Abstract
Retinoic X receptor (RXR) is a master nuclear receptor in the processes of cell development and homeostasis. Unliganded RXR exists in an autorepressed tetramer, and agonists can induce RXR dimerization and coactivator recruitment for activation. However, the molecular mechanisms involving the corepressor recruitment and antagonist-mediated repression of RXR are still elusive. Here we report the crystal structure of RXRα ligand-binding domain (LBD) complexed with silencing mediator for retinoid and thyroid hormone receptors (SMRT) corepressor motif. As the first structural report on the unliganded nuclear receptor bound to the corepressor motif, RXRαLBD-SMRT exhibits a significant structural rearrangement, compared with apoRXRαLBD tetramer. To elucidate further the molecular determinants for RXR repression by its antagonist, we also determine the crystal structure of RXRαLBD-SMRT complexed with the identified antagonist rhein. In the structure, two rhein molecules and two SMRT peptides are in the RXRαLBD tetramer, different from the case in RXRαLBD-SMRT structure, where four SMRT peptides bind to RXRαLBD tetramer. It seems that rhein induces a displacement of SMRT motif by activation function 2 (AF-2) motif binding to the receptor. Combining our current work with the published results, structural superposition of RXRαLBDs in different states reveals that RXR uses an overlapped binding site for coactivator, corepressor, and AF-2 motifs, whereas the AF-2 motif adopts different conformations for agonist or antagonist interaction and coactivator or corepressor recruitment. Taken together, we thus propose a molecular model of RXR repression on the tetramer.
Keywords: Corepressor Transcription, Crystal Structure, Drug Action, Nuclear Receptors, X-ray Crystallography
Introduction
The nuclear receptor retinoic X receptor (RXR)3 is a ligand-regulated transcription factor and is involved in the processes of cell patterning, organogenesis, proliferation, and differentiation (1). The functions of many other nuclear receptors such as retinoid acid receptor, peroxisome proliferator-activated receptor (PPAR), farnesoid X receptor (FXR), liver X receptor (LXR), and thyroid hormone receptor are all dependent on RXR by forming heterodimers with this master receptor (2). Considering the central roles of RXR in multiple biological processes, the molecular mechanism for RXR regulation, especially receptor-ligand and receptor-coregulator interactions, will undoubtedly provide insights into the RXR-involved signaling pathways.
Currently, RXR transcription activation has been characterized in some detail (3, 4). Upon agonist binding to the ligand-binding domain (LBD) of the receptor, the DNA-binding domain (DBD) recognizes the specific response elements, and then the activation function 2 (AF-2) motif conducts conformational changes for recruiting the coactivators to initiate gene expression (3, 4). Without ligand binding, RXR exists dominantly as an autorepressed homotetramer (4). Previous reports on the structures of the apoRXRαLBD tetramer revealed that the AF-2 motif extends from the core structure of the LBD and makes intermolecular interactions with the coactivator-binding site of its neighboring monomer (4, 5). Such contacts thus suggest the molecular mechanism of RXR autorepression and AF-2-mediated inhibition of coactivator binding. However, interactions between RXR and the corepressors such as the nuclear receptor corepressor (N-CoR) and the silencing mediator for retinoid and thyroid hormone receptors (SMRT) were still elusive.
SMRT has been implicated in resistance to thyroid hormone, a human genetic disease characterized by an impaired physiological response to thyroid hormone (6, 7). It also links to several types of leukemia, including acute promyelocytic leukemia, acute myeloid leukemia, and pediatric β-cell acute leukemia (8). In addition, circumstantial evidence associates N-CoR and/or SMRT expression and subcellular localization with some cancers like colorectal carcinoma (9) and endometrial carcinoma (10). Recently, SMRT was reported to promote oxidative phosphorylation in adipose tissue and protect against diet-induced obesity and insulin resistance (11). Our previous study based on isothermal titration calorimetry technology showed that the SMRT corepressor motif peptide (residues 2337TNMGLEAIIRKALMGK2352) bind directly to either apo or antagonist-bound RXRαLBD (5). To reveal the molecular basis for the receptor-corepressor interaction, we determine here the crystal structure of RXRαLBD complexed with the SMRT corepressor motif peptide. Structural analysis indicates that four SMRT peptides bind to the tetrameric RXRαLBD in a conserved binding pocket (12, 13) whereas SMRT binding unexpectedly induces a significant conformational rearrangement of the receptor compared with apoRXRαLBD structure.
To elucidate further the potential molecular determinants for RXR repression by the antagonist, we also determine the crystal structure of RXRαLBD-SMRT complexed with the identified antagonist rhein. Rhein is one of the major bioactive components in rhubarb (Dahuang), a famous traditional Chinese medicine derived from the rhizome of Rheum palmatum and related species. It exhibits anti-tumor, anti-inflammatory and anti-angiogenic activities (14). Rhein was also proved to be effective in the treatment of diabetic nephropathy (15, 16), although no target for rhein had ever been discovered. In our current work, rhein is identified as a selective RXRα antagonist, which could repress all of the tested RXRα-involved homo- or heterodimeric transcription activities. The crystal structure of RXRαLBD-rhein-SMRT reveals that two rhein molecules and two SMRT peptides bind to RXRαLBD tetramer, which is different from the case in RXRαLBD-SMRT, where four SMRT peptides are in RXRαLBD tetramer. These results thus imply that rhein results in a displacement of the SMRT motif by the AF-2 motif at the corepressor-binding pocket. Further structural superposition of RXRαLBDs in its apo, agonist-, and antagonist-bound states reveals that the AF-2 motif adopts different conformations catering to agonist or antagonist interaction, as well as coactivator or corepressor recruitment. Based on all these findings, we thereby propose a potential molecular model for RXR repression on the tetramer.
EXPERIMENTAL PROCEDURES
Protein Purification and Peptide Synthesis
The coding sequence of human RXRαLBD (residues 221–458) was cloned to the vector pET15b, and Escherichia coli strain BL21 (DE3) was used for protein expression. The protein expression was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 25 °C for 6 h. His-tagged RXRαLBD was purified with nickel-nitrilotriacetic acid resin (Qiagen), and the tag was then removed by thrombin (Novagen). The protein was further purified with Superdex 200 (Amersham Biosciences). The SMRT corepressor motif peptide was commercially synthesized with the sequence 2337TNMGLEAIIRKALMG2351.
Cocrystallization
All crystallization experiments were performed by hanging-drop method at 20 °C. RXRαLBD was mixed with the SMRT peptide in a ratio of 1:3. Crystals grew in the condition of 100 mm sodium cacodylate, pH 6.5, 15% PEG 4000, 100 mm magnesium acetate. For the RXRαLBD-rhein-SMRT complex, the ratio of protein to peptide:ligand was 1:3:5. Crystals grew in the condition of 100 mm sodium cacodylate, pH 6.5, 20% PEG 4000, 100 mm magnesium acetate.
Data Collection, Processing, Structure Determination, and Refinement
Diffraction data were collected at BL17U of Shanghai Synchrotron Radiation Facility in China and integrated with HKL2000 (17). Phasing was performed with Molrep (18). Structure refinement was carried out with Refmac5 (18). Model building was manually performed with COOT (19). The statistics of the data collection and structure refinement are summarized in Table 1. Atomic coordinates and structure factors have been deposited in the Protein Data Bank under ID codes 3R29 (RXRαLBD-SMRT) and 3R2A (RXRαLBD-rhein-SMRT).
TABLE 1.
Data collection and refinement statistics
| Statistics | RXRαLBD-SMRT | RXRαLBD-rhein-SMRT |
|---|---|---|
| Data collection | ||
| Space group | P 41 21 2 | P 21 21 21 |
| Cell dimensions | ||
| a, b, c (Å) | 118.22, 118.22, 84.02 | 85.03, 117.58, 117.12 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 44.7-2.9 (3.0-2.9)a | 42.5-3.0 (3.1-3.0) |
| Rsym or Rmerge | 0.092 (0.447) | 0.083 (0.542) |
| I/σI | 49.2 (7.3) | 29.8 (4.1) |
| Completeness (%) | 99.5 (100) | 99.0 (100) |
| Redundancy | 7.5 (7.5) | 3.6 (3.8) |
| Refinement | ||
| Resolution (Å) | 44.8-2.9 | 42.5-3.0 |
| No. of reflections | 13,219 | 22,792 |
| Rwork/Rfree | 0.251/0.292 | 0.247/0.290 |
| No. of atoms | 3,504 | 6,901 |
| B-factors | 65.1 | 70.1 |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.019 | 0.017 |
| Bond angles (°) | 1.239 | 1.109 |
| Ramachandran plot (%) | ||
| Most favored regions | 94.3 | 93.8 |
| Allowed regions | 5.7 | 6.2 |
a Values in parentheses are for highest resolution shell.
Luciferase Assay
Mammalian one-hybrid and transactivation experiments were performed using luciferase assays in HEK293T cells. Transient transfection was conducted using Lipofectamine 2000 (Invitrogen) according to the manufacturer's guideline. To evaluate the selectivity of rhein on different nuclear receptors, GAL4DBD-RXRαLBD, -PPARγLBD, -LXRαLBD, or -FXRLBD was cotransfected with UAS-TK-Luc reporter and Renilla luciferase vector pRL-SV40. For the transactivation assays, pcDNA3.1-RXRα, -PPARγ, -LXRα, or -FXR was cotransfected with pRL-SV40 plus pGL3-pro-RXRE-Luc, -PPRE-Luc, -LXRE-Luc, or FXRE-Luc, respectively. Five hours after transfection, cells were incubated with varied concentrations of rhein and the related positive agonist for another 24 h. RXRα agonist 9-cis-retinoic acid, PPARγ agonist rosiglitazone, LXRα agonist TO901317, and FXR agonist chenodeoxycholic acid were used as positive agonists. Luciferase activities were then measured using the Dual Luciferase Assay System kit (Promega). Results were shown as means ± S.E., n = 3. The figures were prepared with the software Origin.
Surface Plasma Resonance Technology-based Assay
Binding affinity of rhein toward RXRαLBD was determined with BIACORE 3000 instrument (BIACORE) based on our previous report (5). Briefly, RXRαLBD was immobilized onto a CM5 sensor chip according to the standard primary amine-coupling procedures. Rhein was serially diluted and injected into the channels at a flow rate of 20 μl/min for 60 s followed by disassociation for 120 s.
RESULTS
SMRT Binding Induces a Significant Structural Rearrangement of RXR Tetramer
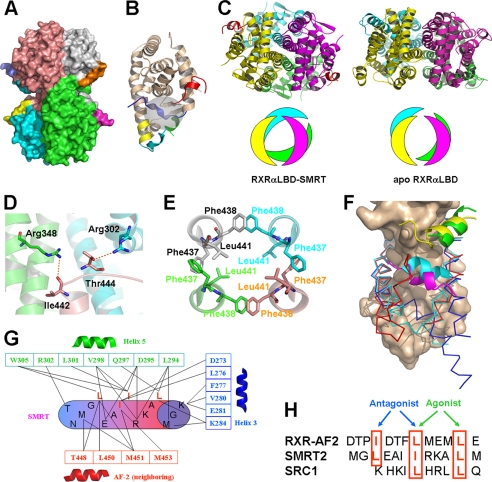
Our determined crystal structure of RXRαLBD-SMRT shows a tetrameric RXRαLBD conformation with one SMRT peptide bound to each receptor (Fig. 1A). The key secondary structures involving RXR activation and ligand-binding pocket are shown in Fig. 1B. Compared with the previously reported apoRXRαLBD tetramer, SMRT binding induces a significant rearrangement of the tetramer (4, 5). The two symmetry dimers in the tetramer rotate about 30° along the tetrameric axis from the position in its apo form (Fig. 1C). As a result, the atom O of Ile442 and OG1 of Thr444 in the C terminus of helix 10 forms two hydrogen bonds with NH1 of Arg346 in helix 7 and NH2 of Arg302 in helix 4 from its two neighboring monomers, respectively (Fig. 1D). Remarkably, the two adjacent phenylalanine residues Phe437 and Phe438 in helix 11 from each monomer of the tetramer form hydrophobic interactions in a hand-in-hand manner (Fig. 1E). Such interactions among the four helices 11 are believed to play a critical role for the tetramer stabilization. Additionally, Leu441 from each helix 11 extends itself inward to further stabilize these four helices hydrophobically (Fig. 1E). As a consequence of the tetrameric rearrangement, SMRT binding results in a much larger tetramer interface (2179.8 Å2) compared with the tetramer of apoRXRαLBD (1475.8 Å2) (5). These findings thus suggest that SMRT-bound RXRαLBD may be a more stable tetramer than apoRXRαLBD tetramer.
FIGURE 1.
Structure of RXRαLBD-SMRT. A, overall architecture of RXRαLBD-SMRT. Each RXRαLBD monomer and SMRT peptide is shown in a distinct color. B, RXRαLBD monomer indicating the key secondary structures for activation. wheat, conserved RXRαLBD core structure; green, N-terminal helix 3; yellow, helix 11; cyan, C-terminal AF-2 motif; blue, neighboring AF-2 motif; red, SMRT. Ligand-binding pockets are also indicated in the gray cycle. C, comparison of the tetramer rotations between the RXRαLBD-SMRT on the left and apoRXRαLBD on the right. Rotations are shown in both ribbon (upper) and model (lower) configurations. D, Ile442 and Thr444 forming two hydrogen bonds with Arg348 and Arg302 of the two neighboring monomers, respectively. E, Phe437 and Phe438 in the four monomers making hydrophobic interactions with each other in a hand-in-hand manner. Leu441 of each monomer extends itself inward to further stabilize helix 11 hydrophobically. F, structural superposition among apoRXRαLBD (blue), SMRT-bound RXRαLBD (red), and agonist-activated RXRαLBD (cyan) structures revealing different conformations of helix 3 and AF-2. The conserved core structure of the LBD is shown as surface; coactivator SRC-1 is in green, corepressor SMRT in yellow, and the AF-2 motif of the neighboring monomer in magenta. G, interactions between the SMRT motif (cylinder) and RXRαLBD (boxed residues, helix 3 in blue, helix 5 in green, and the AF-2 motif of the neighboring monomer in red). H, structure-based sequence alignment of the AF-2 motif, corepressor SMRT, and coactivator SRC1 on the overlapped binding pocket. The key residues are shown in red boxes and are indicated for the agonist or antagonist binding.
Structural superposition of RXRαLBD monomers from tetramers in the presence or absence of SMRT indicates a conserved core structure of LBD, but different helix 3 and AF-2 motif conformations (Fig. 1F). When SMRTs bind to RXRαLBD, the N terminus of helix 3 bends toward the ligand-binding pocket as in the active state upon agonist binding (20) (Fig. 1F). In addition, the AF-2 motif positions itself into the neighboring monomer and is fixed by helices 3′, 4′, and 10′ (Fig. 1A). Due to the rotation along the tetrameric axis, the AF-2 motif thus exhibits different conformations in apo and SMRT-bound RXRαLBD structures.
Compared with other SMRT-bound nuclear receptors, the SMRT corepressor motif in the current structure binds into a conserved corepressor-binding site (12, 13). The key residues Leu2341, Ile2345, and Leu2349 make hydrophobic interactions with Lys284 in helix 3; Leu294, Asp295, Gln297, Val298, Leu301, and Trp305 in helix 5; Thr449, Leu451, and Met452 in the neighboring AF-2 motif (Fig. 1G). Additionally, Ile2344 also interacts with Asp273 and Leu276 in helix 3 and Met452 and Met454 in the neighboring AF-2 motif. Interactions between SMRT and the N terminus of the AF-2 motif indicate that the AF-2 motif stabilized SMRT binding to the receptor. The C terminus of the AF-2 motif in our RXRαLBD-SMRT structure, however, shows no clear electron density due to its flexibility induced by SMRT/AF-2 competitive binding to the receptor (21).
Structure-based sequence alignment of steroid receptor coactivator 1 (SRC-1) coactivator motif, SMRT corepressor motif, and AF-2 motif reveals conserved binding interactions (Fig. 1H). The three critical residues Ile447, Leu451, and Leu455 of the AF-2 motif bound to a similar binding site to Leu2341, Ile2345, and Leu2349 of SMRT, making SMRT/AF-2 competitive binding to the receptor (21). On the other side, 451LMEML455 of the AF-2 motif mimicked the LXXLL motif of coactivator SRC-1 for the AF-2-mediated inhibition against coactivator binding (4, 5). Therefore, it is suggested that RXR uses an overlapped coregulator-binding pocket for coactivator SRC-1, corepressor SMRT and AF-2 motifs, whereas coactivator, corepressor, and AF-2 motifs have to compete with each other for binding into this pocket.
Rhein Is Identified as a Selective RXR Antagonist
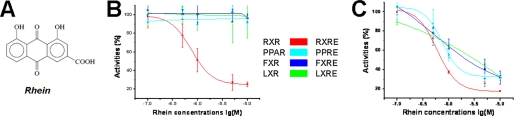
In our previous study, danthron from the traditional Chinese medicine rhubarb was found to be a specific antagonist against RXRα (5). Rhein (Fig. 2A) is another major component of rhubarb extracts. Compared with danthron, rhein has an additional carboxyl group at the C3 site of danthron. Mammalian one-hybrid-based assays are subsequently employed to evaluate the effects of rhein on RXRα transcriptional activities. As shown in Fig. 2B, rhein inhibits the known agonist 9-cis-retinoic acid-induced RXRα transactivation by IC50 value of 0.75 μm. However, this compound exhibits no activities against all other tested nuclear receptors, including PPARγ, FXR, and LXRα (Fig. 2B). Further transactivation assays demonstrate that rhein represses not only the transcription of RXRE mediated by RXRα/RXRα homodimer, but also several kinds of RXRα-involved heterodimers, including PPRE, FXRE, and LXRE, which are mediated by RXRα/PPARγ, RXRα/FXR, and RXRα/LXRα heterodimers, respectively (Fig. 2C). These results thus suggest that rhein exerts its trans-repression activities on RXRα homodimer and RXRα-involved heterodimers by binding RXRα as a good selective target over other nuclear receptors.
FIGURE 2.
Rhein as a selective RXRα antagonist. A, chemical structures of rhein. B, rhein repressing the 9-cis-retinoic acid-induced transcription of GAL4DBD-RXRαLBD by mammalian one hybrid. However, rhein exhibits no activities on the transcription of GAL4DBD-PPARγLBD, GAL4DBD-FXRLBD, and GAL4DBD-LXRαLBD. C, transactivation assays indicating that rhein represses all RXRα-involved dimeric transcription, including RXRE, PPRE, FXRE, and LXRE, mediated by RXRα/RXRα homodimer, RXRα/PPARγ, RXRα/FXR, and RXRα/LXRα heterodimers, respectively.
Rhein Binding Results in a Displacement of SMRT for AF-2 Motif
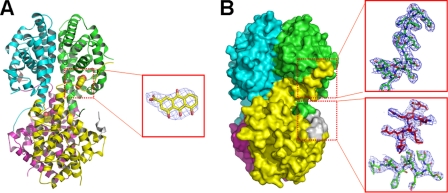
Our previous isothermal titration calorimetry results on danthron binding to RXRαLBD showed that this antagonist bound to the receptor with a stoichiometric ratio of 1:2, and such a binding changed SMRT recruiting ratio from 1:1 to 1:2 (5). Our current crystal structure of RXRαLBD-rhein-SMRT reveals the structural basis behind these biophysical results. In this structure, two rhein molecules and two SMRT peptides are bound in RXRαLBD tetramer (Fig. 3A), different from the case in RXRαLBD-SMRT structure, where four SMRT peptides bind to RXRαLBD tetramer (Fig. 1A). Rhein binds into the hydrophobic ligand-binding pocket with the hydrogen bonds between OAC and OAF of rhein and O of Cys432 in the C terminus of helix 10.
FIGURE 3.
Structure of RXRαLBD-rhein-SMRT. A, overall architecture of RXRαLBD-rhein-SMRT with the electron density of rhein (omit map contoured at 1.0σ level). B, surface of RXRαLBD-rhein-SMRT showing that the AF-2 motif of the neighboring monomer covers the ligand-binding pocket and displaces SMRT from the coregulator-binding site. The electron density map contoured at 1.0σ level shows an unambiguous positioning of the AF-2 motif (in green sticks) and SMRT corepressor motif (in red sticks).
As indicated, the most significant conformational changes upon rhein binding are observed in helix 3 and the AF-2 motif, both of which are crucial for RXR activation. The N terminus of helix 3 (43VTNICQAADKQLF55) shifts outward to adapt itself for ligand binding. Notably, rhein binding redirects the neighboring AF-2 into the SMRT binding site (Fig. 3B). The electron density map shows an unambiguous positioning of SMRT corepressor motif and the C terminus of the AF-2 motif (Fig. 3B). This observation of rhein-induced displacement of SMRT by the AF-2 motif thus confirms the competitive binding of SMRT/AF-2 to the receptor (21). Moreover, such a displacement will undoubtedly contribute to the stabilization of RXR tetramerization by an enlarged tetramer interface. Additionally, by interacting with Ile447 and Leu451 of the neighboring AF-2 motif (Fig. 3A), rhein catches this motif to stabilize the inactive tetramer further, thus totally repressing the RXR-involved transactivation (Fig. 2D).
Multiple Functions of AF-2 Motif
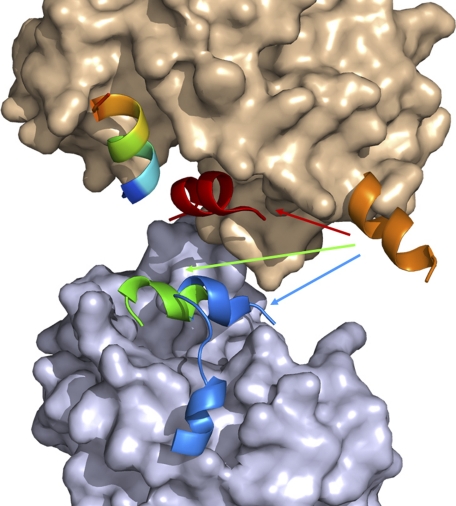
Interestingly, the AF-2 motif adopts different conformations for agonist, antagonist, and coregulator bindings (Fig. 4). In our previous crystal structure of bigelovin-activated RXRαLBD, the AF-2 motif overturned to seal off the ligand-binding pocket, in which Leu451 and Leu455 of the AF-2 motif played essential roles by interacting with the agonist (20). AF-2 overturning also made this motif interact directly with SRC-1 coactivator motif for RXR activation. In the autorepressed apoRXRαLBD tetramer structure, these two leucine residues (Leu451 and Leu455) of the AF-2 motif positioned themselves into the coregulator-binding pocket of the neighboring monomer to resemble the LXXLL motif of coactivator SRC-1 for RXR autorepression (4, 5) (Fig. 1H). While in the presence of corepressor SMRT, the AF-2 motif competes with SMRT for binding to the receptor. Moreover, SMRT-induced tetrameric rearrangement makes the N terminus of AF-2 interact with SMRT, but the C terminus of the AF-2 motif becomes flexible. Upon antagonist binding, Ile447 and Leu451 of the neighboring AF-2 motif interact with the antagonist, followed by a displacement of SMRT by AF-2 binding into the coregulator-binding site (Fig. 2B). Thus, the AF-2 motif allocates different residues for agonist or antagonist interaction in the active or repressive state. It is also indicated that the AF-2 motif competes with both the coactivator and corepressor for RXR binding. As reported, removing of the AF-2 motif converted RXR to a potent transcriptional repressor (22), which might be attributed to the multiple functions of the AF-2 motif on both coregulator competition and recruitment. Therefore, it is concluded that the multifunctional AF-2 motif adopts different conformations to interact with agonist or antagonist and mediates the recruitment of both coactivator and corepressor.
FIGURE 4.
Multiple functions of AF-2 motif. AF-2 exhibits different conformations in apo (orange), SMRT-bound (green), antagonist-bound (blue), and agonist-bound (red) structures. Surface of the receptor is shown in wheat, surface of the neighboring monomer shown in light blue is from the SMRT-bound tetramer, and the coregulator is shown in rainbow.
DISCUSSION
Nuclear receptors comprise a superfamily of transcription factors regulating complex networks in the processes of cell development and metabolism (23). The LBDs of nuclear receptors possess similarities in sequence and structure as well as mechanisms for activation. However, they adopt different conformations for antagonist and corepressor bindings. It was reported that the PPARα AF-2 motif packed loosely against helix 3 (12), and the estrogen-related receptor-γ AF-2 motif extended from the LBD (13), both of which exhibited no effects on SMRT and antagonist binding. In our current crystal structures of RXRαLBD-SMRT and RXRαLBD-rhein-SMRT, RXR adopts alternative ways for the repression, in which AF-2 mediates both SMRT and antagonist binding to their pockets. Additionally, SMRT-repressed and antagonist-repressed RXRs are both in tetramer, different from the dimeric forms for the other nuclear receptors.
Structure-based sequence alignment for the motifs of coactivator, corepressor, and AF-2 shows that RXR employs an overlapped pocket for their bindings (Fig. 1F). In apoRXRαLBD, the AF-2 motif sterically masks this binding site to inhibit the coactivator recruitment. While in the presence of RXR agonist, AF-2 turns to interact with the agonist, leaving the coregulator-binding site for coactivator SRC-1 recruitment (20). Interestingly, our current work indicates that the RXR antagonist is not required for corepressor SMRT recruitment. SMRT bound to the tetrameric RXR in the absence of the antagonist, and SMRT-induced RXR rearrangement further stabilizes the inactive tetramer. Moreover, the RXR antagonist displaces SMRT for AF-2 binding instead of helping SMRT recruitment. Such a displacement benefits the stabilization of the inactive RXR tetramer by enlarged tetrameric interface.
As shown in Fig. 1E, helix 11 plays a critical role in the tetramer stabilization of SMRT-induced RXR rearrangement. Although helix 11 (436LFFFKL441) was reported to help the corepressor SMRT binding to RXR (24), both of our current structures indicated that this helix is too far from the coregulator-binding site to make direct contacts with the corepressor motif.
Another significant discovery from our current structures is about the multiple functions of the AF-2 motif. This motif occupies the coregulator-binding site in the autorepressed RXR structure and plays either a positive role in transcriptional activation by recruiting the coactivator SRC-1 or a negative role in transcriptional repression by recruiting the corepressor SMRT. Previous studies indicated that the AF-2 motif inhibited the coactivator SRC-1 binding (4), whereas the competitive binding of the AF-2 motif and corepressor SMRT to the receptor is also observed in both of our current structures. Additionally, we find that the N terminus of the AF-2 motif helps SMRT binding to the receptor by direct interactions. Structural superposition between agonist- and antagonist-bound RXR structures reveals different residues of the AF-2 motif responsible for agonist or antagonist binding (Fig. 1H). Ile447 and Leu451 are for the antagonist, whereas Leu451 and Leu455 are for the agonist.
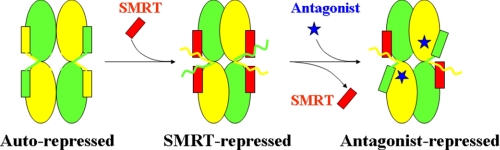
Therefore, based on all of these findings, we propose a potential model of RXR repression on the tetramer (Fig. 5). In the absence of ligand, RXRs exist in equilibrium of the autorepressed tetramers and SMRT-repressed tetramers, with the competitive binding of the AF-2 motif and SMRT corepressor motif to the coregulator-binding site. The RXR antagonist mediates the SMRT/AF-2 motif exchange with an enlarged tetramer interface. In whatever tetrameric conformations, RXRs have no transcriptional activities until the agonist-induced RXR dimerization and coactivator recruitment happen. Therefore, ligand-dependent exchange of coactivator, corepressor, and AF-2 motifs should be the fundamental regulator for RXR functioning.
FIGURE 5.
Proposed model of RXR repression on the tetramer.
This work was supported by State Key Program of Basic Research of China Grants 2010CB912501, 2007CB914304, and 2009CB918502, National High-Tech R&D Program of China Grant 2011AA090071-2, National Natural Science Foundation of China Grants 30925040, 30890044, 30801415, and 10979072, Science Foundation of Shanghai Grant 08431902900, Shanghai Basic Research Project from the Shanghai Science and Technology Commission Grant 11ZR1444500, and Shanghai Rising-Star Program Grant 09QA1406800.
The atomic coordinates and structure factors (codes 3R29 and 3R2A) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- RXR
- retinoic X receptor
- AF-2
- activation function 2
- DBD
- DNA-binding domain
- FXR
- farnesoid X receptor
- LBD
- ligand-binding domain
- LXR
- liver X receptor
- PPAR
- peroxisome proliferator-activated receptor
- rhein
- 4,5-dihydroxy-9,10-dioxoanthracene-2-carboxylic acid
- SMRT
- silencing mediator for retinoid and thyroid hormone receptors
- SRC
- steroid receptor coactivator.
REFERENCES
- 1. McKenna N. J., O'Malley B. W. (2010) Cell 142, 822–822.e1 [DOI] [PubMed] [Google Scholar]
- 2. Germain P., Chambon P., Eichele G., Evans R. M., Lazar M. A., Leid M., De Lera A. R., Lotan R., Mangelsdorf D. J., Gronemeyer H. (2006) Pharmacol. Rev. 58, 760–772 [DOI] [PubMed] [Google Scholar]
- 3. Egea P. F., Mitschler A., Rochel N., Ruff M., Chambon P., Moras D. (2000) EMBO J. 19, 2592–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gampe R. T., Jr., Montana V. G., Lambert M. H., Wisely G. B., Milburn M. V., Xu H. E. (2000) Genes Dev. 14, 2229–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang H., Zhou R., Li L., Chen J., Chen L., Li C., Ding H., Yu L., Hu L., Jiang H., Shen X. (2011) J. Biol. Chem. 286, 1868–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Safer J. D., Cohen R. N., Hollenberg A. N., Wondisford F. E. (1998) J. Biol. Chem. 273, 30175–30182 [DOI] [PubMed] [Google Scholar]
- 7. Yoh S. M., Chatterjee V. K., Privalsky M. L. (1997) Mol. Endocrinol. 11, 470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Karagianni P., Wong J. (2007) Oncogene 26, 5439–5449 [DOI] [PubMed] [Google Scholar]
- 9. Fernández-Majada V., Pujadas J., Vilardell F., Capella G., Mayo M. W., Bigas A., Espinosa L. (2007) Cell Cycle 6, 1748–1752 [DOI] [PubMed] [Google Scholar]
- 10. Uchikawa J., Shiozawa T., Shih H. C., Miyamoto T., Feng Y. Z., Kashima H., Oka K., Konishi I. (2003) Cancer 98, 2207–2213 [DOI] [PubMed] [Google Scholar]
- 11. Fang S., Suh J. M., Atkins A. R., Hong S. H., Leblanc M., Nofsinger R. R., Yu R. T., Downes M., Evans R. M. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 3412–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu H. E., Stanley T. B., Montana V. G., Lambert M. H., Shearer B. G., Cobb J. E., McKee D. D., Galardi C. M., Plunket K. D., Nolte R. T., Parks D. J., Moore J. T., Kliewer S. A., Willson T. M., Stimmel J. B. (2002) Nature 415, 813–817 [DOI] [PubMed] [Google Scholar]
- 13. Wang L., Zuercher W. J., Consler T. G., Lambert M. H., Miller A. B., Orband-Miller L. A., McKee D. D., Willson T. M., Nolte R. T. (2006) J. Biol. Chem. 281, 37773–37781 [DOI] [PubMed] [Google Scholar]
- 14. He Z. H., He M. F., Ma S. C., But P. P. (2009) J. Ethnopharmacol. 121, 313–317 [DOI] [PubMed] [Google Scholar]
- 15. Jia Z. H., Liu Z. H., Zheng J. M., Zeng C. H., Li L. S. (2007) Exp. Clin. Endocrinol. Diabetes 115, 571–576 [DOI] [PubMed] [Google Scholar]
- 16. Gao Q., Qin W. S., Jia Z. H., Zheng J. M., Zeng C. H., Li L. S., Liu Z. H. (2010) Planta Med. 76, 27–33 [DOI] [PubMed] [Google Scholar]
- 17. Otwinowski Z., Minor W. (1997) Methods Enzymo. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 18. Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 19. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 20. Zhang H., Li L., Chen L., Hu L., Jiang H., Shen X. (2011) J. Mol. Biol. 407, 13–20 [DOI] [PubMed] [Google Scholar]
- 21. Ghosh J. C., Yang X., Zhang A., Lambert M. H., Li H., Xu H. E., Chen J. D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5842–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang J., Hu X., Lazar M. A. (1999) Mol. Cell. Biol. 19, 6448–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Renaud J. P., Moras D. (2000) Cell. Mol. Life Sci. 57, 1748–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu X., Li Y., Lazar M. A. (2001) Mol. Cell. Biol. 21, 1747–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]