species present on a tissue without the need for specific probes or staining.
In order to preserve the normal lung architecture, most histological preparation methods require both inflation and embedding of the lung (18). In this study, it was determined that the traditional methods of lung histological preparations, agarose inflation with either formalin fixation paraffin embedding or OCT embedding, were incompatible with MALDI IMS of phospholipids, and, therefore, a new inflation and embedding agent was developed that was MALDI IMS compatible. Using MALDI IMS, phospholipid molecular species that contained either esterified arachidonic acid or docosahexaenoic acid were localized to specific anatomical regions of the lung.
MATERIALS AND METHODS
Reagents
Low-melting-point agarose was obtained from Promega (Madison, WI). Polyvinyl alcohol (PVA) 6-98, 2,5-dihydroxybenzoic acid (DHB), 2,6-dihydroxyacetophenone (DHAP), and sodium azide were purchased from Sigma Aldrich Chemical Co. (St. Louis, MO). OCT was obtained from Ted Pella, Inc. (Redding, CA). HBSS was purchased from Invitrogen (Carlsbad, CA). Polypropylene glycol, average molecular weight 2000 g/mol (PPG 2000), and organic solvents were purchased from Fisher Scientific (Fair Lawn, NJ).
Animals
C57BL6 mice were from the Jackson Laboratory (Bar Harbor, ME). The Institutional Animal Care and Use Committee at the University of Colorado Denver approved all animal experiments.
Preparation of modified OCT compound
A 10% PVA 6-98 solution was made by the addition of PVA 6-98 (10 g) to HBSS (100 ml). The solution was microwaved and then shaken for 16 cycles of 30 s each in order to dissolve the PVA 6-98. This 10% PVA 6-98 solution was allowed to cool to room temperature, then PPG 2000 (8 ml) and sodium azide (100 mg) were added to make the final modified OCT (mOCT): 10% (w/v) PVA 6-98, 8% (v/v) PPG 2000, 0.1% (w/v) NaN3.
Lung inflation and slicing
A solution of 1% low-melting-point agarose was prepared in HBSS by heating in a microwave oven. It was then cooled to 42°C and kept at that temperature until inflation. Following euthanasia of the mice by CO2 asphyxiation, lungs were deflated by puncture of the diaphragm and the front rib cage was removed. The trachea was cannulated using a catheter (18G Safelet Cath; Exelint International, Los Angeles, CA) and lungs were inflated with mOCT or agarose at 20 cm water pressure. The left lung was excised from the mouse, rinsed with HBSS, blotted dry, placed in a disposable cryomold embedded with OCT or mOCT, and then placed in a −20°C freezer. The embedded lung was mounted onto a cutting chock with OCT and sliced at a thickness of 12 μm with a cryostat (Reichert-Jung Cryocut 1800) at −20°C. The lung sections were placed onto glass coverslips and stored for no longer than 1 week at −20°C until matrix application and MALDI imaging. Additionally, another agarose-inflated left lobe was formalin-fixed and paraffin-embedded (FFPE) using standard procedures. The FFPE left lobe was sliced at a thickness of 4 μm with a microtome and placed onto glass coverslips. The coverslips were dipped into xylene twice to remove the paraffin before matrix application. Two slices of an FFPE lung from one mouse, 5 slices of an OCT-embedded lung from one mouse, and at least 15 different slices of an mOCT-inflated and -embedded lung from three different mice were analyzed by MALDI IMS. For the immunofluorescence experiments described below, adjacent sections were collected onto Superfrost slides.
Matrix application
The glass coverslips containing the lung sections were attached to a stainless steel MALDI plate using copper tape, and the DHB matrix was deposited by sublimation (19). The MALDI plate was attached to the condenser using double-sided thermally conductive tape (3M; St. Paul, MN). The conditions during sublimation of the DHB matrix were a pressure of 0.05 Torr, condenser temperature of 15°C (42 V applied to a heating mantle), and 275 mg DHB in the sublimator. The heat was applied for 11 min, which resulted in the deposition of a thin layer of DHB matrix. For negative-ion MALDI, imaging DHAP was used as the matrix and the conditions of sublimation were a pressure of 0.05 Torr, condenser temperature of 13°C (36 V applied to a heating mantle), and 150 mg DHAP in the sublimator. The heat was applied for 13 min, which resulted in the deposition of a thin layer of DHAP matrix. After MALDI imaging was completed, the glass coverslip containing the lung section was carefully removed from the MALDI plate and dipped in methanol to remove the matrix and fix the tissue prior to modified Giemsa staining, which was performed using standard protocols.
MALDI IMS
A quadrupole time-of-flight tandem mass spectrometer with an orthogonal MALDI source (QSTAR XL; Applied Biosystems/MDS Sciex, Thornhill, Ontario, Canada) was used to acquire images. MALDI mass spectra were obtained using a nitrogen laser (337 nm) at a power of 100 μJ and a pulse rate of 20 Hz with an accumulation time of 243 ms per image spot. The MALDI plate was moved at a rate of 12.75 mm/min, and after each horizontal line was completed, the plate was moved vertically 50 μm. The mass spectrometric data were processed using a specialized script for Analyst software (Applied Biosystems/MDS Sciex) at a mass resolution of 0.1 amu, and images were visualized using TissueView software (Applied Biosystems/MDS Sciex). The lateral resolution of this MALDI IMS technique is approximately 50 μm. Collisional activation of selected ions was carried out using a relative collision energy of 40 V, with argon as collision gas.
Lung homogenization, lipid extraction, and ESI-MS
The mouse left lung was homogenized in 1:1 methanol/HBSS (1 ml) and the phospholipids were then extracted according to the method of Bligh and Dyer (20). The extracted phospholipids from lung homogenate were resuspended in 60:20:20 (v/v/v) methanol-acetonitrile-water with 1 mM ammonium acetate for positive- and negative-ion MS analysis. Additionally, the PC lipids were separated by normal-phase HPLC from the other phospholipid classes (21) and resuspended in 60:20:20 (v/v/v) methanol-acetonitrile-water and infused into a Sciex API QTRAP mass spectrometer (PE Sciex; Toronto, Canada) at a flow rate of 5 μl/min. In the negative-ion mode, the experimental parameters used to obtain the product ion spectra were an electrospray voltage of −4,500 V, a declustering potential of −50 V, and a collision energy of −50 V.
Immunofluorescence
Sections adjacent to those analyzed by MALDI were used for dual immunofluorescence staining to aid in differentiation of airways from blood vessels in lung slices. Sections were placed at room temperature overnight, fixed in 10% neutral buffered formalin for 1 h at room temperature, and washed in deionized water. Antigens were retrieved by the microwave method (15 min, 1,200 W) in 0.01 mM citrate buffer (pH 6.0). Slides were cooled to room temperature, washed with deionized water, and equilibrated in PBS for 5 min. Sections were blocked in blocking buffer (5% FBS in PBS) for at least 1 h. Primary and secondary antibodies were diluted in blocking buffer. Antigens were detected with mouse IgG2b anti-acetylated tubulin (Sigma; 1:8,000) and goat IgG anti-thrombomodulin (CD141; R and D Systems, 1:300), overnight at 4°C. Sections were washed with deionized water and equilibrated in 1× PBS for 10 min. Antigen-antibody complexes were detected with 1:500 (in blocking buffer) dilutions of secondary antibodies: Alexa 488-conjugated donkey anti-mouse IgG2b and Alexa 594-conjugated donkey anti-goat IgG (Invitrogen). Slides were counterstained with 1 µg/ml 4',6-diamidino-2-phenylindole (DAPI). Images were acquired using a Zeiss Imager.Z1 fluorescent microscope (Zeiss) equipped with an Axiocam HRc black and white digital camera.
RESULTS
Inflation and embedding of lungs
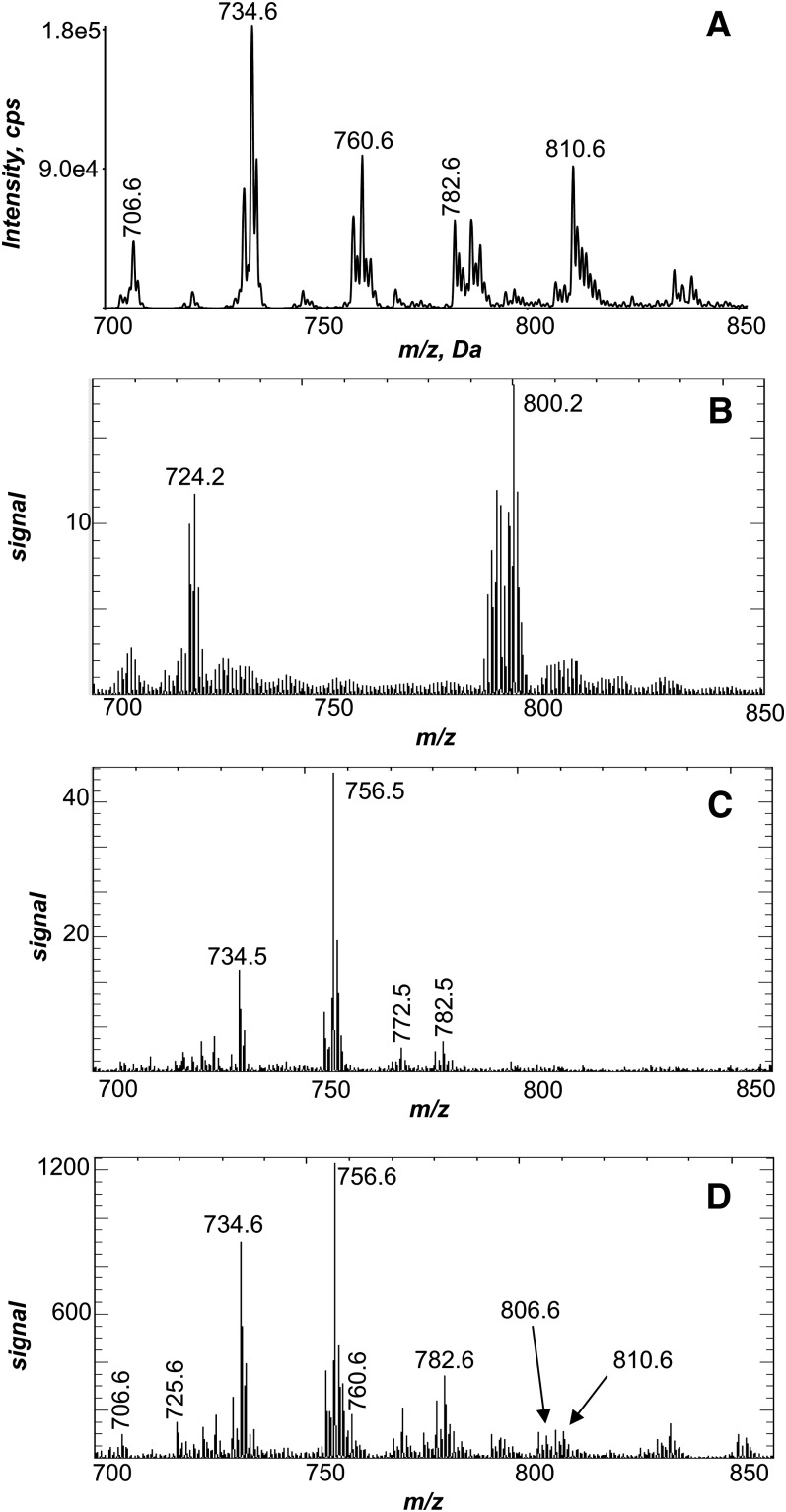
The positive-ion ESI MS of lipids solvent extracted from a mouse lung homogenate (Fig. 1A) indicated numerous PC molecular species present in this tissue as [M+H]+ observed between m/z 700–850. This population of PC molecular species was used to evaluate the initial attempts to image phospholipids in the mouse lung. Lung samples obtained for histological studies typically are inflated using low-melting agarose and either fixed with formalin and embedded in paraffin or embedded with OCT and frozen. The total positive ions obtained as a result of MALDI imaging of a FFPE lung resulted in the absence of phospholipid ion signal (Fig. 1B). For example, ions at m/z 734.6 or 810.6, present as abundant [M+H]+ in the lung homogenate (Fig. 1A), were not abundant ions generated during MALDI imaging of the FFPE lung and, furthermore, did not reveal lung anatomical structure when rendered into images (see supplementary ). The next attempt at MALDI imaging of lung phospholipids was with an agarose-inflated, OCT-embedded lung. The positive-ion MALDI mass spectrum of this preparation (Fig. 1C) indicated the presence of some phospholipids with low signal intensity, including a few of the major ions, such as m/z 734.6, present in the ESI mass spectrum of lung homogenate (Fig. 1A). The MALDI images obtained of phospholipids in the agarose-inflated, OCT-embedded lung were unimpressive, although some definition of the larger airways was observed (see supplementary ).
Fig.1.
Positive-ion electrospray and MALDI mass spectrometric analysis of lung. A: Positive-ion electrospray mass spectrum of the Bligh Dyer extract of a homogenized mouse lung. B: Total positive-ion MALDI mass spectrum of an agarose-inflated FFPE lung. C: Total positive-ion MALDI mass spectrum of an agarose-inflated, OCT-embedded lung. D: Total positive-ion MALDI mass spectrum directly off of an mOCT-inflated and -embedded lung.
OCT contains a benzalkonium salt as a preservative, which can suppress the ionization of analytes during the MALDI process (22), and adducts of benzalkonium with endogenous PC lipids have also been observed as ions in MALDI mass spectra (data not shown). Therefore, a new embedding agent was developed that supported the lung tissue while it was cut on a cryostat and caused minimal interference in the MALDI mass spectrum in the phospholipid mass region (m/z 700–900). The final mOCT composition was 10% PVA 6-98 and 8% PPG 2000, which was based on the OCT composition and a previous study on the formulation of cryoembedding media (23). The positive-ion MALDI mass spectrum of an mOCT-inflated and -embedded lung directly from the tissue (Fig. 1D) indicated the presence of numerous phospholipid molecular species, such as m/z 706.6, 734.6, 760.6, 782.6, and 810.6, which were also present in the positive-ion ESI mass spectrum of lung homogenate (Fig. 1A). Additionally, the MALDI mass spectrum of a lung slice (Fig. 1D) revealed an abundant ion at m/z 756.6 not present in the ESI mass spectrum of the extracted lung lipids. It is most likely that this ion corresponded to a sodiated PC species of m/z 734.6 by the addition of 22 daltons to the [M+H]+ species. Although the ESI mass spectrum of lung homogenate (Fig. 1A) was obtained by controlling the ionic composition of the electrospray solvent, the MALDI ions would be subject to local concentrations of alkali metal ions in the tissue that can generate abundant Na+ in the MALDI plume and adduct of this desorbed PC. In addition, the pulmonary morphology of the lung slices was preserved using mOCT when the stained slices were examined microscopically (see below). In general, the quality of the MALDI images obtained using mOCT were superior when compared with the MALDI images from FFPE and OCT-embedded lungs.
Positive-ion MALDI images
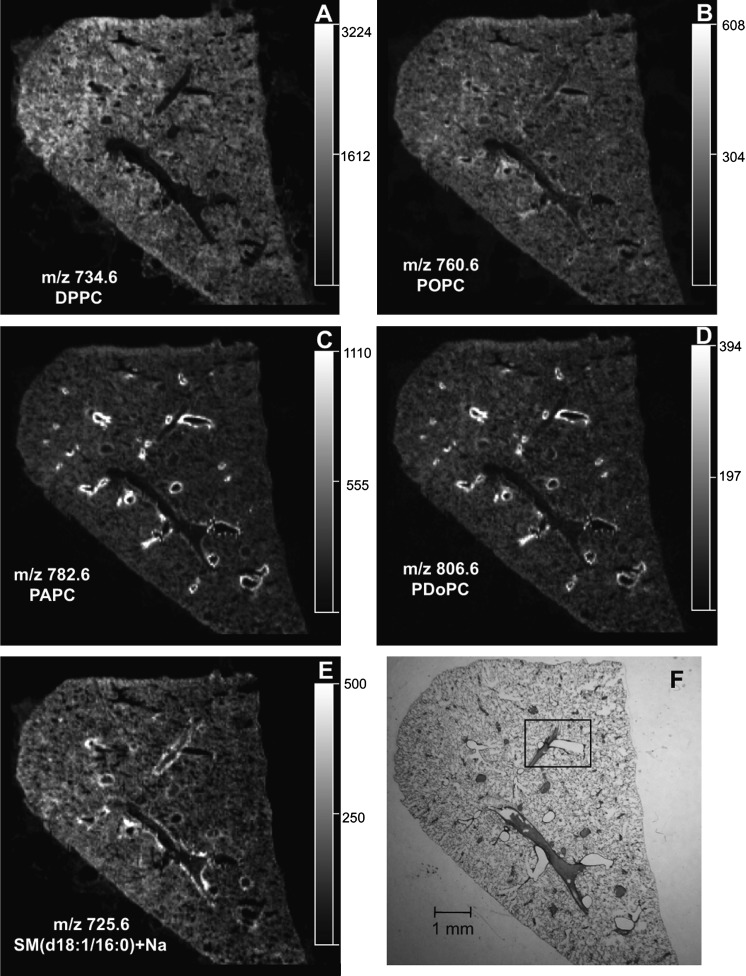
From the mass spectra obtained from each pixel across the lung tissue, an ion of interest can be extracted and an image of that particular ion distribution in the tissue can be visualized. The extracted-ion MALDI image of the most-abundant MALDI ion, m/z 734.6 (Fig. 2A) and its sodiated adduct at m/z 756.6 (data not shown), indicated a relatively uniform intensity in the lung parenchyma, as well as the positions of void regions where airways and vessels occurred when compared with the modified Giemsa stain (Fig. 2F) of this same tissue slice post-MALDI IMS. The positive-ion collision-induced dissociation (CID) mass spectra from the tissue-derived MALDI ion (see supplementary ) and the negative-ion electrospray CID of the corresponding [M-CH3]− ion (see supplementary ) were consistent with the identification of m/z 734.6 as dipalmitoyl-PC (DPPC) (24, 25). Collisional activation of m/z 756.6 yielded product ions at m/z 697.6 (loss of trimethylamine) and m/z 147 (sodiated cyclic 1,2-phosphodiester), characteristic of sodiated adducts of PCs (26) (data not shown). In addition, the positive-ion MALDI images of m/z 706.6, palmitoyl-myristoyl-PC (PMPC) (see supplementary ), and m/z 732.6, palmitoyl-palmitoleoyl-PC (PPoPC), (see supplementary ) had a distribution very similar to that of DPPC.
Fig.2.
The localization of PC and sphingomyelin lipids in lung tissue. Extracted positive-ion MALDI images of (A) DPPC (m/z 734.6), (B) POPC (m/z 760.6), (C) PAPC (m/z 782.6), (D) PDoPC (m/z 806.6), and (E) SM(d18:1/16:0)+Na (m/z 725.6) from an mOCT-inflated and -embedded mouse lung section. F: Modified Giemsa stain of the same lung after MALDI imaging.
The MALDI image of m/z 760.6 (Fig. 2B) revealed a relatively uniform intensity across the lung parenchyma, but a slightly higher intensity along the edges of anatomical structures that appeared to be airways. The ion at m/z 760.6 was identified as palmitoyl-oleoyl-PC (POPC) from positive-ion CID of the ion directly from the tissue (see supplementary ) and the corresponding negative-ion electrospray CID data (see supplementary ).
In sharp contrast, the MALDI images of m/z 782.6 (Fig. 2C) and m/z 806.6 (Fig. 2D) indicated that these phospholipid molecular ions were highly abundant along the edge of the putative airways observed in the modified Giemsa stain (Fig. 2F) and were also present in the alveolar region, albeit with lower abundance. The positive-ion CID data of these ions directly from the tissue on the MALDI plate (see supplementary ) and the negative-ion electrospray CID of the corresponding [M-CH3]− ions (see supplementary ) were consistent with the identification of m/z 782.6 and 806.6 as palmitoyl-arachidonoyl-PC (PAPC) and palmitoyl-docosahexaenoyl-PC, (PDoPC), respectively (24, 25). A similar MALDI image indicating a unique location at the edge of airways was observed in positive-ion mode for m/z 810.6 PC (stearoyl-arachidonoyl-PC) (see supplementary ). Additionally, the positive-ion CID spectra of m/z 782.6, 806.6, and 810.6 indicated that these ions were derived from a mixture of molecular species, owing to the appearance of an ion corresponding to a loss of trimethylamine and a sodiated cyclic 1,2-phosphodiester ion (26) and were probably composed of the sodium adduct of POPC, oleoyl-linoleoyl-PC, and stearoyl-oleoyl-PC, respectively. However, due to the low abundance of these characteristic sodiated PC ions in the positive-ion CID spectra, it was determined that these sodiated adducts were not the major PC species making up the ion abundance at each mass-to-charge ratio.
In addition to airways, blood vessels are also abundant anatomical features in lung tissue. This was readily observed in the modified Giemsa stain (Fig. 2F), in which most of the blood vessels were still filled with blood, since no perfusion was performed and the tissue was not rinsed. Many of the phospholipids observed in the positive-ion MALDI images of the lung did not highlight the blood vessels (Fig. 2A–D). However, the MALDI image of the ion at m/z 725.6 indicated the enrichment of this molecular species at the edges of the pulmonary blood vessels (Fig. 2E). The positive-ion CID spectrum for this ion directly from the tissue on the MALDI plate (see supplementary ) was consistent with the identification of m/z 725.6 as sodiated N-(hexadecanoyl)-sphingenine-phosphocholine [SM(d18:1/16:0)+Na)].
Immunohistochemistry
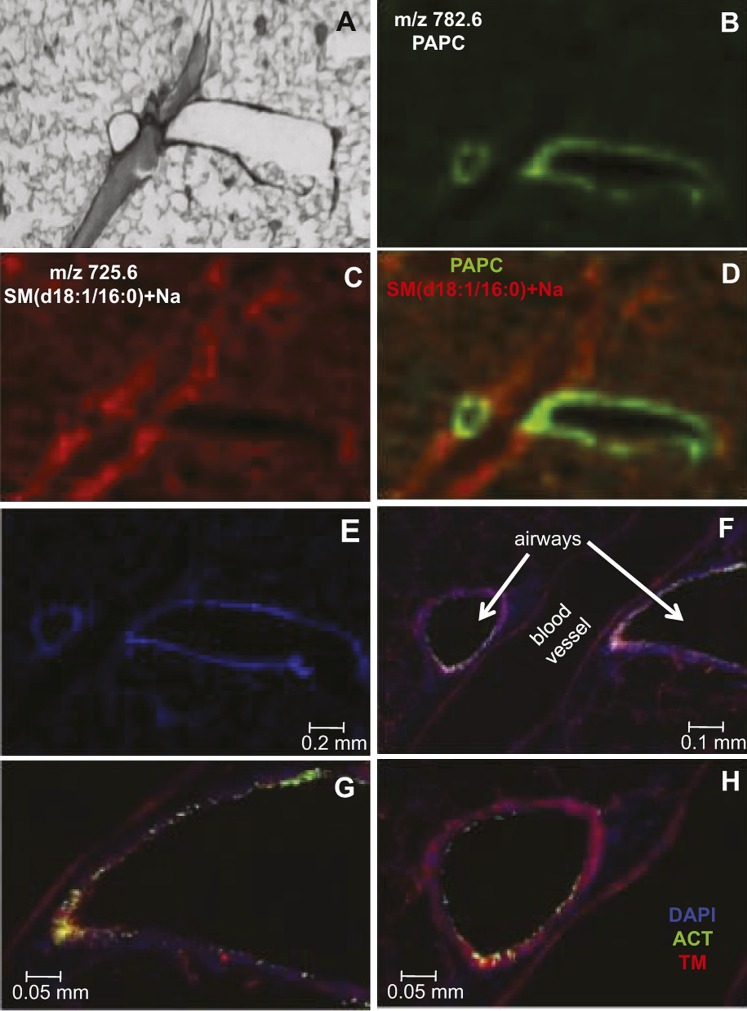
The blood vessels and airways in the lung were unequivocally identified by dual immunofluorescence on a lung section adjacent to that used for the MALDI imaging experiment. The modified Giemsa stain (Fig. 3A), along with the MALDI ion images of PAPC, m/z 782.6 (Fig. 3B), and SM(d18:1/16:0)+Na, m/z 725.6 (Fig. 3C), were magnified at the boxed-in region of the lung tissue shown in Fig. 2F. From these two separate positive-ion MALDI images (Fig. 3B,C), it appeared that PAPC and SM(d18:1/16:0)+Na were not colocalized, and when the positive-ion MALDI images were merged (Fig. 3D), there was a clear difference in the localization of PAPC (green) and SM(d18:1/16:0)+Na (red).
Fig.3.
The anatomical structures in the lung were unequivocally identified by dual immunofluorescence and compared with the MALDI images. A: Enlargement of the boxed part of the modified Giemsa stain of a section of the mOCT-inflated and -embedded mouse lung shown in Fig. 2F. Positive-ion MALDI image of PAPC (m/z 782.6) (B) and SM(d18:1/16:0)+Na (m/z 725.6) (C). D: Merged positive-ion MALDI image of PAPC (green) and SM(d18:1/16:0)+Na (red). E–H: Increasingly magnified images of an adjacent tissue section illustrating the localization of airways by acetylated tubulin (ACT, green), blood vessels by thrombomodulin (TM, red), and cell nuclei by DAPI (blue).
The adjacent lung section was stained to visualize acetylated tubulin (green), specific for airway ciliated cells, and thrombomodulin (red), specific for blood vessels. In addition, DAPI was used to visualize cell nuclei. The DAPI counterstain of this lung section illustrated the lacy appearance of the alveolar region, as well as the characteristic nuclear shape and density of blood vessel and major airway cells (Fig. 3E). Higher magnifications of the immunofluorescence images aided in the identification of two major airways and one large blood vessel (Fig. 3F–H). Additionally, capillaries present in the alveolar region were visualized with the thrombomodulin stain. MALDI IMS and immunofluorescence staining were also performed on adjacent lung sections that contained a main axial airway and a small blood vessel (see supplementary ). From these combined data, it was concluded that both small and major airways had PUFA-containing phospholipids and that both small and large blood vessels contained SM(d18:1/16:0)+Na.
Negative-ion MALDI images
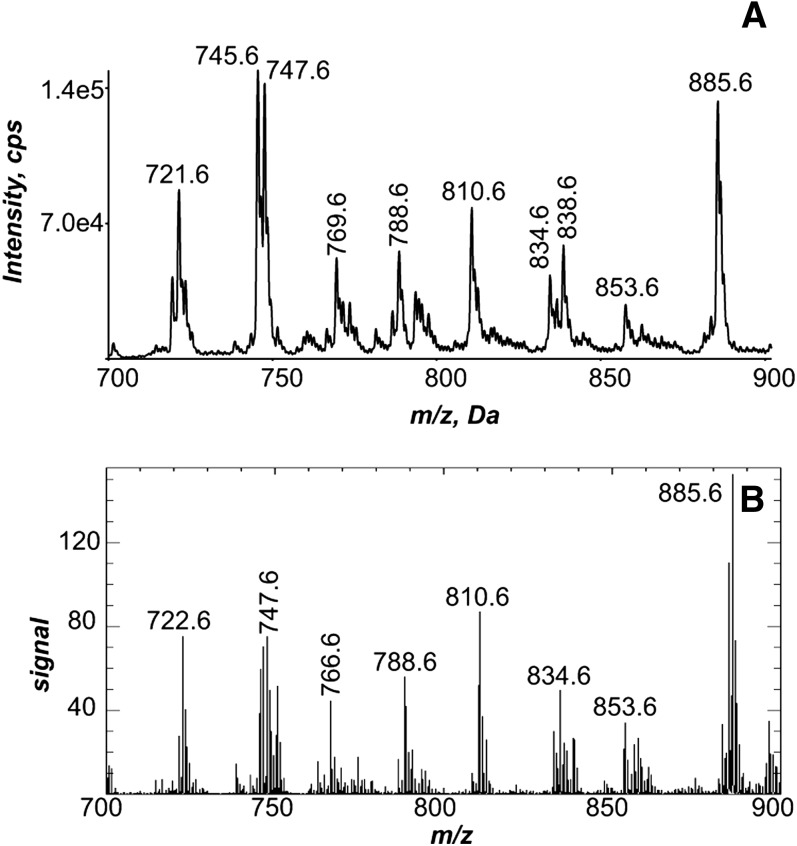
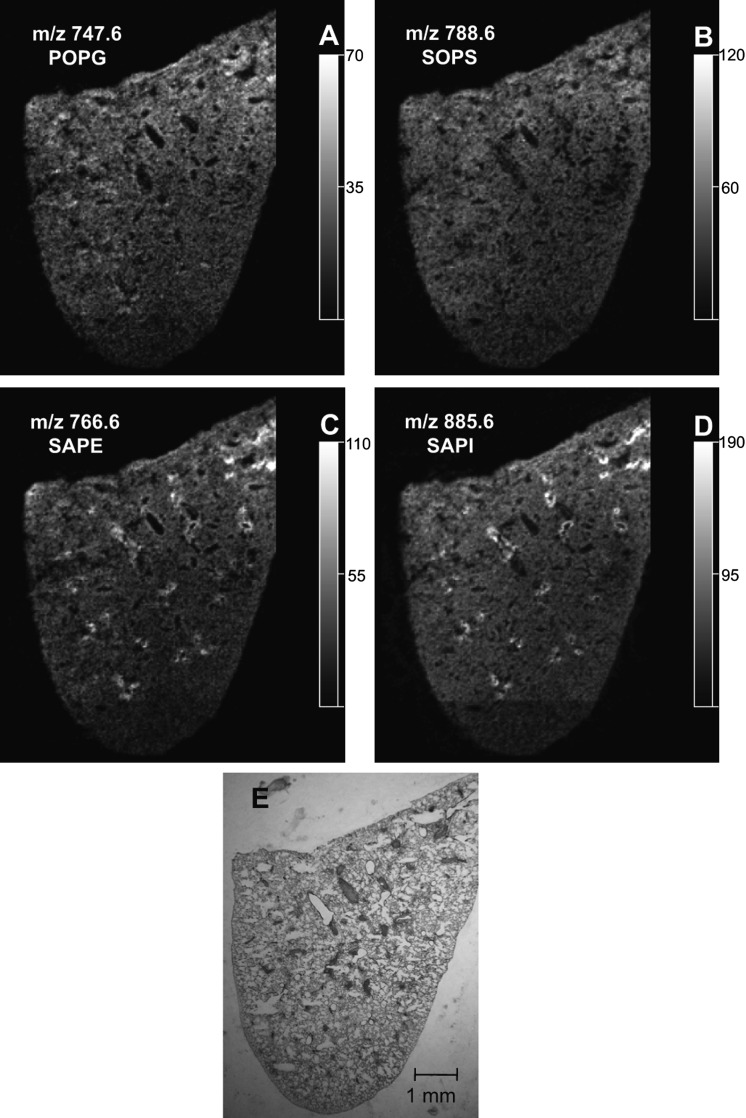
The phospholipid molecular species observed as abundant [M-H]− in the negative-ion ESI mass spectrum of solvent-extracted lipids from the lung homogenate (Fig. 4A) were quite similar to the molecular species present in the negative-ion MALDI mass spectrum of the entire left lung directly from the lung tissue (Fig. 4B). The identity of the phospholipids that generated these negative ions was deduced from collisional activation of each abundant [M-H]− ion in the ESI data (Fig. 4A), as well as the same ion observed in the MALDI IMS (Fig. 4B). For example, the negative-ion MALDI CID spectra obtained directly from the tissue (see supplementary Fig. VIA, B) were consistent with the identification of m/z 747.6 as palmitoyl-oleoyl-PG (POPG) and m/z 788.6 as stearoyl-oleoyl-PS (SOPS). The negative-ion MALDI CID spectra obtained directly from the tissue (see supplementary Fig. VIC, D) were consistent with the identification of m/z 766.6 as stearoyl-arachidonoyl-PE (SAPE) and m/z 885.6 as stearoyl-arachidonoyl-PI (SAPI).
Fig.4.
Negative-ion electrospray and MALDI mass spectrometric analysis of lung. A: Negative-ion electrospray mass spectrum of the Bligh Dyer extract of a homogenized mouse lung. B: Total negative-ion MALDI mass spectrum directly off of an mOCT-inflated and -embedded lung.
The negative-ion MALDI images reveal a unique distribution of phosphatidylglycerol (PG), phosphatidylserine, phosphatidylethanolamine (PE), and phosphatidylinositol (PI) lipid molecular species across the lung slice (Fig. 5). The MALDI images of m/z 747.6 and 788.6 (Fig. 5A, B) indicated that these molecular species are present in the lung parenchyma (Fig. 5E). Additionally, the negative-ion MALDI images of m/z 745.6, palmitoyl-linoleoyl-PG (PLPG) (see supplementary Fig. VIIA), and m/z 810.6, stearoyl-arachidonoyl-PS (see supplementary Fig. VIIB) indicated that these molecular species had a distribution very similar to that of both POPG and SOPS.
Fig.5.
The localization of PG, phosphatidylserine (PS), PE, and PI lipids in lung tissue. Extracted negative-ion MALDI images of (A) POPG (m/z 747.6), (B) SOPS (m/z 788.6), (C) SAPE (m/z 766.6), and (D) SAPI (m/z 885.6) from a section of mouse lung. E: Modified Giemsa stain of the same lung after MALDI imaging.
Additionally, the MALDI images of m/z 766.6 and 885.6 (Fig. 5C, D) indicated that these phospholipids were particularly abundant at the edges of the airways observed in the modified Giemsa stain (Fig. 5E). These molecular species were also present in the parenchyma, although with a much lower signal intensity. Similar MALDI images were observed in negative-ion mode for m/z 722.6, 1-O-hexadecenyl-arachidonoyl-PE (HAPE) (see supplementary Fig. VIIC) and m/z 857.6, palmitoyl-arachidonoyl-PI (see supplementary Fig. VIID). These negative-ion MALDI images of arachidonate-containing phospholipids in the lung were similar to those of the PUFA-containing PC lipids, with a signal highly enriched at the edges of the airways.
DISCUSSION
Most tissues that have been examined by MALDI IMS are nonporous and do not require inflation of the tissue or support by an embedding compound in order to obtain high-quality slices on the cryostat (27). One of the challenges in preparing lung tissue for MALDI IMS is the spongy nature of this organ. Frozen, noninflated lung tissue was very difficult to section on the cryostat, and the resulting sections had compromised morphology (e.g., compressed alveoli); it became clear that inflation of the lung was necessary in order to maintain the lung architecture. However, the two most-common lung preparation methods used for pulmonary histological studies, agarose inflation with FFPE or OCT embedding, did not provide useful MALDI images (see supplementary ). It is thought that the lack of phospholipid signal from an FFPE lung (Fig. 1B) is probably due to the ethanol dehydration step of the FFPE procedure, which would cause the phospholipids to be rinsed away. Additionally, the poor quality of the positive-ion MALDI image of the agarose-inflated, OCT-embedded lung (see supplementary ) was in part due to the presence of OCT, which has been shown to alter the ions observed in mass spectra (22), largely because the benzalkonium ion present as a preservative readily forms adduct ions with phospholipids and otherwise suppresses ion formation of species of interest. In order to achieve high-quality MALDI images of phospholipids in lung, it was necessary to develop new inflation and cryoembedding media. The use of the modified OCT cryoembedding agent, composed of 10% PVA 6-98 and 8% PPG 2000, resulted in MALDI mass spectra of phospholipids directly from the lung tissue (Figs. 1D and 4B) that were quite comparable to the ESI mass spectrum of homogenized lung (Figs. 1A and 4A). The mOCT was essential for obtaining MALDI images that readily indicated the distribution of specific phospholipids in the lung (Figs. 2, 5).
Lung surfactant, made by type II alveolar cells, is a complex mixture composed primarily of phospholipids (80–90%), with PG and PC lipids accounting for 10% and 85%, respectively, of the total phospholipids in pulmonary surfactant (28). The majority of PC in the pulmonary surfactant is present as DPPC (58%), PPoPC (13%), POPC (7.2%), and PMPC (4.7%) (29). Additionally, the PG lipids found in surfactant of most mammals are present as palmitoyl-palmitoleoyl-PG, POPG, and PLPG (30, 31). The MALDI images of these PG and PC surfactant lipids listed above (Figs. 2A, B, and 5A; see supplementary and VIIA) indicated a relatively uniform intensity of these molecular species in the lung parenchyma. The distribution of the surfactant phospholipids in the MALDI images is quite characteristic of surfactant because of its prolific presence in the alveolar region, which is where the surfactant is synthesized, stored, and secreted.
The MALDI images of PUFA-containing PC, PE, and PI lipids (Figs. 2C, D, and 5C, D and supplementary and VIIC, D) indicated that these particular molecular species are very intense at the edges of many airways. These PUFA-containing PC, PE, and PI lipids have been reported to be in very low abundance in pulmonary surfactant (31), and it is reasonable to conclude that the observed signal for PUFA-containing PC, PE, and PI lipids originated from pulmonary cellular membranes. From the unique location of the highest concentration of these phospholipids, it would appear that the epithelial lining of airways would be the most likely cellular origin; however, the spatial resolution of MALDI IMS (∼50 μm) was not adequate to ascertain the exact cellular source of the PUFA-containing PC, PE, and PI lipid signals in the MALDI images of lung tissue. The phospholipid composition of the many pulmonary cell types has not been extensively studied, but limited studies on the phospholipid content of tracheal epithelial cells (32) and human fetal type II alveolar epithelial cells (33) have indicated that the PUFA-containing PC, PE, and PI lipids observed in the MALDI images are present in these cells.
The discovery that arachidonic acid-containing lipids are localized at highest concentrations in specific regions of the lung is valuable, because these lipids are the precursors to leukotriene and prostaglandin production. Incorporation of arachidonic acid into phospholipids occurs through the Lands cycle (34) and is probably mediated by the lysophospholipid acyl transferases MBOAT5 and MBOAT7 (35). It is possible that the arachidonate-containing PI, PC, and PE observed at the edges of the airways by MALDI IMS could be a source of the arachidonic acid that is the substrate for 5-lipoxygenase or cyclooxygenases and the eventual production of lipid mediators that affect airway conductance relevant to both asthma (36) and chronic obstructive pulmonary disease (37). In a similar fashion, docosahexaenoate-containing phospholipids at this unique site might provide the precursor pool of the pro-resolution mediators from subsequent metabolism of this fatty acid (7).
For the first time, it was found that PUFA-containing phospholipids are present at high concentrations in the airways of pulmonary tissue in the mouse. Little is known about how such unique distributions of arachidonate- and docosahexaenoate-containing phospholipids change during alteration of biochemistry within the lung during lung disease or after exposure to environmental toxicants, but the MALDI IMS approach presented here makes it possible to investigate these questions.
Supplementary Material
Footnotes
Abbreviations:
- CID
- collision-induced dissociation
- DAPI
- 4',6-diamidino-2-phenylindole
- DHAP
- 2,6-dihydroxyacetophenone
- DHB
- 2,5-dihydroxybenzoic acid
- DPPC
- dipalmitoyl-PC
- FFPE
- formalin-fixed paraffin-embedded
- HAPE
- 1-O-hexadecenyl-arachidonoyl-PE
- IMS
- imaging MS
- mOCT
- modified OCT
- PAPC
- palmitoyl-arachidonoyl-PC
- PC
- phosphatidylcholine
- PDoPC
- palmitoyl-docosahexaenoyl-PC
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PLPG
- palmitoyl-linoleoyl-PG
- PMPC
- palmitoyl-myristoyl-PC
- POPC
- palmitoyl-oleoyl-PC
- POPG
- palmitoyl-oleoyl-PG
- PPG
- polypropylene glycol
- PPoPC
- palmitoyl-palmitoleoyl-PC
- PS
- phosphatidylserine
- PVA
- polyvinyl alcohol
- SAPE
- stearoyl-arachidonoyl-PE
- SAPI
- stearoyl-arachidonoyl-PI
- SM(d18:1/16:0)+Na
- sodiated N-(hexadecanoyl)-sphingenine-phosphocholine
- SOPS
- stearoyl-oleoyl-PS
This work was supported, in part, by grants from the Heart, Lung, and Blood Institute (HL-25785) and the LipidMAPS Large-Scale Collaborative Grant from the Institute of General Medical Sciences (GM069338) of the National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of seven figures.
REFERENCES
- 1.Stables M. J., Gilroy D. W. 2011. Old and new generation lipid mediators in acute inflammation and resolution. Prog. Lipid Res. 50: 35–51. [DOI] [PubMed] [Google Scholar]
- 2.Numata M., Chu H. W., Dakhama A., Voelker D. R. 2010. Pulmonary surfactant phosphatidylglycerol inhibits respiratory syncytial virus-induced inflammation and infection. Proc. Natl. Acad. Sci. USA. 107: 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griese M. 1999. Pulmonary surfactant in health and human lung diseases: state of the art. Eur. Respir. J. 13: 1455–1476. [DOI] [PubMed] [Google Scholar]
- 4.Samuelsson B., Dahlen S. E., Lindgren J. A., Rouzer C. A., Serhan C. N. 1987. Leukotrienes and lipoxins: structures, biosynthesis, and biological effects. Science. 237: 1171–1176. [DOI] [PubMed] [Google Scholar]
- 5.Lewis R. A., Austen K. F., Soberman R. J. 1990. Leukotrienes and other products of the 5-lipoxygenase pathway: biochemistry and relation to pathobiology in human diseases. N. Engl. J. Med. 323: 645–655. [DOI] [PubMed] [Google Scholar]
- 6.Riccioni G., Bucciarelli T., Mancini B., Di Ilio C., D'Orazio N. 2007. Antileukotriene drugs: clinical application, effectiveness and safety. Curr. Med. Chem. 14: 1966–1977. [DOI] [PubMed] [Google Scholar]
- 7.Serhan C. N., Chiang N. 2008. Endogenous pro-resolving and anti-inflammatory lipid mediators: a new pharmacologic genus. Br. J. Pharmacol. 153 (Suppl.): 200–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schürch S., Lee M., Gehr P. 1992. Pulmonary surfactant: surface properties and function of alveolar and airway surfactant. Pure Appl. Chem. 64: 1745–1750. [Google Scholar]
- 9.Kuronuma K., Mitsuzawa H., Takeda K., Nishitani C., Chan E. D., Kuroki Y., Nakamura M., Voelker D. R. 2009. Anionic pulmonary surfactant phospholipids inhibit inflammatory responses from alveolar macrophages and U937 cells by binding the lipopolysaccharide-interacting proteins CD14 and MD-2. J. Biol. Chem. 284: 25488–25500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenspan P., Mayer E. P., Fowler S. D. 1985. Nile red: a selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 100: 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Goor H., Gerrits P. O., Grond J. 1986. The application of lipid-soluble stains in plastic-embedded sections. Histochemistry. 85: 251–253. [DOI] [PubMed] [Google Scholar]
- 12.Rudolf M., Curcio C. A. 2009. Esterified cholesterol is highly localized to Bruch's membrane, as revealed by lipid histochemistry in wholemounts of human choroid. J. Histochem. Cytochem. 57: 731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossetto V., Spiezia L., Franz F., Salmaso L., Pozza L. V., Gavasso S., Simoni P. 2009. The role of antiphospholipid antibodies toward the protein C/protein S system in venous thromboembolic disease. Am. J. Hematol. 84: 594–596. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda K., Taguchi R. 2010. Highly sensitive localization analysis of gangliosides and sulfatides including structural isomers in mouse cerebellum sections by combination of laser microdissection and hydrophilic interaction liquid chromatography/electrospray ionization mass spectrometry with theoretically expanded multiple reaction monitoring. Rapid Commun. Mass Spectrom. 24: 2957–2965. [DOI] [PubMed] [Google Scholar]
- 15.Cornett D. S., Reyzer M. L., Chaurand P., Caprioli R. M. 2007. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat. Methods. 4: 828–833. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs B., Süss R., Schiller J. 2010. An update of MALDI-TOF mass spectrometry in lipid research. Prog. Lipid Res. 49: 450–475. [DOI] [PubMed] [Google Scholar]
- 17.Hankin J. A., Murphy R. C. 2010. Relationship between MALDI-MSI intensity and measured quantity of selected phospholipids in rat brain sections. Anal. Chem. 82: 8476–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myung J. K., Choe G., Chung D. H., Seo J. W., Jheon S., Lee C., Chung J. 2008. A simple inflation method for frozen section diagnosis of minute precancerous lesions of the lung. Lung Cancer. 59: 198–202. [DOI] [PubMed] [Google Scholar]
- 19.Hankin J. A., Barkley R. M., Murphy R. C. 2007. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 18: 1646–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 21.Berry K. A., Murphy R. C. 2005. Analysis of cell membrane aminophospholipids as isotope-tagged derivatives. J. Lipid Res. 46: 1038–1046. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz S. A., Reyzer M. L., Caprioli R. M. 2003. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J. Mass Spectrom. 38: 699–708. [DOI] [PubMed] [Google Scholar]
- 23.Cocco C., Melis G. V., Ferri G. L. 2003. Embedding media for cryomicrotomy: an applicative reappraisal. Appl. Immunohistochem. Mol. Morphol. 11: 274–280. [DOI] [PubMed] [Google Scholar]
- 24.Pulfer M., Murphy R. C. 2003. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 22: 332–364. [DOI] [PubMed] [Google Scholar]
- 25.Murphy R. C. 2002. Tables of molecular and product ions. In Mass Spectrometry of Phospholipids. Illuminati Press, Denver, CO: 26–53. [Google Scholar]
- 26.Murphy R. C., Hankin J. A., Barkley R. M. 2009. Imaging of lipid species by MALDI mass spectrometry. J. Lipid Res. 50 (Suppl.): 317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonnell L. A., Heeren R. M. 2007. Imaging mass spectrometry. Mass Spectrom. Rev. 26: 606–643. [DOI] [PubMed] [Google Scholar]
- 28.Veldhuizen R., Nag K., Orgeig S., Possmayer F. 1998. The role of lipids in pulmonary surfactant. Biochim. Biophys. Acta. 1408: 90–108. [DOI] [PubMed] [Google Scholar]
- 29.Bernhard W., Wang J. Y., Tschernig T., Tümmler B., Hedrich H. J., von der Hardt H. 1997. Lung surfactant in a cystic fibrosis animal model: increased alveolar phospholipid pool size without altered composition and surface tension function in cftrm1HG/m1HGU mice. Thorax. 52: 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang C. J., Postle A. D., Orgeig S., Possmayer F., Bernhard W., Panda A. K., Jürgens K. D., Milsom W. K., Nag K., Daniels C. B. 2005. Dipalmitoylphosphatidylcholine is not the major surfactant phospholipid species in all mammals. Am. J. Physiol. Regul. Integr. Comp. Physiol. 289: R1426–R1439. [DOI] [PubMed] [Google Scholar]
- 31.Postle A. D., Heeley E. L., Wilton D. C. 2001. A comparison of the molecular species compositions of mammalian lung surfactant phospholipids. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 129: 65–73. [DOI] [PubMed] [Google Scholar]
- 32.Holtzman M. J., Grunberger D., Hunter J. A. 1986. Phospholipid fatty acid composition of pulmonary airway epithelial cells: potential substrates for oxygenation. Biochim. Biophys. Acta. 877: 459–464. [DOI] [PubMed] [Google Scholar]
- 33.Postle A. D., Gonzales L. W., Bernhard W., Clark C. T., Godinez M. H., Godinez R. I., Ballard P. L. 2006. Lipidomics of cellular and secreted phospholipids from differentiated human fetal type II alveolar epithelial cells. J. Lipid Res. 47: 1322–1331. [DOI] [PubMed] [Google Scholar]
- 34.Lands W. E. M. 1960. Metabolism of glycerolipids. II. The enzymatic acylation of lysolecithin. J. Biol. Chem. 235: 2233–2237. [PubMed] [Google Scholar]
- 35.Gijón M. A., Riekhof W. R., Zarini S., Murphy R. C., Voelker D. R. 2008. Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. J. Biol. Chem. 283: 30235–30245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hallstrand T. S., Henderson W. R., Jr 2010. An update on the role of leukotrienes in asthma. Curr. Opin. Allergy Clin. Immunol. 10: 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kilfeather S. 2002. 5-Lipoxygenase inhibitors for the treatment of COPD. Chest. 121 (Suppl.): 197–200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.