Abstract
Certain ritonavir resistance mutations impair HIV infectivity through incomplete Gag processing by the mutant viral protease. Analysis of the mutant virus phenotype indicates that accumulation of capsid-spacer peptide 1 precursor protein in virus particles impairs HIV infectivity and that the protease mutant virus is arrested during the early postentry stage of HIV infection before proviral DNA synthesis. However, activation of the target cell can rescue this defect, implying that specific host factors expressed in activated cells can compensate for the defect in ritonavir-resistant HIV. This ability to rescue impaired HIV replication presented a unique opportunity to identify host factors involved in postentry HIV replication, and we designed a functional genetic screen so that expression of a given host factor extracted from activated T cells would lead directly to its discovery by rescuing mutant virus replication in nonactivated T cells. We identified the cellular heat shock protein 90 kDa α (cytosolic), class B member 1 (HSP90AB1) as a host factor that can rescue impaired replication of ritonavir-resistant HIV. Moreover, we show that pharmacologic inhibition of HSP90AB1 with 17-(allylamino)-17-demethoxygeldanamycin (tanespimycin) has potent in vitro anti-HIV activity and that ritonavir-resistant HIV is hypersensitive to the drug. These results suggest a possible role for HSP90AB1 in postentry HIV replication and may provide an attractive target for therapeutic intervention.
Keywords: Drug Resistance, Heat Shock Protein, HIV, Protease Inhibitor, siRNA, Viral Protease, Viral Replication, Virus, Capsid, Uncoating
Introduction
Certain mutations in HIV protease (PR)2 that confer resistance to ritonavir (RTV), in particular I54V and V82A, result in virus particles with incompletely processed Gag and impaired replication capacity in vitro and in vivo. In vitro analysis indicates that the loss in infectivity results from decreased proteolytic activity of the mutant HIV PR, and RTV-resistant virus particles display an accumulation of uncleaved Gag precursor molecules, specifically capsid-spacer peptide 1 (CA-SP1) and nucleocapsid-spacer peptide 2 (NC-SP2) (1). Mutational analysis of Gag and studies with the HIV maturation inhibitor bevirimat (2) indicate that uncleaved CA-SP1 by itself is sufficient to impair viral replication. This is because CA maturation structurally modifies the HIV core in preparation for host factor-mediated uncoating in the infected cell (3). Physical changes in CA affect overall core stability and reduce viral infectivity (4). Recent studies by Müller et al. (5) have shown that even low amounts (<5%) of Gag processing intermediates can display a transdominant negative effect on HIV infectivity with the maturation cleavage between CA and SP1 being of particular importance for the negative effect. In a previous study, we found that although HIV with RTV resistance PR mutations is minimally impaired for replication in mitogen-activated peripheral blood T cells it is highly impaired for replication in human thymus both in the SCID-hu Thy/Liv mouse model and in thymic organ culture (6). Our results suggest that the unprocessed Gag molecules disrupt CA maturation (and thus prevent proper HIV core uncoating) and that cellular activation is the intrinsic difference between thymocytes and mitogen-activated T cells in determining the replication capacity of RTV-resistant HIV.
Successful core uncoating is the major rate-limiting step in productive retroviral infection and is controlled largely by cellular proteins (4, 7). For example, the host factor cyclophilin A (CypA) interacts with HIV CA to promote uncoating of the viral core (8); alternatively, various retroviral cores can be inactivated through contact between CA and host restriction factor Ref1, Lv1, or Trim5α (9). Studies by Yamashita et al. (10) provided evidence that the CA protein is a dominant viral determinant for infection of nondividing cells. More recently, these authors showed that tripartite motif family proteins and CypA modulate the ability of HIV to infect nondividing cells through their interaction with the CA protein (11). Uncoating and subsequent steps in early stage HIV infection remain unclear but are thought to occur through active recruitment of cytoplasmic proteins by the viral CA core (12, 13), and functional gene silencing screens have revealed that various cellular pathways, enzyme complexes, and cytoskeleton proteins are involved in each step leading to proviral DNA integration (14–17). Although there was minimal overlap among the screens in candidate HIV dependence genes, the consensus from these studies was that host factors associated with postentry HIV replication are cell-specific but often localize to common cellular pathways (18). These interesting observations led us to hypothesize that highly impaired replication of RTV-resistant HIV in thymocytes, as opposed to minimal impairment in peripheral T lymphocytes, is a result of differences in the cytoplasmic environment between the target cells rather than limitations in the PR mutant.
The characteristic cone-shaped HIV core is formed exclusively from CA molecules that initially arrange into a dense hexagonal lattice of uncleaved CA-SP1 subunits (19). In this immature conformation, CA hexamers are stabilized from “below” by SP1 bundles (20). Ultimately, proteolysis at the CA-SP1 junction enables the densely packed CA subunits to rearrange into distinct CA hexamers, which then polymerize into a stable mature HIV core lattice. In addition, this morphological transition prepares the HIV core for postentry events by exposing local motifs in CA to interacting host factors within the infected cell (21).
Three-dimensional analysis of released virus particles has revealed that the number of Gag molecules is double the number of CA molecules needed for formation of the cone-shaped capsid shell, indicating that ∼50% of CA in the mature virion may not be part of the capsid (22). High resolution images of mature retroviral cores show a range of core polymorphisms, which include amorphous shapes, multiple and nested cores, and incompletely closed shells, suggesting that viable retroviral cores do not have a uniquely specified structure; rather a range of CA structures may be acceptable (23). Moreover, thin section electron microscopy (EM) analysis reveals that virus particles with minor amounts of intermediate Gag cleavage products do not appear to block maturation because the cone-shaped core is present in most particles. However, subtle alterations in virion morphology that are not easily detected by EM may be functionally relevant (5). Consequently, RTV-resistant HIV particles might contain intact cores that assume a defective structure because of uncleaved CA-SP1, thereby precluding uncoating in thymocytes that lack the appropriate CA-interacting host factors.
In this report, we confirm that uncleaved CA-SP1 severely impairs RTV-resistant HIV replication in thymocytes and nonactivated T cells and that activation of T cells dramatically reverses this defect. The PR mutant is arrested at the postentry stage of uncoating in nonactivated cells, and this defect presented an opportunity for identification of host factors that rescue RTV-resistant HIV replication. Thus, we designed a genetic screen so that expression of a given host factor would lead directly to its discovery by virtue of its ability to rescue a fluorescent RTV-resistant reporter virus. Using the PR mutant fluorescent reporter, we sought to identify host factors with the greatest positive effect on HIV replication by sorting the most brightly fluorescent infected cells. Through this screen, we identified the cellular heat shock protein 90 kDa α (cytosolic), class B member 1 (HSP90AB1) as a factor that independently rescues RTV-resistant HIV replication. Moreover, we show that pharmacologic inhibition and RNA interference of HSP90AB1 blocks HIV replication in primary human T cells, underscoring the advantage of screening for HIV-interacting host proteins that promote virus replication. The ability of HSP90AB1 to rescue RTV-resistant HIV suggests that this host factor is involved in early postentry virus replication possibly by interacting with the HIV CA protein and represents a novel target for new antiviral therapies.
EXPERIMENTAL PROCEDURES
Cells and Viruses
293T and P4-Magi cell lines were cultured at 37 °C in Dulbecco's modified Eagle's medium (HyClone) supplemented with 10% fetal bovine serum (FBS) (HyClone), and Jurkat E6-1, Jurkat E6-1 CypA−/−, CEM T4, and Sup T1 were cultured at 37 °C in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS. All cell lines were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. PBMC were obtained from six HIV-seronegative donors and maintained in RPMI 1640 medium supplemented with 10% FBS, 1 μg/ml PHA-P (Invitrogen), and 5% human IL-2 (Roche Applied Science) for 3 days before inoculation. The infectious HIV proviral DNA construct pNL4-3 was used in all studies, and the I54V, V82A, and A71V mutations in HIV PR and A431V in the NC-SP2 cleavage junction were generated by oligonucleotide-directed single strand mutagenesis. Briefly, the Gag-Pol region of pNL4-3 was excised using the SwaI restriction endonuclease and cloned into the pBluescript cloning vector (Stratagene). The I54V, A71V, and V82A mutations were created individually or in combination within the protease gene using the QuikChange II site-directed mutagenesis kit (Stratagene), and similarly, the A431V mutation was introduced in the NC-SP2 cleavage junction. All mutations were verified by sequencing. The respective mutations were rebuilt into the parental NL4-3 by transferring the SpeI-Age I fragment from the intermediate cloning vector. The wild-type (WT) HIV-RFP and HIV PRI54V/V82A-RFP fluorescent reporter virus constructs were created by inserting the coding sequence of dTomato red fluorescent protein (RFP) (24) between the env and nef regions of pNL4-3. Initially, the BamHI-KpnI fragment was excised from pNL4-3 and cloned into the pBluescript vector. The end of gp41 gene was amplified by PCR to generate a BglII recognition site at the 3′-end of the amplicon, and the start of the nef gene was amplified with primers terminating the PCR product in a KpnI recognition sequence. The RFP coding sequence was excised from dTomato by first digesting with EcoRI and subsequently blunting the 3′-end with mung bean nuclease. The 5′-start of RFP was digested with BamHI and ligated to the 3′-end of the gp41 PCR product, and the 3′-end of RFP was blunt end-ligated to the 5′-start of the nef gene PCR product. The three-piece ligation product was excised from the intermediate pBluescript cloning vector and rebuilt into the parental NL4-3 clone to produce an infectious molecular clone with the RFP open reading frame beginning 14 nucleotides downstream of the gp41 termination codon and ending one nucleotide upstream of the nef initiation sequence. The lentiviral green fluorescent protein (GFP) reporter system was used for VSV-G pseudotyping experiments as described previously (25). The I54V and V82A PR mutations were introduced in the psPAX2 Gag-Pol packaging construct by site-directed mutagenesis, and recombinant virus particles were generated by transient transfection in 293T cells using calcium phosphate (Invitrogen). The VSV-G envelope was derived from the pMD2G plasmid, and the HIV Env was derived from the pDOL construct (National Institutes of Health AIDS Research and Reference Reagent Program). Vector combinations used to generate virus particles included cotransfecting 3 μg of each envelope plasmid, 8 μg of the packaging plasmid with and without the PR mutations, and 10 μg of the GFP reporter plasmid. Two days after transfection, the culture medium was clarified by filtration (0.45 μm) and frozen in aliquots under liquid nitrogen. The virus stocks were titrated as described in the following section.
HIV Stock Virus Preparation and Titration
All mutant and WT virus stocks were generated by transient transfection of 293T cells using Lipofectamine 2000 (Invitrogen). Two days after transfection, the culture medium was clarified by filtration (0.45 μm) and frozen in aliquots under liquid nitrogen. 50% tissue culture infectious doses (TCID50) were determined with serial half-log dilutions of each virus stock in quadruplicate wells of PHA-activated PBMC (1 × 105 cells/well) and calculated as a reciprocal of the dilution at which 50% of the wells contained detectable (≥50 pg/ml) HIV p24 measured by ELISA (PerkinElmer Life Sciences).
Western Blot Analysis
Virus particles were concentrated from clarified culture medium by ultracentrifugation through a 20% sucrose cushion in phosphate-buffered saline (PBS) at 130,000 × g for 2 h at 4 °C. The pellet was slowly dissolved in Laemmli loading buffer (Sigma). High molecular weight viral proteins were separated on 8–20% linear gradient polyacrylamide gels, and smaller viral proteins were resolved on 15% gels. The viral proteins were electroblotted onto Immobilon PVDF membrane (Millipore) and incubated individually with specific primary antibodies. The following primary antibodies were obtained from the AIDS Reagent Program: CA (catalog number 6521), MA (catalog number 4811), reverse transcriptase (RT) (catalog number 11338), integrase (IN) (catalog number 7374), PR (catalog number 4105), Nef (catalog number 709), Vif (catalog number 6459), Vpr (catalog number 3951), gp120 (catalog number 2343), and gp41 (catalog number 7623). The NC and p6 primary antibodies were obtained from the National Cancer Institute AIDS Vaccine Program. Peroxidase-conjugated secondary antibodies (Pierce) were used, and immune complexes were visualized by enhanced chemiluminescence (Pierce).
In Vitro Infectivity Assay
P4-Magi indicator cells were seeded in a 12-well plate and inoculated at a multiplicity of infection (m.o.i.) of 0.005 with WT and mutant stock viruses. At 48 h after inoculation, cells were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside to detect infected cells, and the percentage of blue cells was determined microscopically.
In Vivo Infectivity Assay
Implantation of fragments of human fetal thymus and liver under the kidney capsule to create SCID-hu Thy/Liv mice and inoculation of the Thy/Liv implants with HIV were carried out as described previously (6). Implanted mice were inoculated with 1,000 TCID50 of HIV, and Thy/Liv implants were collected after 21 days and processed for viral RNA quantification by branched DNA assay and p24 ELISA. Protocols were approved by the University of California, San Francisco Institutional Animal Care and Use Committee.
Infectivity Assay in Activated Cells
Jurkat E6-1 cells were treated with increasing concentrations of phorbol 12-myristate 13-acetate (Sigma) for 24 h. Cells were resuspended in fresh growth medium, inoculated with stock virus at an m.o.i. of 0.005, and cultured for 12 days. Culture supernatant was assayed every 2 days for HIV p24 by ELISA. Additionally, Jurkat E6-1 cells were activated with CD3- and CD28-coated magnetic beads as recommended by the manufacturer (Dynal) for 24 h. Cells were resuspended in fresh growth medium, inoculated with stock virus at an m.o.i. of 0.005, and cultured for 8 days. Culture supernatant was assayed for HIV p24 by ELISA.
Virus-Cell Fusion Assay
The BlaM-Vpr HIV fusion assay was performed as described previously (26). WT HIV-BlaM-Vpr and HIV PRI54V/V82A-BlaM-Vpr chimeric viruses were produced by triple transfection of 293T cells, titrated, and used to inoculate Jurkat E6-1 cells at a range of m.o.i. values for 2 h at 37 °C. Cells were then loaded with the CCF2-AM fluorogenic substrate (Invitrogen) overnight at room temperature. Cells were centrifuged and resuspended in PBS. The extent of virus-cell fusion was determined by fluorescence of the cleaved CCF2-AM using a three-laser BD FACSAria system (BD Biosciences).
Quantitative Assay for Intracellular HIV Reverse Transcription
WT HIV and HIV PRI54V/V82A virus stocks pretreated with DNase I and 10 mm MgCl2 for 1 h to remove contaminating plasmid DNA were used to inoculate Jurkat E6-1 cells at an m.o.i. of 0.005. Cells were collected at intervals after inoculation, and total cellular DNA was extracted with the DNeasy kit (Qiagen). The extent of proviral DNA synthesis was quantified by real time PCR using the RU5 primers and probe set to detect early reverse transcripts and the MH531-MH532 primers and LTR-P probe to detect full-length proviral DNA as described previously (27, 28). Reaction mixtures included the Ex Taq master mix (TaKaRa), 100 ng of DNA, a 200 nm concentration of each primer, and a 100 nm concentration of probe. Thermal cycling conditions were 2 min at 50 °C, 10 min at 95 °C, and 40 cycles of 95 °C for 15 s and 60 °C for 60 s. Control Jurkat E6-1 cells were pretreated with 50 μm azidothymidine for 4 h before inoculation.
Endogenous Reverse Transcriptase Assay and Total Viral RNA Content
WT HIV and HIV PRI54V/V82A virus (500 TCID50) were pretreated with DNase I and 10 mm MgCl2 for 1 h to remove contaminating plasmid DNA and concentrated as described above. The concentrated virus was resuspended in 10 μl of PBS and diluted with 40 μl of endogenous RT buffer (5 mm MgCl2; 50 mm NaCl; 10 mm dithiothreitol; 0.5 mm dATP, dCTP, and dGTP; 50 mm Tris (pH 8); 0.015% Triton X-100; 10 μm [α-32P]TTP). The reaction mixtures were incubated at 37 °C for 30 min and terminated with 0.5% SDS and 25 mm EDTA. Samples were spotted on DE81 filters, washed three times in 2× SSC, and dried, and 32P was quantified by liquid scintillation counting. Control samples were treated with 25 μm nevirapine. Total RNA was extracted from the same concentrated virus samples using the QIAmp Viral RNA kit (Qiagen), dissolved in an ice-cold solution of 10 mm NaOH and 1 mm EDTA, and spotted on a nitrocellulose membrane. The membrane was prehybridized overnight at 42 °C in hybridization buffer (50% formamide, 5× SSC, 1× Denhardt's solution, 50 mm NaHPO4 (pH 6.5), 250 μg/ml denatured salmon sperm DNA) followed by incubation with a random primed HIV-specific 32P-labeled probe (Stratagene) at 42 °C for 16 h. The membrane was washed three times in 2× SSC and dried, and the amount of hybridized probe was measured by liquid scintillation counting.
Construction of Retroviral cDNA Expression Library
Total RNA was extracted from CD3- and CD28-activated Jurkat E6-1 cells using the RNeasy kit (Qiagen) and subjected to consecutive rounds of mRNA purification using the Oligotex mRNA purification kit (Qiagen). First strand synthesis was carried out on 5 μg of purified mRNA using a hybrid XhoI-oligo(dT) primer (Stratagene), and the resulting cDNAs had 5′-EcoRI and 3′-XhoI cohesive ends for directional ligation into the retroviral vector. The retroviral cDNA expression library was constructed using the pFB retroviral vector (Stratagene), and the multiple cloning region was replaced with a PCR fragment containing a Kozak sequence, ATG start codon, and FLAG tag sequence followed by the EcoRI and XhoI recognition sequences. Two additional constructs were generated by inserting A and AA nucleotides immediately before the EcoRI site to ensure in-frame expression of the cDNA library. The coding sequence for the GFP derived from pAcGFP1-1 (Clontech) was inserted into the modified pFB vector using the EcoRI and XhoI sites and served as a stuffer fragment to reduce the background of parental vector in the cDNA library. Furthermore, the modified pFB-GFP construct was used as a reference virus to calculate the titer of the retroviral cDNA expression library (29). The cDNA was ligated directionally into the modified pFB plasmid by replacing the stuffer fragment, and the reaction products were used to transform XL-10 GOLD Escherichia coli (Stratagene) by electroporation. The bacterial colonies were amplified in semisolid 2× LB-agarose (Stratagene) to reduce the potential for underrepresentation of particular clones due to overgrowth of faster growing colonies.
Production and Titration of Retroviral cDNA Expression Library
To generate a library of transducing viruses, 20 μg of the pFB cDNA expression plasmid was transiently transfected into the AmphoPack-293 packaging cell line (Clontech) using Lipofectamine 2000 (Invitrogen). The culture supernatant was harvested 48 h after transfection, clarified by low speed centrifugation, and filtered (0.2 μm), and aliquots were stored under liquid nitrogen. Because the pFB transducing vector does not carry a reporter, the titer of the retroviral cDNA expression library was determined by a one-step quantitative RT-PCR using a calibrated RNA standard curve generated with the pFB-GFP reference virus. The m.o.i. of the pFB-GFP virus was determined by flow cytometry, and the titer of the retroviral cDNA-expressing library was normalized to the RNA titer of the reference GFP-transducing virus (30).
Genetic Screen
Jurkat E6-1 cells were transduced with the cDNA-expressing retroviral library at an m.o.i. of 0.1 in multiple sets to ensure even representation of the cDNA library and cultured for 24 h. Transduced cells were inoculated with the HIV PRI54V/V82A-RFP reporter virus at an m.o.i. of 0.05 and cultured for an additional 24 h. Cells were resuspended in PBS, and red fluorescent cells were sorted on a FACSVantage DiVa cell sorting system (BD Biosciences). The RFP-sorted cells were examined microscopically to confirm that all the cells expressed red fluorescence, and total cellular DNA was extracted with the DNeasy kit (Qiagen). The cDNA insert was recovered by PCR amplification using primers specific to the modified retroviral vector that flanked the cDNA sequence. The extracted cDNAs were recloned into the modified pFB vector and subjected to a second round of selection where only cells with a high red fluorescence were sorted (0.01–0.1% of total RFP-positive cells) after which cDNA inserts were amplified using a reduced number of PCR cycles to ensure even representation of each individual cDNA. PCR products were recloned into the modified pFB retroviral vector plasmid, bacterial colonies representing single cDNAs were picked, and transducing retroviral particles were generated individually as described above. Jurkat E6-1 cells were transduced at an m.o.i. of 0.1 with individual cDNA-expressing retroviral particles and then inoculated with the HIV PRI54V/V82A-RFP reporter virus as described above. The cells were assayed 24 h after inoculation by flow cytometry, and cDNA clones that supported replication of the RTV-resistant reporter virus were identified by expression of the RFP in infected cells. The DNA sequence of selected cDNA clones was aligned with the NCBI Human Genome Project database using the BLAST human sequence search engine.
Cloning and Expression of HSP90AB1
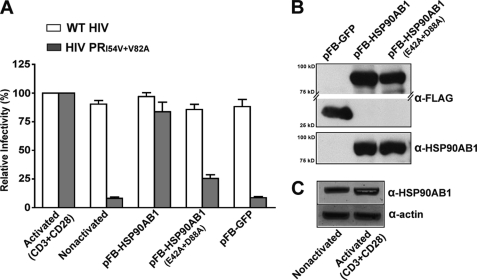
The full-length protein-coding sequence of HSP90AB1 was PCR-amplified from the cDNA clone isolated through the genetic screen and inserted immediately downstream of the FLAG sequence in the pFB retroviral expression vector (Stratagene). In addition, the E42A and D88A mutations (31) were introduced in the native HSP90AB1 sequence by oligonucleotide-directed single strand mutagenesis. Transducing retrovirus stocks carrying the native and mutant HSP90AB1 cDNAs were generated and titrated as described above. Jurkat E6-1 cells were transduced with the cDNA-expressing retrovirus at an m.o.i. of 0.1 and cultured for 24 h. The pFB-GFP reference virus was included as a negative control and used to determine recombinant protein expression. A fraction of the cells were resuspended in fresh growth medium, inoculated with WT and mutant stock viruses at an m.o.i. of 0.005, and cultured for 8 days. Culture supernatant was assayed for HIV p24 by ELISA. The remaining cells were lysed in CelLytic M solution (Sigma), and recombinant proteins were immunoprecipitated using anti-FLAG M2 affinity gel (Sigma). Immunoprecipitated protein from an equal number of transduced cells was separated on a 10% polyacrylamide gel matrix and subjected to Western blot analysis using the anti-FLAG M5 clone antibody (Sigma) and anti-HSP90AB1 antibody (Abcam) as described above. The endogenous expression level of HSP90AB1 was assayed on 10 μg of protein extracted from nonactivated and CD3- and CD28-activated Jurkat E6-1 cells. Cellular HSP90AB1 protein levels were quantified on a Typhoon Trio imager (GE Healthcare). Equal protein loading was confirmed by probing for actin (Abcam).
In Vitro Antiviral Activity Assay
Antiviral activity of 17-(allylamino)-17-demethoxygeldanamycin (Sigma) was assayed in PHA-activated PBMC as described previously (32). Briefly, PBMC were inoculated in bulk with each virus at an m.o.i. of 0.001 for 2 h, cells were washed, and 1 × 105 cells in 100 μl were seeded in triplicate wells of a 96-well plate. Wells were treated with 100 μl of serial half-log dilutions of the drug or with medium alone, and supernatants were assayed at day 7 for p24 by ELISA. Parallel cellular toxicity of the drug was determined on uninfected cells by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (Sigma) on day 7. The 50% inhibition of virus replication (IC50) and 50% cytotoxic concentration (CC50) were calculated as the mean of three independent assays. Similarly, the antiviral activity of an siRNA against HSP90AB1 was assayed. The sense and antisense strands of the siRNA were synthesized separately (Integrated DNA Technologies) with the following modifications: HSP90AB1 sense, 5-UUCAGGGCAUUGCUAUUUAdT*dT*-3; HSP90AB1 antisense, 5-UAAAUAGCAAUGCCCUGAAdT*dT*chol-3; scrambled sense, 5-GGUUAUAAUUUCGUUCGACdT*dT*-3; scrambled antisense, 5-GUCGAACGAAAUUAUAACCdT*dT*chol-3. * designates substitution of the phosphodiester bond with a phosphorothioate bond, and chol indicates cholesterol conjugation. The two RNA strands were annealed in equimolar amounts, and WT HIV- and HIV PRI54V/V82A-infected PBMC were treated with quarter-log dilutions of the siRNA duplex. Inhibition of viral replication and cellular toxicity were assayed after 7 days, and results represent the mean of three independent assays.
RESULTS
RTV Resistance Affects Cleavage Efficiency of HIV Protease
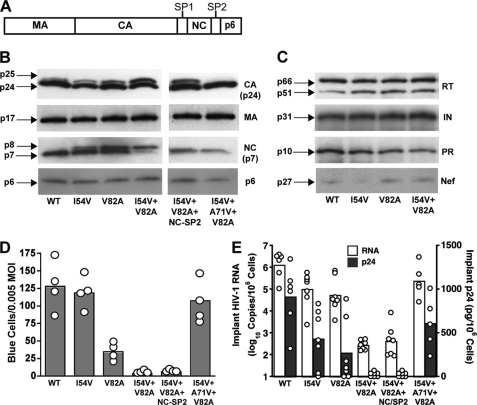
Sequential cleavage of the Gag polyprotein by HIV PR occurs during virus maturation and is essential for infectious virus particle formation (Fig. 1A). In our previous report, HIV gag and pol sequences isolated from patients receiving RTV (and other PR inhibitor) monotherapy revealed that the I54V and V82A drug resistance mutations in HIV PR led to decreased viral fitness. We introduced the I54V and V82A mutations into the PR-coding sequence of the NL4-3 clone of HIV and analyzed individual viral proteins by Western blot. Cleavage of the MA and p6 proteins was not affected by the RTV-resistant HIV PR; however, a significant reduction in cleavage at the CA-SP1 and NC-SP2 junctions was observed in the presence of both PR mutations (Fig. 1B).
FIGURE 1.
Impaired replication of RTV-resistant HIV results from uncleaved CA-SP1. A, schematic of the protein domain organization in the HIV Gag polyprotein precursor. Proteolytic cleavage of the Gag polyprotein during HIV maturation results in the MA (p17), CA (p24), NC (p7), and p6 proteins. B, Western blot analysis of Gag cleavage products from purified WT HIV and mutant virus particles carrying the indicated RTV resistance mutations in the protease. The right panel shows PR mutant virus particles including compensatory substitutions in the NC-SP2 cleavage junction and the A71V mutation in HIV protease. Insufficient cleavage of Gag is indicated by the presence of p25 (CA-SP1) and p8 (NC-SP2) protein bands. The Western blot is representative of three independent purified virus preparations. C, Western blot analysis of Pol polyprotein and Nef precursor protein cleavage products from wild-type and RTV-resistant HIV particles. Complete cleavage of the Pol polyprotein is indicated by mature RT, IN, and PR proteins. Molecular mass of mature viral proteins is indicated on the left, and the panel is representative of three independent purified virus preparations. D, triplicate wells of P4-Magi cells were inoculated with each virus at an m.o.i. of 0.005 and stained for β-galactosidase activity after 48 h. The columns represent mean number of infected blue cells from four (open circles) independent experiments. E, SCID-hu Thy/Liv mice were inoculated with 1,000 TCID50 of each virus, and Thy/Liv implants were collected 21 days after inoculation. Viral replication was assessed by branched DNA assay for HIV RNA and by ELISA for p24. The columns represent means, and open circles represent individual animals from the same cohort.
In vivo resistance to RTV occurs sequentially with the V82A primary mutation located in the PR active site frequently preceding the I54V secondary mutation (33). This combination confers greater resistance to RTV in vivo but decreases the in vitro activity of HIV PR. Eventually, resistance to RTV in vivo results from compensatory mutations within HIV PR and in Gag target sites to counteract reduced activity of the mutant PR (34, 35). We introduced the naturally occurring A431V compensatory mutation into the NC-SP2 cleavage junction (1); this facilitated cleavage of NC-SP2 but did not alter partial cleavage at the CA-SP1 junction by the RTV-resistant HIV PR. Furthermore, we introduced an additional substitution in PR (A71V) commonly observed in patients failing RTV monotherapy (36) and showed that this substitution restored enzymatic activity to the RTV-resistant PR at both the CA-SP1 and NC-SP2 junctions (Fig. 1B). The HIV PR is also responsible for cleavage of the Gag-Pol polyprotein and Nef precursor protein, and we confirmed that the I54V and V82A PR substitutions did not affect proteolysis of these additional substrates (Fig. 1C).
Uncleaved CA-SP1 (p25) but Not Uncleaved NC-SP2 (p8) Impairs RTV-resistant HIV
To determine whether insufficient cleavage of CA-SP1 and NC-SP2 affected replication of RTV-resistant HIV, we challenged P4-Magi indicator cells with the different PR mutants at equal m.o.i. values (Fig. 1D). The I54V PR mutation alone did not affect virus replication, whereas significant replication impairment was observed with the V82A mutation. The combined effect of I54V and V82A PR mutations severely impaired RTV-resistant HIV replication. The NC-SP2 compensatory substitution did not rescue infectivity of the PR mutant, however, suggesting that uncleaved CA-SP1, but not NC-SP2, impairs replication of RTV-resistant HIV. The compensatory A71V PR substitution rescued infectivity of RTV-resistant HIV, confirming that RTV resistance impacts virus replication.
More importantly, we confirmed impaired replication of RTV-resistant HIV in human thymus implants of SCID-hu Thy/Liv mice. We inoculated SCID-hu Thy/Liv mice with 1,000 TCID50 of each virus. The Thy/Liv implants were collected 21 days after inoculation for quantification of HIV RNA by branched DNA assay and p24 by ELISA. The in vivo pattern of RTV-resistant HIV replication was similar to that seen in vitro: the combination of I54V and V82A PR mutations impaired replication of RTV-resistant HIV (HIV PRI54V/V82A) in human thymocytes, and compensatory cleavage of NC-SP2 did not rescue infectivity of the PR mutant (Fig. 1E).
Taken together, our results indicate that the presence of uncleaved CA-SP1 is the primary cause for impaired replication of RTV-resistant HIV in human thymocytes. This defect, however, does not adversely affect RTV-resistant HIV replication in activated peripheral T cells. We speculate that, as a result of uncleaved CA-SP1, the RTV-resistant HIV core lattice assumes an alternate conformation that cannot undergo proper uncoating in thymocytes. If so, host factors up-regulated in activated cells may make the difference between the presence and the absence of efficient viral replication.
Impaired Replication of RTV-resistant HIV Is Rescued by Cellular Activation
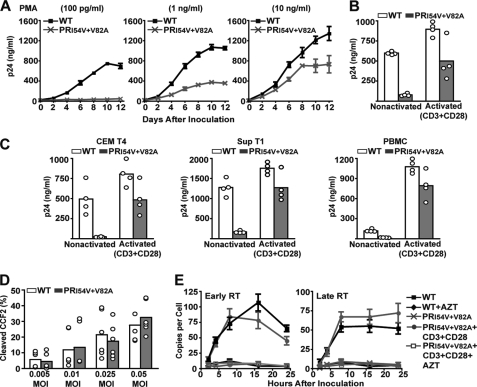
Our earlier studies indicated that expression of specific host factors in PHA-activated PBMC rescued the impaired replication of RTV-resistant HIV. To validate this hypothesis, we screened CD4+ T cell lines for their ability to show a differential capacity to support PR mutant virus replication in the presence or absence of activation. At base line, Jurkat T cells failed to support HIV PRI54V/V82A virus replication when inoculated at the same m.o.i. as that of WT HIV. Replication of this PR mutant was supported when Jurkat T cells were first activated with phorbol 12-myristate 13-acetate, and the amount of replication increased in a dose-dependent manner (Fig. 2A). Replication of the HIV PRI54V/V82A mutant increased dramatically in response to phorbol 12-myristate 13-acetate activation, indicating a direct influence of select host factors on replication of RTV-resistant HIV. We confirmed the role of activation on RTV-resistant HIV replication by activating Jurkat T cells through the T cell receptor using a mixture of antibodies against CD3 and CD28 (Fig. 2B). In addition, activation of CEM T4 cells, Sup T1 cells, and human PBMC specifically through T cell receptor-ligand binding effectively rescued RTV-resistant HIV replication (Fig. 2C). These results confirm that host factors unique to activated human T cells override the transdominant negative effects of uncleaved CA-SP1 and facilitate replication of RTV-resistant HIV.
FIGURE 2.
Cellular activation rescues impaired replication of RTV-resistant HIV. A, Jurkat E6-1 cells treated with increasing concentrations of phorbol 12-myristate 13-acetate (PMA) for 24 h were inoculated at an m.o.i. of 0.005. The cells were cultured for 12 days, and results represent mean supernatant HIV p24 levels ±S.E. (error bars) from six independent experiments. B, Jurkat E6-1 cells were activated with a mixture of CD3 and CD28 antibodies for 24 h, inoculated at an m.o.i. of 0.005, and cultured for 8 days. The columns represent mean supernatant HIV p24 levels from four (open circles) independent experiments. C, CEM T4, Sup T1, and PBMC were activated with a mixture of CD3 and CD28 antibodies for 24 h, inoculated at an m.o.i. of 0.005, and cultured for 8 days. The columns represent mean supernatant HIV p24 levels from four (open circles) independent experiments. D, wild-type HIV-BlaM-Vpr and HIV PRI54V/V82A-BlaM-Vpr chimeric viruses at a range of m.o.i. values were used to inoculate nonactivated Jurkat E6-1 cells for detection of CCF2 substrate cleavage by flow cytometry. Each m.o.i. was analyzed in triplicate, and the columns represent mean percentage of cleaved CCF2 from six (open circles) independent experiments. E, activated and nonactivated Jurkat E6-1 cells were inoculated at an m.o.i. of 0.005, and cellular DNA was extracted over a 24-h period. Early and late proviral reverse transcripts were quantified by TaqMan real time PCR in triplicate, and the results represent mean ± S.E. (error bars) of six independent experiments. AZT, azidothymidine.
Replication of RTV-resistant HIV Is Arrested after Entry and before Reverse Transcription
To determine whether uncleaved CA-SP1 prevented fusion of the RTV-resistant virus to the target cell, we inoculated nonactivated Jurkat T cells with HIV PRI54V/V82A-BlaM-Vpr chimeric virus using a fluorescence fusion assay (26). In comparison with WT HIV-BlaM-Vpr chimeric virus, we did not observe fusion defects, suggesting that replication of RTV-resistant virus is impaired at a postentry stage of the virus life cycle (Fig. 2D). We also inoculated Jurkat T cells with WT HIV and HIV PRI54V/V82A and quantified early and late proviral reverse transcripts over a 24-h time course by real time PCR. The absence of both early and late RTV-resistant proviral transcripts in nonactivated Jurkat T cells suggested that RTV-resistant HIV infection was arrested soon after entry, most likely during uncoating (Fig. 2E). However, full-length RTV-resistant proviral transcripts were detected in CD3- and CD28-activated Jurkat T cells (Fig. 2E), indicating functional reverse transcription by the PR mutant. In addition, HIV PRI54V/V82A virus was tested for endogenous RT activity and the ability to package viral genomic RNA. No significant differences in RT activity or viral RNA content were observed (Table 1), confirming that RTV-resistant HIV is not defective in RNA packaging or formation of a functional ribonucleoprotein-RT complex within the virion. Taken together, these results suggest that defective uncoating of RTV-resistant HIV cores in nonactivated T cells precludes reverse transcription of proviral DNA, resulting in dead-end infection. In contrast, cellular activation provides appropriate host factors that facilitate uncoating of the PR mutant virus core, leading to productive proviral DNA synthesis of RTV-resistant HIV.
TABLE 1.
RT activity is expressed as counts/25 μl of spotted reaction mixture. Results represent the mean ± S.E. of six independent RT assays. Viral RNA content was calculated from four independent RNA extractions, and results represent the mean ± S.E. of eight spots (two per sample)
| Virus | Endogenous RT activity | Viral RNA content |
|---|---|---|
| cpm × 104/500 TCID50 | cpm × 102/500 TCID50 | |
| Wild-type HIV | 2,204 ± 271 | 387 ± 56 |
| HIV PRI54V/V82A | 1,626 ± 678 | 426 ± 84 |
| Wild-type HIV + nevirapine | 75 ± 26 | Not determined |
| HIV PRI54V/V82A + nevirapine | 113 ± 18 | Not determined |
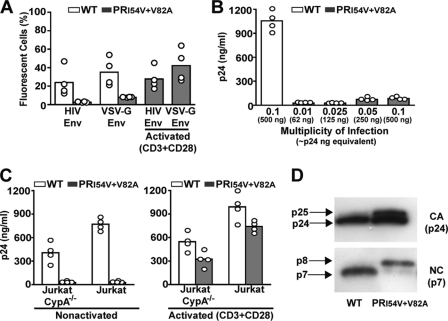
Pseudotyping of RTV-resistant HIV Does Not Rescue Impaired Replication
Infectivity of HIV can be influenced by the route of entry, and pseudotyping with envelope proteins from other viruses (e.g. VSV-G) enables some mutant virus to bypass early postentry blocks (37). Using GFP reporter viruses, RTV-resistant HIV was targeted for alternate entry through the endosomal pathway by pseudotyping with the VSV-G envelope, but this failed to rescue infectivity to RTV-resistant HIV in nonactivated Jurkat T cells (Fig. 3A). The pseudotyped RTV-resistant HIV, however, responded predictably to cellular activation (Fig. 3A). In addition, we inoculated nonactivated Jurkat T cells with PR mutant at increasing m.o.i. values to determine whether host restriction factors are responsible for impaired replication of RTV-resistant HIV. A common feature of retroviral postentry blocks is that host restriction factors can be saturated by virus overload (38), but increasing the m.o.i. to 0.1 (equivalent to 500 ng of p24) did not rescue RTV-resistant HIV replication in nonactivated Jurkat T cells (Fig. 3B).
FIGURE 3.
Characteristics of impaired RTV-resistant HIV replication. A, WT HIV-GFP and HIV PRI54V/V82A-GFP reporter viruses at an m.o.i. of 0.1 with HIV Env and WT HIV-GFP and HIV PRI54V/V82A-GFP reporter viruses at an m.o.i. of 0.01 pseudotyped with VSV-G Env were used to inoculate nonactivated and CD3- and CD28-activated Jurkat E6-1 cells. The columns represent mean GFP fluorescence from four (open circles) independent infection experiments. B, nonactivated Jurkat E6-1 cells inoculated with HIV PRI54V/V82A virus at increasing m.o.i. values were cultured for 8 days. The columns represent mean supernatant HIV p24 levels from four (open circles) independent experiments. C, CD3- and CD28-activated Jurkat E6-1 and Jurkat E6-1 CypA−/− cells were inoculated at an m.o.i. of 0.005 and cultured for 8 days. The columns represent mean supernatant HIV p24 levels from four (open circles) independent experiments. D, Jurkat E6-1 cells were activated with antibodies against CD3 and CD28 for 24 h, inoculated at an m.o.i. of 0.05, and cultured for 8 days. Virus particles were purified from culture supernatants, and CA and NC viral proteins were detected by Western blot analysis. Results are representative of three independently purified virus preparations.
CypA Does Not Rescue Replication of RTV-resistant HIV in Activated T Cells
To determine whether overexpression of CypA contributed to the rescue of RTV-resistant HIV in activated T cells, we activated CypA-null Jurkat T cells (CypA−/−) (39) with antibodies against CD3 and CD28 and then inoculated the cells with RTV-resistant HIV. Replication of the PR mutant in CD3- and CD28-activated CypA−/− Jurkat T cells was comparable with levels observed in activated Jurkat T cells, indicating that the necessary host factor is not CypA (Fig. 3C).
Cellular Activation Does Not Repair Maturation Defect of RTV-resistant HIV
In addition, we observed that cellular activation did not enhance maturation of RTV-resistant HIV by facilitating CA-SP1 cleavage in virus particles released from activated Jurkat T cells. We inoculated CD3- and CD28-activated Jurkat T cells with RTV-resistant HIV and harvested virus particles from the culture supernatant. Western blot analysis of the RTV-resistant HIV proteins confirmed that virus particles released from activated T cells contained uncleaved CA-SP1 and NC-SP2 subunits (Fig. 3D) that were unable to infect nonactivated Jurkat T cells.
Genetic Screen to Identify HIV-interacting Host Factors
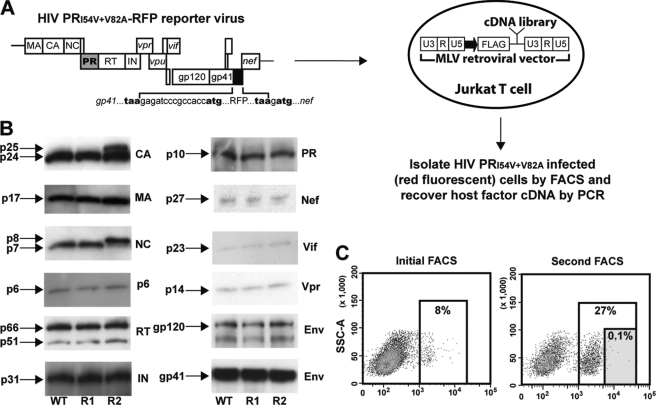
Given the above evidence, we reasoned that host factors expressed in activated T cells interact with the uncleaved CA-SP1 subunit to rescue RTV-resistant HIV replication. To identify such factors, we designed a genetic screen using a cDNA library generated from CD3- and CD28-activated Jurkat T cells (Fig. 4A). The cDNA library was introduced into nonactivated Jurkat T cells using a modified retroviral vector, and transduced cells were then inoculated with RTV-resistant HIV carrying the gene for RFP inserted into the viral RNA genome such that expression of the reporter would occur only upon productive infection and integration of RTV-resistant HIV. We predicted that transduced Jurkat T cells that supported productive infection of the RTV-resistant RFP HIV would express a permissive host factor encoded by the unique cDNA present in the infected cell. Infected cells with high RFP expression (i.e. those enabling integration and replication of RTV-resistant HIV) were enriched by fluorescence-activated cell sorting (FACS), and the cDNA responsible for rescue of the replication-impaired RTV-resistant RFP reporter virus was extracted and further characterized.
FIGURE 4.
Strategy of genetic screen. A, schematic of the genetic screen to identify host factors that rescue RTV-resistant HIV replication. B, Western blot analysis of WT HIV-RFP (R1) and HIV PRI54V/V82A-RFP (R2) reporter virus proteins compared with the protein profile of WT HIV. Defective cleavage of Gag in the HIV PRI54V/V82A-RFP reporter virus is indicated by the presence of p25 and p8 proteins. Panels represent individual viral proteins of the Pol polyprotein, accessory gene products, and surface (gp120) and transmembrane (gp41) Env proteins in mature virus particles. Shown are representative data from one of three independent experiments. C, Jurkat T cells infected with the HIV PRI54V/V82A-RFP reporter virus were sorted by flow cytometry and gated based on the intensity of red fluorescence. In the second round of FACS, total RFP cells were gated into two populations, and cells with the highest fluorescence intensity (0.01–0.1%) were sorted. SSC-A, side scatter area.
The WT and PR mutant reporter viruses were constructed so that the highly sensitive RFP would be expressed independently of the viral Nef protein in infected cells, and the presence of the reporter gene did not affect virus particle formation (Fig. 4B). Bright red fluorescence was detected 12–18 h after inoculation with the WT HIV-RFP reporter virus, and we were able to reproduce the CD3 and CD28 antibody activation-dependent rescue of RTV-resistant HIV with the HIV PRI54V/V82A-RFP virus. The cDNA library was constructed for directional gene expression under control of the retroviral promoter, and expressed host factor proteins were preceded by an N-terminal FLAG tag. The high complexity retrovirus-based cDNA library was transfected into an amphotropic packaging cell line to generate a high titer transducing virus stock. The recombinant virus stock was used to transduce nonactivated Jurkat T cells at a low m.o.i. of 0.1 to limit the number of cDNAs expressed in individual cells. We achieved >85% transduction efficiency of the cDNA library in nonactivated Jurkat T cells and confirmed protein expression using an anti-FLAG antibody.
Nonactivated Jurkat T cells transduced with the retrovirus-based cDNA library were cultured briefly to allow adequate expression of the gene products and then inoculated with the HIV PRI54V/V82A-RFP virus. Jurkat T cells infected by the PR mutant reporter virus were sorted for bright red fluorescence by flow cytometry. The host factor-expressing cDNAs were PCR-amplified from genomic DNA of the sort-purified Jurkat T cells using retrovirus-specific primers and subjected to an additional round of selection in the genetic screen (Fig. 4C). The second round of screening yielded a greater number of RTV-resistant HIV-infected red fluorescent cells, and we sorted the very brightest (0.01–0.1%) RFP-positive cells. Selection of brightly red fluorescent cells ensured isolation of host factors that strongly interacted with RTV-resistant HIV and reduced false-positive events that can occur with routine manipulation of target cells. Host factor cDNAs were amplified using a limited number of PCR cycles to ensure even representation of each clone. The PCR products were recloned into the modified retroviral vector, and individual bacterial colonies, representing single cDNA clones, were tested independently with the HIV PRI54V/V82A-RFP virus. Through the second round of selection we identified a cDNA clone, which aligned with the human HSP90AB1 gene (GeneID 3326), that consistently supported RTV-resistant HIV replication when expressed in nonactivated Jurkat T cells.
Validation of HSP90AB1 in RTV-resistant HIV Replication
To confirm the influence of HSP90AB1 in RTV-resistant HIV replication, we generated a full-length expression clone of the host factor. We also introduced the inactivating E42A and D88A mutations into the native HSP90AB1 sequence to prevent ATP binding and hydrolysis (31). Jurkat T cells transduced with recombinant retrovirus carrying the native and mutant forms of HSP90AB1 were inoculated with WT and RTV-resistant HIV, and the extent of virus replication was determined in relation to HIV replication in CD3- and CD28-activated Jurkat T cells (Fig. 5A). Replication of RTV-resistant HIV in Jurkat T cells expressing the native HSP90AB1 protein was comparable with mutant virus replication in CD3- and CD28-activated cells, whereas the inactivated mutant form of HSP90AB1 failed to rescue RTV-resistant HIV replication. Overexpression of HSP90AB1 in Jurkat T cells did not adversely affect replication of WT HIV, and to eliminate the possible influence of retrovirally mediated gene transfer on RTV-resistant HIV replication, Jurkat T cells were simultaneously transduced with a GFP-expressing retroviral vector. Furthermore, we immunoprecipitated the FLAG-tagged recombinant proteins from an equal number of transduced Jurkat T cells to ensure comparable expression of both native and mutant HSP90AB1 proteins (Fig. 5B).
FIGURE 5.
HSP90AB1 can rescue RTV-resistant replication. A, nonactivated Jurkat E6-1 cells transduced with retrovirus vectors expressing native and mutant HSP90AB1 and GFP were inoculated with WT and RTV-resistant HIV at an m.o.i. of 0.005. Results are expressed as relative infectivity (%) of WT and RTV-resistant HIV replication in CD3- and CD28-activated Jurkat T cells and represent mean supernatant HIV p24 levels ±S.E. (error bars) for six independent transduction experiments. B, immunoprecipitation of recombinant proteins from transduced Jurkat E6-1 cells. Equal volumes of resolved proteins were probed with anti-FLAG- and anti-HSP90AB1-specific antibodies. Results are representative of three independently immunoprecipitated preparations. C, protein lysate from nonactivated and CD3- and CD28-activated Jurkat E6-1 cells probed with anti-HSP90AB1- and -actin-specific antibodies. Protein levels were quantified on a Typhoon Trio imager, and the mean ± S.E. increase in HSP90AB1 expression in CD3- and CD28-activated Jurkat T cells was 2.0 ± 0.5-fold. Results are representative of three independently prepared cell lysates.
Cell activation through external stimuli often results in the overexpression of several cellular proteins, and we accordingly observed a mean 2-fold increase in HSP90AB1 protein production in CD3- and CD28-activated Jurkat T cells (Fig. 5C). Taken together, these results suggest that up-regulation of HSP90AB1 is necessary for RTV-resistant HIV replication.
RTV-resistant HIV Is Hypersensitive to HSP90AB1 Inhibition in Vitro
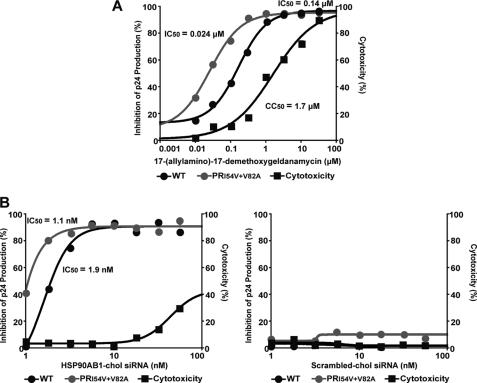
The abundant HSP90AB1 chaperone protein mediates conformational maturation of several cellular proteins and is believed to stabilize deregulated signaling proteins in oncogenesis. We examined the in vitro antiviral activity of 17-(allylamino)-17-demethoxygeldanamycin, a pharmacologic inhibitor of HSP90AB1 (40) that is currently in clinical trials to treat metastatic solid tumors and lymphomas (41). 17-(Allylamino)-17-demethoxygeldanamycin had potent activity against WT HIV NL4-3 in PHA-activated PBMC, and RTV-resistant HIV (as predicted by our hypothesis) was hypersensitive to the drug with a mean IC50 7-fold lower than for WT HIV (0.024 versus 0.17 μm, p < 0.05 by paired Student's t test) (Fig. 6A). We confirmed that inhibition of HSP90AB1 in activated PBMC does indeed prevent HIV replication by using a cholesterol-conjugated siRNA to knock down HSP90AB1 expression (Fig. 6B).
FIGURE 6.
RTV-resistant HIV is hypersensitive to HSP90AB1 inhibition in vitro. A, PHA-activated PBMC inoculated at an m.o.i. of 0.001 were treated with serial half-log dilutions of 17-(allylamino)-17-demethoxygeldanamycin, and the IC50 was calculated after 7 days. The CC50 of the drug was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. The graph represents one of three independent assays. The mean ± S.E. IC50 for WT HIV was 0.17 ± 0.03 μm, the mean IC50 for the PR mutant was 0.024 ± 0.001 μm, and the mean CC50 was 1.9 ± 0.26 μm. B, PHA-activated PBMC inoculated at an m.o.i. of 0.001 were treated with serial quarter-log dilutions of a cholesterol (chol)-conjugated siRNA duplex against HSP90AB1 (left) and a control scrambled siRNA (right). The IC50 was calculated after 7 days, and the CC50 of the siRNA was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. The graph represents one of three independent assays. The mean ± S.E. IC50 for WT HIV was 2 ± 0.06 nm, the mean IC50 for the PR mutant was 1.4 ± 0.19 nm, and the CC50 was <65 nm. No significant inhibition of WT HIV and PR mutant virus replication or cytotoxicity was observed with the cholesterol-conjugated scrambled siRNA duplex.
DISCUSSION
The emergence of drug resistance is not always beneficial to HIV and sometimes comes at the cost of decreased viral fitness. In the case of RTV-resistant HIV, the PR mutant has highly impaired infectivity in thymocytes and nonactivated T cells (1, 6). Nonetheless, we show here that loss of RTV-resistant HIV fitness can be rescued in activated T cells, suggesting that the variation in PR mutant virus replication is a result of the target cell environment rather than limitations in the PR mutant. In this study, we used the model of RTV-resistant HIV replication to identify host factors expressed in activated T cells that rescue impaired replication of the PR mutant.
The temporal cleavage of Gag by HIV PR is critical for virus maturation, and RTV resistance affects cleavage efficiency of mutant PR specifically at the Gag spacer peptide junctions. As a result, uncleaved CA-SP1 accumulates in virus particles and impairs RTV-resistant HIV replication in human thymocytes and nonactivated T cells. A similar replication defect has been observed with the maturation inhibitor bevirimat, which binds to SP1 and prevents CA lattice formation (42). Bevirimat causes a dose-dependent inhibition of CA-SP1 cleavage and severely impairs HIV infectivity in human thymocytes (32). Furthermore, RTV-resistant HIV is hypersensitive to the maturation inhibitor, indicating that high levels of uncleaved CA-SP1 in a virus particle adversely affect its ability to infect target cells.
The short SP1 peptide attached to the C terminus of CA enables HIV cores to assemble into immature (SP1-present) or mature (SP1-absent) lattices. The accumulation of uncleaved CA-SP1 in virus particles most likely affects stability of the HIV core structure, which is rapidly degraded in the infected cell. Alternatively, uncleaved CA-SP1 subunits might transform the HIV core into an unusually stable structure that cannot undergo obligatory postentry modifications. However, cellular activation rescues RTV-resistant HIV replication, suggesting that the PR mutant is infectious but in need of specific uncoating factors that are underrepresented in human thymocytes and nonactivated T cells. In contrast, activated cells express unique host factors that compensate for uncleaved CA-SP1 and rescue RTV-resistant HIV replication. Although defective core stability resulting from uncleaved CA-SP1 is the favored explanation for impaired infectivity of RTV-resistant HIV, we cannot ignore the possibility that only half of the total CA molecules assemble into the viral core. Consequently, RTV-resistant HIV particles may contain an intact core irrespective of uncleaved CA-SP1. Nevertheless, we show here that the presence of uncleaved CA-SP1 in virus particles precludes reverse transcription of the viral genome in thymocytes and nonactivated T cells and that replication of RTV-resistant HIV is arrested during the early postentry stage of the virus life cycle.
The intent of this study was to identify HIV-uncoating host factors by exploiting this easily reversible model of RTV-resistant HIV replication. A genetic screen was designed on the premise that RTV-resistant HIV replicates specifically in activated T cells; therefore, expression of cellular genes from activated T cells in nonactivated T cells should potentially support RTV-resistant HIV replication. The search for host factors, carried out under stringent conditions, revealed that HSP90AB1 can rescue RTV-resistant HIV replication.
Identification of HSP90AB1 as a potential uncoating host factor validated our selection strategy and reveals an exciting avenue of HIV postentry modification by cellular chaperones. Mammalian HSP90AB1 is one of two cytosolic isoforms in the heat shock protein 90 family that collectively regulate cell homeostasis by stabilizing a diverse clientele of proteins (43). Heat shock protein 90 chaperones in particular have been linked to replication of several viruses where they are essential for picornavirus capsid maturation and activate the viral polymerase complexes of several RNA viruses (44). A recent study by Vozzolo et al. (45) demonstrates that heat shock protein 90 interacts with the HIV DNA promoter sequence and regulates viral gene expression. However, this study also suggests that heat shock protein 90 is involved in the early postentry stage of HIV replication. Furthermore, HSP90AB1 is intimately coupled with the ubiquitin-proteasome pathway, and recent studies indicate that ubiquitin ligases and components of the proteasome play a structural role in HIV uncoating and reverse transcription (46). Overall, a link between virus evolution and chaperone dependence has been hypothesized where cellular chaperones are proposed to counteract the destabilizing effects of viral capsid mutations that continuously emerge in response to immune surveillance (47). Such a buffering mechanism would allow for greater flexibility in viral protein structure. If this is the case, then up-regulation of HSP90AB1 in activated T cells might compensate for the transdominant negative effects of uncleaved CA-SP1 and rescue RTV-resistant replication. Then again, the anti-HIV effect of 17-(allylamino)-17-demethoxygeldanamycin and the fact that RTV-resistant HIV is hypersensitive to HSP90AB1 inhibition suggest an association between the cellular chaperone and HIV core uncoating.
It remains to be determined whether HSP90AB1 directly influences HIV uncoating or whether this cellular protein regulates other host factors required for viral replication. However, virus-host protein interactions by nature are refractory to the development of drug resistance and are therefore attractive targets for antiviral therapy.
Acknowledgments
We thank Mary Beth Moreno, José Rivera, Sofiya Galkina, Barbara Sloan, Valerie Stepps, and David Chung for expert technical assistance.
This work was supported, in whole or in part, by a National Institutes of Health grant from NIAID under Contract N01-AI-70002. This work was also supported by California HIV/AIDS Research Program Grant ID09-SF-051 (to C. A. S.) and in part by the University of California, San Francisco AIDS Research Institute and the Harvey V. Berneking Living Trust.
- PR
- protease
- RTV
- ritonavir
- CA-SP1
- capsid-spacer peptide 1
- NC-SP2
- nucleocapsid-spacer 2
- CypA
- cyclophilin A
- HSP90AB1
- heat shock protein 90 kDa α (cytosolic), class B member 1
- CA
- capsid
- NC
- nucleocapsid
- PBMC
- peripheral blood mononuclear cells
- PHA
- phytohemagglutinin
- VSV-G
- vesicular stomatitis virus G
- TCID50
- 50% tissue culture infectious dose
- MA
- matrix
- IN
- integrase
- m.o.i.
- multiplicity of infection
- CC50
- 50% cytotoxic concentration.
REFERENCES
- 1. Mammano F., Petit C., Clavel F. (1998) J. Virol. 72, 7632–7637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou J., Huang L., Hachey D. L., Chen C. H., Aiken C. (2005) J. Biol. Chem. 280, 42149–42155 [DOI] [PubMed] [Google Scholar]
- 3. Vogt V. M. (1996) Curr. Top. Microbiol. Immunol. 214, 95–131 [DOI] [PubMed] [Google Scholar]
- 4. Forshey B. M., von Schwedler U., Sundquist W. I., Aiken C. (2002) J. Virol. 76, 5667–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Müller B., Anders M., Akiyama H., Welsch S., Glass B., Nikovics K., Clavel F., Tervo H. M., Keppler O. T., Kräusslich H. G. (2009) J. Biol. Chem. 284, 29692–29703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stoddart C. A., Liegler T. J., Mammano F., Linquist-Stepps V. D., Hayden M. S., Deeks S. G., Grant R. M., Clavel F., McCune J. M. (2001) Nat. Med. 7, 712–718 [DOI] [PubMed] [Google Scholar]
- 7. Goff S. P. (2007) Nat. Rev. Microbiol. 5, 253–263 [DOI] [PubMed] [Google Scholar]
- 8. Luban J., Bossolt K. L., Franke E. K., Kalpana G. V., Goff S. P. (1993) Cell 73, 1067–1078 [DOI] [PubMed] [Google Scholar]
- 9. Goff S. P. (2004) Mol. Cell 16, 849–859 [DOI] [PubMed] [Google Scholar]
- 10. Yamashita M., Perez O., Hope T. J., Emerman M. (2007) PLoS Pathog. 3, 1502–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamashita M., Emerman M. (2009) J. Virol. 83, 9835–9843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goff S. P. (2001) J. Gene Med. 3, 517–528 [DOI] [PubMed] [Google Scholar]
- 13. Nisole S., Saïb A. (2004) Retrovirology 1, 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brass A. L., Dykxhoorn D. M., Benita Y., Yan N., Engelman A., Xavier R. J., Lieberman J., Elledge S. J. (2008) Science 319, 921–926 [DOI] [PubMed] [Google Scholar]
- 15. König R., Zhou Y., Elleder D., Diamond T. L., Bonamy G. M., Irelan J. T., Chiang C. Y., Tu B. P., De Jesus P. D., Lilley C. E., Seidel S., Opaluch A. M., Caldwell J. S., Weitzman M. D., Kuhen K. L., Bandyopadhyay S., Ideker T., Orth A. P., Miraglia L. J., Bushman F. D., Young J. A., Chanda S. K. (2008) Cell 135, 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou H., Xu M., Huang Q., Gates A. T., Zhang X. D., Castle J. C., Stec E., Ferrer M., Strulovici B., Hazuda D. J., Espeseth A. S. (2008) Cell Host Microbe 4, 495–504 [DOI] [PubMed] [Google Scholar]
- 17. Yeung M. L., Houzet L., Yedavalli V. S., Jeang K. T. (2009) J. Biol. Chem. 284, 19463–19473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bushman F. D., Malani N., Fernandes J., D'Orso I., Cagney G., Diamond T. L., Zhou H., Hazuda D. J., Espeseth A. S., König R., Bandyopadhyay S., Ideker T., Goff S. P., Krogan N. J., Frankel A. D., Young J. A., Chanda S. K. (2009) PLoS Pathog. 5, e1000437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. von Schwedler U. K., Stemmler T. L., Klishko V. Y., Li S., Albertine K. H., Davis D. R., Sundquist W. I. (1998) EMBO J. 17, 1555–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wright E. R., Schooler J. B., Ding H. J., Kieffer C., Fillmore C., Sundquist W. I., Jensen G. J. (2007) EMBO J. 26, 2218–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Steven A. C., Heymann J. B., Cheng N., Trus B. L., Conway J. F. (2005) Curr. Opin. Struct. Biol. 15, 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carlson L. A., Briggs J. A., Glass B., Riches J. D., Simon M. N., Johnson M. C., Müller B., Grünewald K., Kräusslich H. G. (2008) Cell Host Microbe 4, 592–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Butan C., Winkler D. C., Heymann J. B., Craven R. C., Steven A. C. (2008) J. Mol. Biol. 376, 1168–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004) Nat. Biotechnol. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- 25. Salmon P., Trono D. (2007) Curr. Protoc. Hum. Genet. Chapter 12, Unit 12.10 [DOI] [PubMed] [Google Scholar]
- 26. Cavrois M., De Noronha C., Greene W. C. (2002) Nat. Biotechnol. 20, 1151–1154 [DOI] [PubMed] [Google Scholar]
- 27. Julias J. G., Ferris A. L., Boyer P. L., Hughes S. H. (2001) J. Virol. 75, 6537–6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Butler S. L., Hansen M. S., Bushman F. D. (2001) Nat. Med. 7, 631–634 [DOI] [PubMed] [Google Scholar]
- 29. Yu H., Kwon Y. J. (2008) Methods Mol. Biol. 433, 1–16 [DOI] [PubMed] [Google Scholar]
- 30. Cornetta K., Pollok K. E., Miller A. D. (2008) CSH Protoc. 10.1101/pdb.top29 [DOI] [PubMed] [Google Scholar]
- 31. Chadli A., Bruinsma E. S., Stensgard B., Toft D. (2008) Biochemistry 47, 2850–2857 [DOI] [PubMed] [Google Scholar]
- 32. Stoddart C. A., Joshi P., Sloan B., Bare J. C., Smith P. C., Allaway G. P., Wild C. T., Martin D. E. (2007) PLoS One 2, e1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rhee S. Y., Liu T. F., Holmes S. P., Shafer R. W. (2007) PLoS Comput. Biol. 3, e87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anderson J., Schiffer C., Lee S. K., Swanstrom R. (2009) Handb. Exp. Pharmacol. 189, 85–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yin P. D., Das D., Mitsuya H. (2006) Cell. Mol. Life Sci. 63, 1706–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mammano F., Trouplin V., Zennou V., Clavel F. (2000) J. Virol. 74, 8524–8531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aiken C. (1997) J. Virol. 71, 5871–5877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cowan S., Hatziioannou T., Cunningham T., Muesing M. A., Gottlinger H. G., Bieniasz P. D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11914–11919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Braaten D., Luban J. (2001) EMBO J. 20, 1300–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Powers M. V., Workman P. (2007) FEBS Lett. 581, 3758–3769 [DOI] [PubMed] [Google Scholar]
- 41. Goetz M. P., Toft D., Reid J., Ames M., Stensgard B., Safgren S., Adjei A. A., Sloan J., Atherton P., Vasile V., Salazaar S., Adjei A., Croghan G., Erlichman C. (2005) J. Clin. Oncol. 23, 1078–1087 [DOI] [PubMed] [Google Scholar]
- 42. Sakalian M., McMurtrey C. P., Deeg F. J., Maloy C. W., Li F., Wild C. T., Salzwedel K. (2006) J. Virol. 80, 5716–5722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen B., Piel W. H., Gui L., Bruford E., Monteiro A. (2005) Genomics 86, 627–637 [DOI] [PubMed] [Google Scholar]
- 44. Kamal A., Boehm M. F., Burrows F. J. (2004) Trends Mol. Med. 10, 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vozzolo L., Loh B., Gane P. J., Tribak M., Zhou L., Anderson I., Nyakatura E., Jenner R. G., Selwood D., Fassati A. (2010) J. Biol. Chem. 285, 39314–39328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Isaacson M. K., Ploegh H. L. (2009) Cell Host Microbe 5, 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Geller R., Vignuzzi M., Andino R., Frydman J. (2007) Genes Dev. 21, 195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]