Abstract
Serum coagulation factor X (FX) is proposed to play a major role in adenovirus tropism, promoting transduction by bridging the virus to cell-surface heparan sulfate proteoglycans (HSPGs). Both murine FX and human FX increased transduction by Ad.CMVfLuc, an adenovirus vector, in murine hepatocyte-like cells and human hepatocarcinoma cells. In contrast, only hFX increased transduction of several non-hepatic cancer cell lines and Chinese hamster ovary (CHO) cells. Not only was mFX unable to promote transduction in these cells, it competitively blocked hFX-enhanced transduction. Competition and HSPG digestion experiments suggested mFX- and hFX-enhanced transduction in hepatocyte-derived cells, and hFX-enhanced transduction in epithelial cancer cells were dependent on HSPGs. Ad·hFX-mediated transduction of CHO mutants unable to produce HSPGs was also curtailed. Hepatocyte-derived cells expressed substantially more HSPGs than the cancer cell lines. Dose-response curves and heparin-Sepharose binding suggested Ad·hFX has greater affinity for HSPGs than does Ad·mFX. In coagulation factor-depleted mice hFX also had enhanced ability, compared with mFX, to reconstitute hepatic adenovirus transduction. The results suggest that differences in Ad·hFX and Ad·mFX affinity to HSPGs may result in differences in their ability to enhance adenovirus transduction of many cells. These findings may have implications for murine models of adenovirus vector targeting.
Keywords: Adenoviruses, Blood Coagulation Factors, Gene Therapy, Glycoprotein, Heparan Sulfate
Introduction
Adenovirus (Ad)3 belongs to the Adenoviridea family of double-stranded DNA viruses (1). There are 55 known adenovirus serotypes, subdivided into 7 subgroups (A–G). Adenovirus serotype 5 (Ad5), which belongs to subgroup C, has been extensively studied as a vector for gene therapy, oncolytic therapy, and vaccine production (2).
Infection of cells in culture by Subgroup C adenoviruses, including Ad5, was thought to be predominantly, if not exclusively, initiated by binding of the virus fiber knob protein to the cellular Coxsackie and adenovirus receptor (3); entry is subsequently facilitated by interaction between the RGD-containing peptide on the virus penton base and cell-surface integrins (4). The Coxsackie and adenovirus receptor was also thought to be the cellular receptor responsible for the very efficient liver transduction that follows intravenous adenovirus injection into mice (5). However, modifications of the Ad5 fiber/knob that abolish Coxsackie and adenovirus receptor binding fail to reduce liver transduction (6). Recent studies suggest that binding of serum factors, including blood coagulation factor X, to the adenovirus capsid facilitates hepatic transduction in mice by bridging the virus to cell-surface receptors (7–10). Warfarin, which depletes FX, FIX, and other vitamin K-dependent serum factors, abrogates liver transduction and transduction by adenovirus vectors. Purified hFX, which binds with high affinity to the adenovirus hexon protein (8, 10), reconstitutes adenovirus liver transduction in warfarin-treated mice (11). These results suggest that liver transduction, in large part, is dependent on Ad-FX interactions in blood and is Coxsackie and adenovirus receptor-independent.
HSPGs have been suggested to be the receptors for adenovirus-FX complexes (8–11). HSPGs are glycoproteins that consist of a protein core post-translationally modified to contain one or more heparan sulfate chains, a type of sulfated glycosaminoglycan (12). HSPGs are found in abundance on virtually all mammalian cell plasma membranes, where they interact with a multitude of ligands, e.g. chemokines and cytokines, growth factors, lipoproteins, and proteases (13). HSPGs have also been proposed as receptors for several other pathogens, including adeno-associated virus (14, 15) and herpes simplex virus (16, 17). Because intravenous adenovirus comes in contact with FX in the blood, HSPGs may play a major role in adenovirus transduction of many cell types in vivo.
In this report, we compare the roles of murine FX (mFX) and human FX (hFX) on adenovirus transduction of hepatocyte/hepatoma-derived cell culture lines and on transduction of cell culture lines derived from non-hepatic epithelial tumors. Both mFX and hFX increase adenovirus transduction of hepatocyte/hepatoma-derived cells; however, only hFX, but not mFX, enhances adenovirus transduction of the epithelial tumor cell lines. HSPGs are necessary for mFX- and hFX-enhanced adenovirus transduction in all cells examined. Competition and heparin-column binding experiments suggest that the differences between mFX- and hFX-enhanced adenovirus transduction result from a greater affinity of the Ad·hFX complex for heparan sulfate. In vivo, as observed in the cell culture experiments, adenovirus liver transduction was more effectively restored by hFX reconstitution than by mFX reconstitution in warfarin-treated mice.
EXPERIMENTAL PROCEDURES
Virus and Reagents
Ad.CMVfLuc, a non-replicating adenovirus in which the cytomegalovirus promoter drives firefly luciferase expression (18), was propagated on 293A cells (Invitrogen) and purified by two sequential cesium chloride gradient centrifugation steps and dialysis against 3% sucrose. The virus particle titer (particles/ml) was determined by measuring absorbance at 260 nm. Viral infectious-unit titers (IU/ml) were determined with the Adeno-X Rapid Titer kit (Clontech, Mountain View, CA).
Human and murine Factors X were purchased from Hematologic Technologies (Essex Junction, VT). Polyhistidine-tagged forms of heparinase I, II, and III were purified by Ni2+-affinity chromatography and gel filtration. Heparin, warfarin, and lactoferrin from human milk were obtained from Sigma. Heparin-Sepharose was obtained from GE Healthcare.
Cells and Culture Conditions
HepG2 cells (human hepatocellular carcinoma), AML12 cells (murine hepatocyte-derived cell line), and HT29 cells (human colorectal adenocarcinoma) were from the American Type Culture Collection (ATCC HB-8065, CRL-2254, and HTB-38). MC38 cells (murine colon carcinoma) were provided by Dr. Jeffrey Schlom (National Institutes of Health). LS174T cells (human colon adenocarcinoma) were provided by Dr. Anna Wu (UCLA), and WAS17.6 cells (murine breast cancer) were obtained from Dr. Luisa Iruela-Arispe (UCLA).
HepG2 cells were maintained in ATCC-formulated Eagle's minimum essential medium (ATCC) supplemented with 10% FBS and 1% penicillin-streptomycin. AML12, HT29, LS174T, and MC38 cells were grown in DMEM containing 10% FBS and 1% penicillin-streptomycin, 0.1 mm nonessential amino acids, and 1.0 mm sodium pyruvate. WAS17.6 cells were maintained in DMEM containing 10% FBS and 1% penicillin-streptomycin. Chinese hamster ovary cells (CHO-K1, ATCC CCL61) and CHO mutants in glycosaminoglycan biosynthesis (pgsA-745, pgsD-677, pgsE-606, and pgsF-17) were cultured in Ham's F-12 medium containing 1% penicillin-streptomycin and 8% FBS. All media and supplements were obtained from Invitrogen. All cells were maintained and grown at 37 °C in 5% CO2/95% air.
In Vitro Adenovirus Vector Transduction Protocols
Ad.CMVfLuc transductions were performed in 24-well plates, seeded with 5 × 104-1 × 105 cells per well 48 h earlier. Virus particles (3 × 108 particles/well) were diluted in serum-free Opti-MEM (Invitrogen, 200 μl/well) and incubated with the cells at 37 °C. After 90 min, virus was removed, and medium was replaced with complete culture medium (including 10% FBS). Following transduction (times as indicated in the figure legends and/or under “Results”) cells were washed with PBS and lysed in Passive Lysis Buffer (Promega, Madison, WI) for 15 min at room temperature. Luciferase activity was determined from cell lysates, using a luciferase assay system kit (Promega) and Luminat LB9501 instrumentation.
In some experiments the serum-free Opti-MEM medium was supplemented with hFX or mFX; concentrations are indicated in the figure legends and/or under “Results.” In experiments that include heparin or lactoferrin blocking reagents, these reagents were added to the culture medium containing virus and coagulation factors immediately before the medium was added to the cells. Heparin and lactoferrin concentrations are indicated in the figure legends and/or under “Results.”
To remove heparan sulfate from the cell surface, a mixture of recombinant heparin lyase I (2 milliunits/ml), II (2 milliunits/ml), and III (10 milliunits/ml) prepared in serum-free Opti-MEM was added to the cells. After 15 min at 37 °C, the medium containing heparinases was removed, cells were washed with PBS, and medium containing virus was added to the cells. As for other experiments, the virus was removed after 90 min and replaced with complete culture medium. Control experiments, performed following procedures described previously (19), showed that heparinase digestion removed >90% of cell-surface heparan sulfate, judged by reduction of binding of biotinylated FGF2 (data not shown) (17).
Heparan Sulfate Proteoglycan Analysis of Cultured Cells
To quantify and characterize heparan sulfate present in the cell lines, AML12, HepG2, MC38, LS174T, and CHO cells were cultured in 150-mm plates. Once the cells reached densities equivalent to that used for virus transduction, they were washed with PBS and treated with 0.05% trypsin/0.53 mm EDTA for 5 min at 37 °C to cleave HSPGs from the cell surface. At least 108 cells for each cell line were used for enzyme digestion. The cells were counted, then sedimented at 300 × g (10 min). Cell pellets were washed with PBS. Trypsin supernatants containing cell-surface heparan sulfate and cell pellets were analyzed separately; disaccharide compositions, sulfate groups, and total amounts of heparan sulfate were determined by glycan reductive isotope labeling LC/MS (20).
Heparin-Sepharose Chromatography and Immunoblotting
To determine Ad·FX binding to heparin, BioSpin columns (Bio-Rad) were prepared with heparin-Sepharose beads and equilibrated with wash buffer (20 mm Hepes, pH 7.0/150 mm NaCl/2.5 mm CaCl2). Ad.CMVfLuc (2 × 1010 particles) was mixed with either hFX or mFX (10 μg), with 100 μg of BSA as carrier protein, in a final volume of 200 μl of wash buffer and loaded onto the column. The column was washed with wash buffer, and then eluted stepwise with buffers containing increasing concentrations of NaCl. For immunoblotting, adenovirus proteins in the eluates were separated on SDS-PAGE gels and transferred onto nitrocellulose membranes. The membranes were blocked with 5% milk proteins in 1× PBS and 0.5% Tween 20, then probed with primary rabbit anti-adenovirus 5 antibody (kindly provided by Dr. Arnold Berk, UCLA), followed by HRP-conjugated secondary antibody. Proteins were visualized using ECL reagent (Pierce). For quantification, membranes were scanned with Typhoon instrumentation at the Biological Chemistry Imaging Facility at UCLA according to the manufacturer's instructions, and bands were graphed as relative volume densities.
mFX- and hFX-mediated Adenovirus Hepatic Transduction in Warfarinized Mice
All animal experiments were conducted according to guidelines of the UCLA animal care committee. Female C57BL/6 mice (8–10 weeks old, ∼30 g), were obtained from Jackson Laboratories. Warfarin (300 μg/mouse, in peanut oil) was administered subcutaneously 3 days and again 1 day prior to Ad.CMVfLuc·FX injection (11). Groups of 3–5 mice were injected intravenously via the tail vein with human or murine FX and Ad.CMVfLuc (3 × 1010 particles/mouse). Livers were removed 3 days later, homogenized in Passive Lysis Buffer (Promega), and analyzed for luciferase activity, using a luciferase assay system kit (Promega) and Luminat LB9501 instrumentation. Luciferase activities were normalized against protein content.
Statistical Analysis
All experiments were performed a minimum of three times, each time with triplicate samples. Data are expressed as means ± S.D. Data were compared between groups with an unpaired Student's t test or one-way analysis of variance, as appropriate. To compare luciferase activities in livers of mice reconstituted with either mFX or hFX, we first performed a log transformation of luciferase measurements. Next, a two-way analysis of variance model was conducted for luciferase activity, which included the main effects of FX dose and treatment type (mFX versus hFX). Individual dose levels were compared between treatment groups by t test. All p values less than p < 0.05 were considered significant. Statistical analyses were performed using GraphPad Instat version 3.01 and S-plus version 8 (TIBCO, Palo Alto, CA).
RESULTS
hFX, but Not mFX, Increases Adenovirus Transduction in Non-hepatic Epithelial Tumor Cell Lines
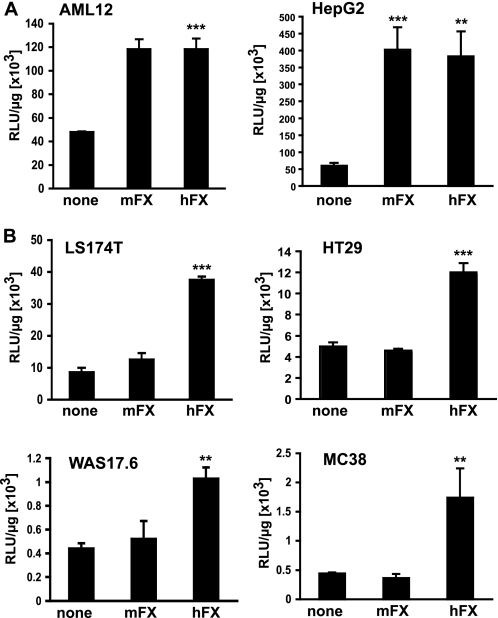
Adenovirus transduction of hepatocytes in vivo has recently been postulated to be facilitated by initial FX binding to the viral hexon protein and subsequent FX-mediated bridging of the virus to cell-surface hepatocyte HSPGs (8–10). Consistent with these findings, both human and murine FX at physiological concentrations increased by 2- to 4-fold the transduction of murine AML12 hepatocytes and human hepatocarcinoma HepG2 cells in cell culture by Ad.CMVfLuc (an adenovirus vector expressing firefly luciferase from the CMV promoter) (Fig. 1A). In contrast, adenovirus transduction of human (LS174T colon adenocarcinoma and HT29 colorectal adenocarcinoma) and murine (MC38 murine colon cancer and WAS17.6 murine breast cancer) epithelial tumor cell lines was increased by hFX, but not mFX (Fig. 1B).
FIGURE 1.
Adenovirus transduction in the presence of mFX and hFX in liver-derived cells and in non-hepatic epithelial cancer cells. A, Ad.CMVfLuc transduction of AML12 murine hepatocyte-derived cells and HepG2 human hepatocellular carcinoma cells in the presence of mFX or hFX (10 μg/ml). B, Ad.CMVfLuc transduction of human LS174T (colorectal carcinoma) and HT29 (colorectal carcinoma) and murine MC38 (colon cancer) and WAS17.6 (breast cancer) cell lines in the presence of mFX or hFX (10 μg/ml). Luciferase activity was analyzed in the cell lysates 48 h post transduction. All data are expressed as relative luciferase units (RLU) per microgram of protein. Values are means ± S.D. (statistics are indicated relative to transduction in serum-free medium; ***, p < 0.001; **, p < 0.01).
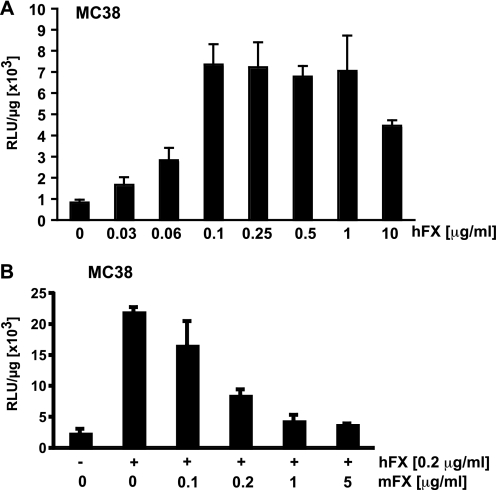
The difference in mFX- versus hFX-mediated adenovirus transduction of non-hepatic epithelial tumor cells might result either from differences in mFX and hFX interactions with the virus capsid or, alternatively, differences in interaction of the Ad·hFX and Ad·mFX complexes with cells. However, Kalyuzhniy et al. (10) demonstrated, by surface plasmon resonance analysis, that hFX and mFX bind with equal affinities to the Ad5 hexon protein, suggesting that the observed difference between mFX and hFX in enhancing adenovirus transduction might result from different interactions with their respective cell-surface receptors. To investigate this difference in the context of Ad.CMVfLuc transduction of non-hepatic epithelial tumor cells, we first ascertained the minimum amount of hFX required to maximally enhance MC38 cell transduction by the virus and then determined if/how efficiently mFX could interfere with hFX-mediated enhancement of Ad.CMVfLuc transduction. Maximal transduction enhancement by hFX was observed at 0.1 μg/ml (Fig. 2A), ∼1% of the level of hFX present in serum (8–10 μg/ml). Murine FX reduced hFX-mediated enhancement of MC38 cell transduction by adenovirus in a dose-dependent manner (Fig. 2B). A 1:1 ratio of mFX:hFX reduced hFX-mediated transduction by half, suggesting that (i) as reported (10), mFX and hFX bind to the adenovirus capsid with equivalent affinities and (ii) the difference in mFX and hFX in mediating transduction of the various cell lines reflects a difference in the recognition of cell-surface receptors by Ad·mFX and Ad·hFX complexes.
FIGURE 2.
hFX-mediated adenovirus transduction of MC38 murine colorectal cancer cells in the presence of increasing amounts of mFX. A, Ad.CMVfLuc transduction of MC38 cells in the presence of increasing hFX concentrations. B, MC38 cells were transduced with Ad.CMVfLuc in the presence of hFX (0.2 μg/ml) and increasing amounts of mFX. Luciferase activity was analyzed in the cell lysates 24 h post transduction. Data are expressed as relative luciferase units (RLU) per microgram of protein. Values are means ± S.D.
FX-enhanced Adenovirus Transduction of Cultured Cells Is Mediated by Cell Surface Heparan Sulfate Proteoglycans
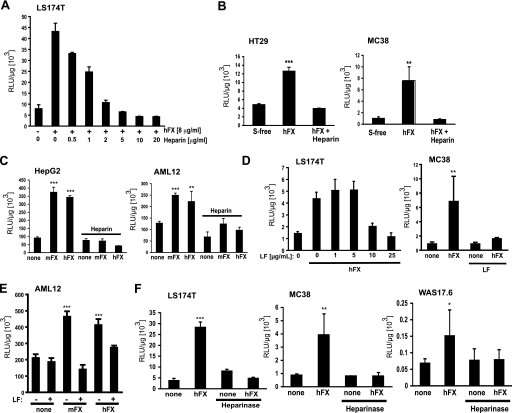
HSPGs have been proposed as cellular receptors for Ad·FX-mediated transduction (8–11). Whether HSPGs play a role in hFX and/or mFX-mediated adenovirus transduction in the cell lines used in this study was investigated by three approaches: (i) by using heparin as a soluble receptor analog for competitive inhibition of Ad.CMVfLuc transduction, (ii) by blocking adenovirus transduction with lactoferrin, a ligand that binds to heparan sulfate (21), and (iii) by enzymatic removal of heparan sulfate from the cell surface with heparin lyases prior to adenovirus vector transduction.
Heparin, a highly sulfated form of heparan sulfate (12), blocked hFX-enhanced Ad.CMVfLuc transduction in murine and human cancer cell lines (Fig. 3, A and B). Heparin also effectively blocked both hFX- and mFX-enhanced Ad.CMVfLuc transduction in hepatocyte/hepatoma-derived cells (Fig. 3C). Like heparin (Fig. 3, A–C), lactoferrin efficiently blocked Ad·hFX-mediated transduction of epithelial tumor cells (Fig. 3D). Lactoferrin also blocked both hFX-mediated and mFX-mediated increased transduction of hepatocyte/hepatoma-derived cells (Fig. 3E). To further confirm a role for heparan sulfate in Ad·hFX transduction, we pretreated human and murine cancer cell lines and hepatocyte/hepatoma-derived cells with enzymes that digest cell-surface HSPGs and then examined hFX-facilitated and mFX-facilitated Ad.CMVfLuc transduction. Heparin lyases I, II, and III cleave distinct linkages found in heparan sulfate, leaving behind the protein cores of proteoglycans while removing sulfated glycosaminoglycan chains. Treatment of MC38, WAS17.6, and LS174T cells and non-hepatocyte epithelial tumor cells with a mixture consisting of the three lyases prior to adenovirus transduction decreased hFX-enhanced transduction to baseline values (Fig. 3F). Heparin lyase pretreatment also led to an abrogation of FX-enhanced transduction by both Ad.CMVfLuc·hFX and Ad.CMVfLuc·mFX in human HepG2 and murine AML12 hepatocyte/hepatoma-derived cells (data not shown). The other common sulfated glycosaminoglycans present on cell membranes, in addition to heparan sulfate, are chondroitin sulfate and dermatan sulfate (12). Pretreatment with chondroitinase ABC, which degrades both chondroitin sulfate and dermatan sulfate, but does not degrade heparan sulfate, had no effect on FX-mediated enhancement of adenovirus transduction (data not shown).
FIGURE 3.
The role of heparan sulfate in FX-mediated adenovirus transduction of cancer cell lines and hepatocyte/hepatoma-derived cell lines. A, heparin blocks hFX-mediated enhancement of adenovirus transduction of LS174T human colorectal cancer cells in a dose-dependent fashion. B, heparin (10 μg/ml) blocks hFX-mediated enhancement of adenovirus transduction of HT29 (human colorectal) and MC38 (murine colon) cancer cell transduction. C, heparin (10 μg/ml) blocks hFX-mediated and mFX-mediated enhancement of adenovirus transduction of HepG2 human hepatocellular carcinoma cells and AML12 murine hepatocyte-derived cells. D, lactoferrin (LF) blocks hFX-mediated enhancement of adenovirus transduction of LS174T (human) and MC38 (mouse) colon cancer cells. For MC38 cells, lactoferrin concentration was 25 μg/ml. E, lactoferrin (25 μg/ml) blocks both mFX- and hFX-mediated enhancement of adenovirus transduction of AML12 hepatocyte-derived cells. F, heparinase pretreatment prevents hFX-mediated adenovirus transduction of MC 38 cells, WAS17.6 cells, and LS174T cells. FX concentration was 10 μg/ml. Cells were harvested 24 h (A) or 48 h (B–F) after Ad.CMVfLuc addition, and lysates were analyzed for luciferase activity. Values are means ± S.D. relative to transduction in serum-free medium (***, p < 0.001; **, p < 0.01; *, p < 0.05).
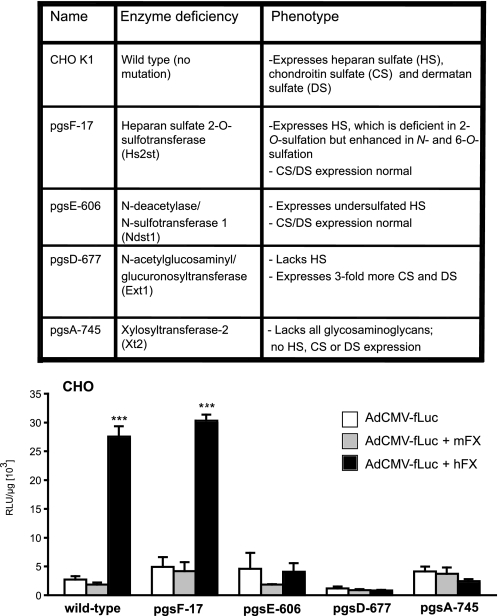
To corroborate data suggesting that Ad·FX complexes bind to cell-surface heparan sulfate, we examined a set of CHO cells genetically modified to lack specific enzymes of glycosaminoglycan biosynthesis (22) (inserted table, Fig. 4). Wild-type and mutant CHO cells were transduced with Ad.CMVfLuc in the presence or absence of hFX or mFX (Fig. 4). hFX strongly enhanced adenovirus transduction of wild-type CHO cells; in contrast, as observed for non-hepatic epithelial cancer cell lines, mFX failed to increase Ad.CMVfLuc transduction in these (or any other) CHO cells. hFX was unable to enhance Ad.CMVfLuc transduction in mutant CHO cells that lack all glycosaminoglycans (pgsA-745), lack only heparan sulfate but produce more chondroitin sulfate (pgsD-677), or express globally undersulfated heparan sulfate (pgsE-606). However, hFX enhanced adenovirus transduction of pgsF-17 cells, which lack the 2-O-sulfotransferase; hFX does not appear to require 2-O-sulfation of cell-surface heparan to facilitate adenovirus binding. These results confirm experiments suggesting that hFX mediates increased adenovirus transduction of cells (CHO, human epithelial cancer cells, and human and mouse hepatocyte/hepatoma-derived cell lines) primarily, if not exclusively, through heparan sulfate.
FIGURE 4.
hFX-mediated enhancement of adenovirus transduction of CHO cells mutated in heparan sulfate biosynthesis. The table identifies the enzymes mutated in the CHO lines and their resulting phenotypes. All cell lines were transduced with Ad.CMVfLuc in the absence of FX or in the presence of physiological (10 μg/ml) amounts of mFX or hFX. Luciferase activity was measured in cell lysates 48 h after virus addition. Values are means ± S.D., relative to wild-type CHO cell transduction in the absence of FX (***, p < 0.001).
Human FX Displays a Stronger Affinity than Murine FX for Heparan Sulfate
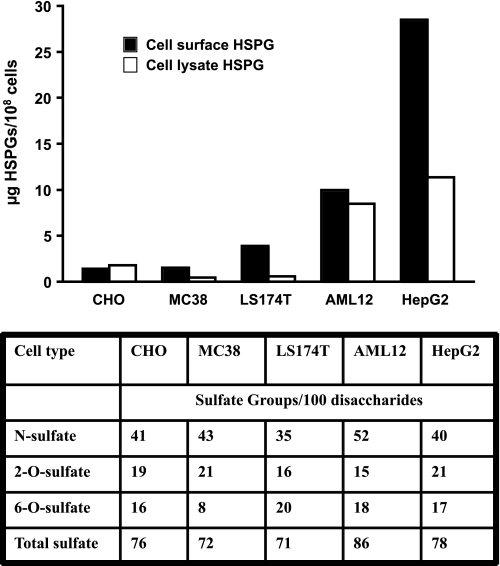
mFX was able to increase adenovirus transduction in HepG2 cells and AML12 cells (Fig. 1), but not in the non-hepatic derived epithelial cancer cell lines (Fig. 1) or in CHO cells (Fig. 4). To understand the basis for differential facilitation of adenovirus transduction of these cell lines by hFX versus mFX, we compared the composition of heparan sulfate derived from cell-surface proteoglycans. The density and arrangement of sulfate groups on heparan sulfate, which varies across cell types and tissues, can be assessed quantitatively by determining the disaccharide composition of the chains after heparin lyase treatment and LC/MS (20). Although some variation in the content of O-sulfate and N-sulfate groups was noted, the differential susceptibility of the cell lines to Ad.CMVfLuc transduction did not correlate with the content of O-sulfate or N-sulfate, or with the total number of sulfate groups per chain (table of Fig. 5). However, hepatocyte-derived AML12 and HepG2 human hepatoma cells express 2-fold and 8-fold more heparan sulfate on their cell surface, respectively, than do CHO or non-hepatic epithelial cancer cells (Fig. 5). These data suggest that mFX may mediate increased adenovirus transduction only on cells that exhibit a relatively high amount of heparan sulfate on their cell surface; lower cell-surface heparan sulfate levels may be sufficient for hFX-mediated adenovirus transduction, but not for mFX-mediated transduction.
FIGURE 5.
Heparan sulfate proteoglycan content of cell lines. CHO cells, MC38 colon cancer cells, LS174T colon cancer cells, AML12-hepatocye-derived cells, and HepG2 hepatoma-derived cells were trypsinized extensively to remove HSPGs from the cell surface. The cells were then washed and counted, and the heparan sulfate concentrations in both the lysed cell pellets and the cell surface/trypsin-supernatants were determined. The table describes, for heparan sulfate liberated from the cell surface, the percentages of N-, 2-O-, and 6-O-sulfation, and the average number of sulfate groups per disaccharide subunit.
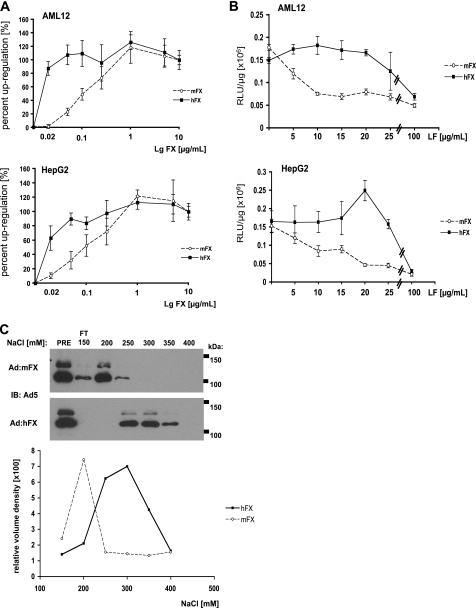
If the Ad·hFX complex can bind more effectively than the Ad·mFX complex to cell-surface HSPGs, then adenovirus with limiting amounts of hFX bound should more effectively transduce cells with high levels of HSPG than will adenovirus with limiting amounts of bound mFX (recall that hFX and mFX bind to the Ad5 with equal affinities (10)). Indeed, substantially less hFX is required to obtain maximal Ad.CMVfLuc-enhanced transduction of AML12 and HepG2 cells, when compared with mFX (Fig. 6A); the ED50 for hFX-mediated enhancement of Ad.CMVfLuc transduction for both AML12 and HepG2 cells was <0.02 mg/ml, whereas the ED50 for mFX-enhanced transductions of both cell lines was ∼0.1 mg/ml.
FIGURE 6.
The adenovirus·hFX complex has greater affinity for heparan sulfate than does the adenovirus·mFX complex. A, AML12 and HepG2 cells were transduced with Ad.CMVfLuc plus increasing concentrations of hFX or mFX. Values are averages of the percentage of the maximum FX-enhanced transduction (“0” is luciferase activity of cells transfected with virus alone; 100% is luciferase activity of cells transfected with virus plus 10 μg mFX or hFX/ml). B, AML12 and HepG2 cells were transduced with Ad.CMVfLuc in the presence of hFX or mFX (10 μg/ml) and increasing amounts of lactoferrin. For A and B, luciferase specific activity was measured in cell lysates 48 h after virus addition. Values are means ± S.D. C, Ad·mFX and Ad·hFX were loaded onto heparin-Sepharose columns in buffer containing 150 mm NaCl. The columns were washed with this buffer (flow through, FT), then eluted with buffer containing increasing NaCl concentrations. Eluate samples were subjected to electrophoresis, and membranes were probed with antibody to adenovirus 5 (upper panel). Lower panel, quantification of immunoblots.
To obtain corroborating evidence, we limited the amount of available heparan sulfate binding sites on cell surfaces of hepatocyte/hepatoma-derived cells by competing Ad.CMVfLuc·hFX and Ad.CMVfLuc·mFX transduction with lactoferrin, which binds to heparin and heparan sulfate (21, 23). AML12 and HepG2 cells were transduced with Ad.CMVfLuc in the presence of physiological amounts of mFX or hFX, along with increasing amounts of lactoferrin. Lactoferrin was much more effective at reducing the efficacy of mFX-promoted Ad.CMVfLuc transduction than it was at reducing the efficacy of hFX-promoted transduction; i.e. hFX much more effectively promoted transduction of cells with limited availability of heparan sulfate binding sites than did mFX (Fig. 6B). Similarly, partial removal of cell-surface heparan sulfate with heparin lyases was more effective at limiting mFX-mediated Ad.CMVfLuc transduction compared with hFX-mediated adenovirus transduction (data not shown).
Finally, we directly compared the binding of Ad·hFX and Ad·mFX to heparin, using heparin-Sepharose chromatography. Ad.CMVfLuc·hFX and Ad·mFX were applied to the columns, washed with buffer containing 150 mm NaCl, and eluted with buffers containing increasing NaCl concentrations. Eluate samples were then subjected to Western blot analysis to detect adenovirus proteins. The bulk of Ad·mFX eluted at 200 mm NaCl, whereas Ad·hFX eluted between 250 and 350 mm NaCl (Fig. 6C). These results suggest that Ad·hFX has a greater affinity to heparin than does Ad·mFX.
Human FX Is More Effective than Murine FX in Mediating Adenovirus Liver Transduction
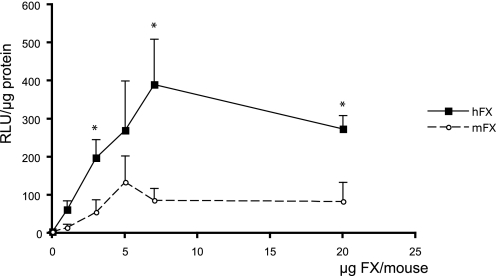
Warfarin inhibits the vitamin K-dependent carboxylation of multiple coagulation factors, including FX, leading to FX depletion from blood. We compared the efficacy of hFX and mFX in reconstituting Ad.CMVfLuc transgene expression in warfarin-treated mice. Treated animals were injected with Ad.CMVfLuc in the presence of increasing amounts of either mFX or hFX. Three days later, hepatic viral transduction efficacy was assessed by analyzing luciferase activity in liver extracts. As observed for cultured hepatocyte/hepatoma cells in culture, hFX is able to more effectively promote hepatic adenovirus transduction than is mFX (Fig. 7). A two-way analysis of variance demonstrated a significant treatment effect difference (p < 0.001) for hFX versus mFX across all dose levels. These results demonstrate hFX has an increased ability to mediate adenovirus transduction in murine liver in vivo, as well as in cultured cells.
FIGURE 7.
Adenovirus liver transduction of warfarin-treated mice reconstituted with exogenous human FX or murine FX. C57BL/6 mice were pretreated with warfarin and injected intravenously with Ad.CMVfLuc (3 × 1010 particles/mouse) along with mFX or hFX. Hepatic luciferase transgene expression was analyzed 3 days later. Both dose and treatment effects (mFX versus hFX) are statistically significant; p < 0.02 and p < 0.001, respectively. Individual mFX versus hFX concentrations were compared using the Student's t test, n = 5–8 mice per group (*, p < 0.05).Values are means ± S.E.
DISCUSSION
Both hFX and mFX increase adenovirus transduction of hepatocyte/hepatoma-derived cell lines in cell culture (Fig. 1). However, for a number of non-hepatic, epithelial cancer cell lines, only hFX can increase adenovirus transduction. The difference in mFX versus hFX enhancement of adenovirus transduction of epithelial tumor-derived cells is not due to species incompatibilities; hFX increased adenovirus transduction for both human and murine cell lines. In contrast, mFX failed to increase adenovirus transduction of either murine or human non-hepatic epithelial-derived tumor cell lines. Moreover, mFX competitively inhibited hFX-mediated increased cell transduction, at equimolar concentrations, suggesting that mFX and hFX interact with the adenovirus hexon protein binding site with equivalent stoichiometry and affinity, consistent with findings by Kayuzhniy et al. (10). The data suggest that the inability of mFX to increase adenovirus cell transduction in non-hepatic epithelial cancer cells is likely to be due to differential cell-surface receptor use by Ad·mFX and Ad·hFX complexes or, alternatively, to a different/weaker interaction of the Ad·mFX complex with a common cell-surface receptor.
hFX-mediated increased transduction of human and murine cancer cell lines was blocked by both heparin and lactoferrin and was prevented by enzymatic removal of heparan sulfate from the cell surface (Fig. 3). These three independent protocols all suggest that HSPGs are the receptor (or a contributing and essential part of a receptor complex) responsible for the hFX-mediated increase in adenovirus transduction of cultured epithelial cancer cells. Additional insight into the specificity of hFX-enhanced adenovirus transduction was obtained by studying CHO cell mutants. hFX augmented transduction of wild-type CHO cells, which express HSPGs on their cell surface (22). In contrast, CHO pgsD-677 cells, which lack heparan sulfate expression, do not support hFX-increased adenovirus transduction. In pgsD cells, the level of chondroitin/dermatan sulfate increases nearly 3-fold (22). Thus the loss of hFX-mediated enhancement of transduction in the pgsD-677 mutant implies that elevated levels of chondroitin/dermatan sulfate cannot substitute for heparan sulfate as an adenovirus·hFX receptor. CHO pgsF cells contain a mutation affecting the HS-specific uronyl 2-O-sulfotransferase and lack all 2-O-sulfated uronic acids in the heparan sulfate chains (19, 20). In the absence of this enzyme, glucosamine N-sulfation and 6-O-sulfation are enhanced, resulting in comparable or somewhat higher net negative charge along the chains relative to heparan sulfate produced by wild-type CHO cells. Thus, alternative arrangements of sulfate groups along the heparan chain may serve as an Ad·hFX heparan binding site. Consistent with this idea, reduction of overall sulfation of the chains in pgsE-606 cells significantly diminished transduction. These results are in part consistent with a study by Bradshaw et al. (9) who showed that pretreatment of HepG2 and SKOV3 cells with sodium chlorate, a selective inhibitor of sulfation, abrogates FX-mediated adenovirus transduction. Those authors, however, did not observe increased adenovirus transduction in CHO pgsF-17 cells, suggesting that FX-mediated transduction might be dependent on 2-O-sulfated residues rather than a function of sulfation/charge in general. A definitive explanation for this discrepancy is not available, but perhaps differences in cell culture conditions, viral stocks, or multiplicity of infection may have contributed to these findings.
The inability of mFX to mediate adenovirus transduction via HSPG-containing receptors present on non-hepatic epithelial cancer cells and CHO cells raises the question of how mFX increases adenovirus transduction in hepatocyte/hepatoma-derived cells such as HepG2 and AML12. The heparan sulfate chains on hepatocytes are more highly sulfated than chains produced in other cell types (20), with regions of the chains resembling therapeutic heparin in charge density, due to the presence of trisulfated disaccharides. However, HepG2 cells have overall levels of sulfation similar to CHO and cancer cell lines (Fig. 5, table). Moreover, the level of trisulfated disaccharide present on HepG2 cells is substantially less than that present on normal liver or isolated hepatocytes (20, 24), consistent with the observation that tumor lines often express more undersulfated heparan sulfate than their normal counterparts (25). However, both AML12 and HepG2 cells expressed substantially greater amounts of cell-surface heparan sulfate than do the cancer cell lines examined (Fig. 5). CHO heparan sulfate expression is in the same range as that observed for the epithelial cancer cell lines, consistent with the inability of mFX to increase transduction in CHO cells. These data suggest that FX-mediated transduction may depend on HSPG density or, alternatively, specific species of HSPGs expressed uniquely by hepatocytes and hepatocyte-derived cell lines. Partial blockade of binding sites with lactoferrin mimics the effect of reduced heparan sulfate density, relative to hepatocyte/hepatoma derived cells, present both on epithelial cancer cells and on CHO cells; mFX-mediated adenovirus transduction decreased rapidly with increasing amounts of lactoferrin, for both AML12 and HepG2 cells. In contrast, hFX-mediated transduction was unaffected over a large dose range. The data support the hypothesis that reduced cell-surface HSPG levels contribute to the inability of mFX to enhance Ad transduction of the non-hepatocyte derived cells.
hFX mediates adenovirus transduction in hepatocyte/hepatoma cultures more effectively than mFX at levels below the physiological concentration of 10 μg/ml (Fig. 6A). These results extrapolate to an in vivo context: hFX more effectively reconstitutes the ability of systemically administered adenovirus to transduce liver in vivo in warfarin-treated, FX-depleted mice than does mFX (Fig. 7). The difference in transduction efficacy does not reflect differences in binding of mFX to the virion, based on competition experiments (Fig. 2) and surface plasmon resonance experiments (10), but instead appears to reflect intrinsic differences in the ability of mFX and hFX to promote adenovirus binding to heparan sulfate. hFX interacts with heparin (26). By comparing the structure of hFX to thrombin (another serine protease in the coagulation pathway that binds to heparin), a putative heparin-binding site has been mapped to a series of arginine and lysine residues that reside on the surface of the FX protein (27). Comparison of mFX and hFX sequences shows a high degree of conservation, including the putative residues involved in heparin binding. However, hFX contains two additional lysine residues contiguous to the proposed heparin binding site, suggesting the possibility that the additional charge interactions of hFX with sulfate residues in heparan sulfate increases the affinity of hFX relative to mFX. Consistent with this suggestion, heparin-Sepharose chromatography demonstrated that Ad·hFX has greater heparin affinity than Ad·mFX.
In reviewing the literature, we find that studies reporting FX-enhanced adenovirus transduction in cultured cell lines other than hepatocytes either used hFX or did not specify whether hFX or mFX was used. Jonsson et al. (28) analyzed the effect of FIX and FX on adenovirus transduction of human epithelial cells. Although the authors did not specify from which species the factors were derived, they most likely used human coagulation factors, because they also reported the effects of human saliva, plasma, and tear fluid on adenovirus transduction of human epithelial cells. FX increased adenovirus transduction of A549 human lung cancer cells by over 5-fold, and the increased transduction was blocked by heparin competition and eliminated by pretreatment of the cells with heparin lyases (28). Increased transduction in the presence of FX was also reported in A431 (epithelial carcinoma) (29), in BxPC3 (pancreatic carcinoma) cells (29), and in SKOV3 (ovarian carcinoma) cells (8, 9).
Several laboratories have demonstrated that adenovirus unable to bind FX is effectively untargeted from liver in vivo (8, 10, 11, 30–33). Vigant et al. (33) reported that one such virus, with a modified hexon hypervariable region 5, was as effective as wild-type adenovirus in transducing subcutaneous murine melanoma and murine lung carcinoma tumors in mice following intratumoral injection. Their data suggest that, in vivo, endogenous mFX does not play a role in adenovirus transduction of tumor cells, in contrast to its ability to enhance adenovirus liver transduction. These data are consistent with our results; we found that mFX did not enhance adenovirus transduction of many non-hepatic epithelial tumor cells. In contrast, Gimenez-Alejandre et al. (29) report that, following intravenous administration, adenovirus transduction of subcutaneous human A549 or A431 xenograft tumors is reduced in mice by prior warfarin injection, suggesting that mFX (and/or other warfarin-inhibited blood components) can mediate adenovirus-enhanced transduction of some epithelial tumors in mice. In this latter study the specific role of mFX, versus other blood factors, in enhancing adenovirus transduction of liver and of tumor cells was, however, not investigated by FX reconstitution experiments.
One of the major goals in using adenovirus either for transgene or oncolytic therapies, or as a vector for non-invasive imaging, is to modify viral tropism in vivo, untargeting “undesirable” cells and retargeting the vector to specific, desired tissues or cell types (34). Clearly, to develop untargeting and retargeting strategies for therapeutic adenovirus vectors, it is important to understand the mechanisms by which adenovirus infects cells. Our findings demonstrate that hFX may have an overall increased ability to mediate adenovirus transduction compared with mFX, including mediating transduction of a wider variety of cells other than hepatocytes. These results are likely to be of importance in using animal models of adenovirus targeting and in interpreting the results of such experiments, as well as in extrapolating conclusions from these models to clinical trials.
Acknowledgments
We thank Ding Xu for providing heparin lyase preparations and advice on heparin-Sepharose chromatography, and Art Catapang for technical assistance. We also thank Sotirios Tetradis for helpful comments.
This work was supported, in whole or in part, by National Institutes of Health Grants P50 CA086306-08 (to H. R. H.) and HL57345 and GM33063 (to J. D. E.).
- Ad
- adenovirus
- Ad5
- adenovirus serotype 5
- hFX
- human coagulation factor X
- mFX
- murine coagulation factor X
- HSPG
- heparan sulfate proteoglycan.
REFERENCES
- 1. Russell W. C. (2009) J. Gen. Virol. 90, 1–20 [DOI] [PubMed] [Google Scholar]
- 2. Kay M. A., Glorioso J. C., Naldini L. (2001) Nat. Med. 7, 33–40 [DOI] [PubMed] [Google Scholar]
- 3. Bergelson J. M., Cunningham J. A., Droguett G., Kurt-Jones E. A., Krithivas A., Hong J. S., Horwitz M. S., Crowell R. L., Finberg R. W. (1997) Science 275, 1320–1323 [DOI] [PubMed] [Google Scholar]
- 4. Wickham T. J., Mathias P., Cheresh D. A., Nemerow G. R. (1993) Cell 73, 309–319 [DOI] [PubMed] [Google Scholar]
- 5. Tomko R. P., Xu R., Philipson L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3352–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nicklin S. A., Wu E., Nemerow G. R., Baker A. H. (2005) Mol. Ther. 12, 384–393 [DOI] [PubMed] [Google Scholar]
- 7. Shayakhmetov D. M., Gaggar A., Ni S., Li Z. Y., Lieber A. (2005) J. Virol. 79, 7478–7491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Waddington S. N., McVey J. H., Bhella D., Parker A. L., Barker K., Atoda H., Pink R., Buckley S. M., Greig J. A., Denby L., Custers J., Morita T., Francischetti I. M., Monteiro R. Q., Barouch D. H., van Rooijen N., Napoli C., Havenga M. J., Nicklin S. A., Baker A. H. (2008) Cell 132, 397–409 [DOI] [PubMed] [Google Scholar]
- 9. Bradshaw A. C., Parker A. L., Duffy M. R., Coughlan L., van Rooijen N., Kahari V. M., Nicklin S. A., Baker A. H. (2010) PLoS Pathog. 6, e1001142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kalyuzhniy O., Di Paolo N. C., Silvestry M., Hofherr S. E., Barry M. A., Stewart P. L., Shayakhmetov D. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5483–5488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parker A. L., Waddington S. N., Nicol C. G., Shayakhmetov D. M., Buckley S. M., Denby L., Kemball-Cook G., Ni S., Lieber A., McVey J. H., Nicklin S. A., Baker A. H. (2006) Blood 108, 2554–2561 [DOI] [PubMed] [Google Scholar]
- 12. Esko J. D., Kimata K., Lindahl U. (2009) In Essentials of Glycobiology, 2nd Ed., (Varki A., Cummings R., Esko J. D., Freeze H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E. eds) pp. 229–248, Cold Spring Harbor Laboratory Press, New York: [PubMed] [Google Scholar]
- 13. Bishop J. R., Schuksz M., Esko J. D. (2007) Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 14. Perabo L., Goldnau D., White K., Endell J., Boucas J., Humme S., Work L. M., Janicki H., Hallek M., Baker A. H., Büning H. (2006) J. Virol. 80, 7265–7269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Summerford C., Samulski R. J. (1998) J. Virol. 72, 1438–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. WuDunn D., Spear P. G. (1989) J. Virol. 63, 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shukla D., Liu J., Blaiklock P., Shworak N. W., Bai X., Esko J. D., Cohen G. H., Eisenberg R. J., Rosenberg R. D., Spear P. G. (1999) Cell 99, 13–22 [DOI] [PubMed] [Google Scholar]
- 18. Liang Q., Yamamoto M., Curiel D. T., Herschman H. R. (2004) Mol. Imaging Biol. 6, 395–404 [DOI] [PubMed] [Google Scholar]
- 19. Bai X., Esko J. D. (1996) J. Biol. Chem. 271, 17711–17717 [DOI] [PubMed] [Google Scholar]
- 20. Lawrence R., Olson S. K., Steele R. E., Wang L., Warrior R., Cummings R. D., Esko J. D. (2008) J. Biol. Chem. 283, 33674–33684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ji Z. S., Mahley R. W. (1994) Arterioscler. Thromb. 14, 2025–2031 [DOI] [PubMed] [Google Scholar]
- 22. Zhang L., Lawrence R., Frazier B. A., Esko J. D. (2006) Methods Enzymol. 416, 205–221 [DOI] [PubMed] [Google Scholar]
- 23. Zou S., Magura C. E., Hurley W. L. (1992) Comp. Biochem. Physiol. B. 103, 889–895 [DOI] [PubMed] [Google Scholar]
- 24. Tátrai P., Egedi K., Somorácz A., van Kuppevelt T. H., Ten Dam G., Lyon M., Deakin J. A., Kiss A., Schaff Z., Kovalszky I. (2010) J. Histochem. Cytochem. 58, 429–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Esko J. D., Rostand K. S., Weinke J. L. (1988) Science 241, 1092–1096 [DOI] [PubMed] [Google Scholar]
- 26. Rezaie A. R. (2000) J. Biol. Chem. 275, 3320–3327 [DOI] [PubMed] [Google Scholar]
- 27. Padmanabhan K., Padmanabhan K. P., Tulinsky A., Park C. H., Bode W., Huber R., Blankenship D. T., Cardin A. D., Kisiel W. (1993) J. Mol. Biol. 232, 947–966 [DOI] [PubMed] [Google Scholar]
- 28. Jonsson M. I., Lenman A. E., Frängsmyr L., Nyberg C., Abdullahi M., Arnberg N. (2009) J. Virol. 83, 3816–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gimenez-Alejandre M., Cascallo M., Bayo-Puxan N., Alemany R. (2008) Hum. Gene Ther. 19, 1415–1419 [DOI] [PubMed] [Google Scholar]
- 30. Short J. J., Rivera A. A., Wu H., Walter M. R., Yamamoto M., Mathis J. M., Curiel D. T. (2010) Mol. Cancer Ther. 9, 2536–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alba R., Bradshaw A. C., Coughlan L., Denby L., McDonald R. A., Waddington S. N., Buckley S. M., Greig J. A., Parker A. L., Miller A. M., Wang H., Lieber A., van Rooijen N., McVey J. H., Nicklin S. A., Baker A. H. (2010) Blood 116, 2656–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Waddington S. N., Parker A. L., Havenga M., Nicklin S. A., Buckley S. M., McVey J. H., Baker A. H. (2007) J. Virol. 81, 9568–9571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vigant F., Descamps D., Jullienne B., Esselin S., Connault E., Opolon P., Tordjmann T., Vigne E., Perricaudet M., Benihoud K. (2008) Mol. Ther. 16, 1474–1480 [DOI] [PubMed] [Google Scholar]
- 34. Noureddini S. C., Curiel D. T. (2005) Mol. Pharm. 2, 341–347 [DOI] [PubMed] [Google Scholar]