Abstract
Rtt109 is a yeast histone acetyltransferase (HAT) that associates with histone chaperones Asf1 and Vps75 to acetylate H3K56, H3K9, and H3K27 and is important in DNA replication and maintaining genomic integrity. Recently, mass spectrometry and structural studies of Rtt109 have shown that active site residue Lys-290 is acetylated. However, the functional role of this modification and how the acetyl group is added to Lys-290 was unclear. Here, we examined the mechanism of Lys-290 acetylation and found that Rtt109 catalyzes intramolecular autoacetylation of Lys-290 ∼200-times slower than H3 acetylation. Deacetylated Rtt109 was prepared by reacting with a sirtuin protein deacetylase, producing an enzyme with negligible HAT activity. Autoacetylation of Rtt109 restored full HAT activity, indicating that autoacetylation is necessary for HAT activity and is a fully reversible process. To dissect the mechanism of activation, biochemical, and kinetic analyses were performed with Lys-290 variants of the Rtt109-Vps75 complex. We found that autoacetylation of Lys-290 increases the binding affinity for acetyl-CoA and enhances the rate of acetyl-transfer onto histone substrates. This study represents the first detailed investigation of a HAT enzyme regulated by single-site intramolecular autoacetylation.
Keywords: Chromatin Regulation, Enzyme Mechanisms, Epigenetics, Histone Acetylase, Histone Modification, Asf1, Autoacetylation, K56 Acetylation, Rtt109, Vps75
Introduction
Compaction of the eukaryotic genome is achieved by the packaging of DNA into chromatin. The fundamental unit of chromatin, the nucleosome, is composed of 147 base pairs of DNA wrapped around an octamer of histones containing two each of H2A, H2B, H3, and H4 (1). The covalent modification of histones, such as phosphorylation, acetylation, and methylation regulates DNA replication, repair, and transcription (2–6). Histone acetyltransferases (HATs),2 a particular class of modification enzymes, catalyze the transfer of the acetyl group from acetyl-CoA onto the ϵ-amine group of lysines on histone substrates (7–9). There are three distinct HAT families that are classified according to their primary sequence and structural homology. These include the GNAT (Gcn5-related N-acetyltransferase), MYST (MOZ, Ybf2/Sas3, Sas2, and Tip60) and p300/CBP/Rtt109 families (7,9,10). Despite the divergence in sequence homology among the families, these HAT enzymes contain structurally similar core catalytic domains and utilize direct lysine substrate attack mechanisms (sequential) to carry out acetyl transfer (7,9–11).
Rtt109 (regulator of Ty1 transposition gene product 109) is a unique fungal-specific HAT enzyme (also named KAT11) that shares little sequence homology with other HAT enzymes. First identified to regulate the mobility of the Ty1 transposon element, Rtt109 HAT activities are important in a number of nuclear processes including DNA replication and maintaining genome integrity (12–17). Unique from other HAT enzymes, Rtt109 requires association with histone chaperones to direct substrate specificity and enhance catalysis (15, 18–20). Histone chaperones bind histones to prevent unfavorable histone:DNA interactions and function in the assembly and disassembly of nucleosomes (21, 22). Rtt109, with the histone chaperone Asf1 (anti-silencing function 1), acetylates lysine 56 (Lys-56) on the histone H3 core domain (12, 14, 15, 19). Asf1 is thought to form a transient complex with Rtt109 and stimulate activity by properly presenting H3-H4 dimers for Lys-56 acetylation (23–27). This mark occurs on newly synthesized H3 in Saccharomyces cerevisiae and Schizosaccharomyces pombe during S-phase and is required for the assembly of H3-H4 dimers onto DNA during cell replication (12, 15, 28). H3K56 acetylation also promotes histone assembly at sites of DNA damage and is enriched at the promoters of transcriptionally active genes (14, 17, 29–32). Distinct from the activities associated with Asf1, Rtt109 forms a high affinity complex with the NAP1 family histone chaperone Vps75 (Vacuolar protein sorting 75) (11, 18, 33–35). The Rtt109-Vps75 complex acetylates H3K9 and H3K27, which are overlapping functions with the HAT Gcn5 (18, 20, 34, 36–38). Similar to H3K56 acetylation, the acetylation of H3K9 and Lys-27 occurs on newly synthesized histones (18, 36). Importantly, Vps75 stimulates Rtt109 HAT activity by ∼200-fold and stabilizes the catalytically active conformation of Rtt109 (11, 18–20). Thus, distinct pathways exist for the regulation of Rtt109 by two histone chaperones.
Recently solved structures demonstrated that Rtt109 is a structural ortholog to the mammalian acetyltransferase p300(39–41). Notably, Rtt109 structures revealed electron density consistent with an acetyl group attached to lysine 290, a residue located near the proposed active site of Rtt109 (39–41). Mass spectrometric analyses showed Lys-290 is mostly acetylated in affinity purified TAP-tagged Rtt109 from yeast and Rtt109 recombinantly expressed and purified from bacteria (39, 41). Rtt109 Lys-290 mutants show decreased H3 acetylation in vitro and loss of Rtt109-dependent functions in vivo (40, 41). However, installation of a thiocarbamate analog of acetyl lysine at position 290 exhibited only an ∼4-fold increase in activity above a catalytically impaired K290C mutant(42). Thus, the functional role of Lys-290 acetylation remains unclear. Also, the mechanism of Lys-290 acetylation has not been established.
Here, we performed a detailed biochemical investigation on the mechanism of Rtt109 acetylation. We determined that Rtt109 catalyzes intramolecular autoacetylation at Lys-290. Additionally, we found that autoacetylation functions to stimulate Rtt109 by increasing the binding affinity for acetyl-CoA and enhancing the rate of acetyltransfer. This study represents the first detailed biochemical investigation of a HAT enzyme regulated by single-site intramolecular autoacetylation.
EXPERIMENTAL PROCEDURES
Chemical Reagents
Acetyl-CoA, DTT, NAD+ were purchased from Sigma-Aldrich. Tris-Cl, P81 cellulose discs, and sodium chloride were available from Fisher. Most other reagents were of the highest grade available and purchased either from Fisher or Sigma-Aldrich. [14H]acetyl-CoA (50–60 mCi/mmol) and [3H]acetyl-CoA (2–25 Ci/mmol) were obtained from Moravek and acrylamide/bisacrylamide was purchased from Bio-Rad. Site-directed mutagenesis kits from Agilent technologies were utilized to introduce point mutations at Lys-290 to Rtt109.
Protein Purification
Expression and purification of co-expressed recombinant His-Vps75-His-Rtt109 and His-Vps75 were performed as detailed in Berndsen et al. (18). In short, His-Rtt109 and His-Rtt109-His-Vps75 were purified over a nickel affinity column (GE, 5 ml), followed by purification over an SP-column (GE, 5 ml). His-Vps75 and His-Hst2 were purified over a nickel affinity column. H3 protein from Xenopus laevis were recombinantly purified as previously described in Luger et al. (1). Concentrations of co-purified Rtt109-Vps75 complex were determined from densitometry of Rtt109, according to previously described methods, as Vps75 was always present in ∼2-fold excess (11).
Preparation of Deacetylated Rtt109/Rtt109-Vps75
Deacetylation of Rtt109 was carried out in 10-ml reactions containing 50 mm Tris, pH 7.5, 1 mm DTT, 5 mm NAD+, 10 μm Hst2, and 10 μm Rtt109 at 25 °C for 30 min. In some preparations, 20 μm His-Vps75 was added to the reaction to stabilize Rtt109 after the deacetylation reaction. It should be that noted the presence Vps75 interfered with Hst2 deacetylation (data not shown), which made it necessary to add Vps75 following deacetylation. 10 ml of Sp-Column wash buffer (50 mm NaPO4, pH 6.5, 1 mm BME) was subsequently added to the reaction. This mixture was then run over an SP-column (5 ml) and proteins eluted with a linear ionic gradient (0–700 mm NaCl) over 20 column volumes. This purification method achieves purification of deacetylated Rtt109 or deacetylated Rtt109-Vps75, as Hst2 and NAD+ do not bind to the SP-column.
Autoacetylation of Rtt109 Assays
Rtt109 autoacetylation assays were performed in reactions containing 50 mm Tris, pH 7.5, 1 mm DTT, [14H]-acetyl-CoA or [3H]-acetyl-CoA, and deacetylated Rtt109 or deacetylated Rt109-Vps75 enzyme complex at 25 °C. The reactions were analyzed by one of two methods. [14C]- or [3H]-acetylated products were quantified by filter binding assay, as previously described (9, 11). Alternatively, 20 μl of [14C]-acetylated products were quenched with 10 μl of 5× SDS gel loading buffer, resolved on SDS-PAGE gels, gels dried, and the radioactivity visualized by phosphorimage analysis (Molecular Dynamics, GE).
Histone Acetyltransferase Assays
Rtt109-Vps75 histone acetyltransferase activity assays were performed in reactions containing 50 mm Tris, pH 7.5, 1 mm DTT, acetyl-CoA, full-length H3 or H3 peptide (amino acid sequence ARTKQTARKSTGGKAPRKQL), and Rtt109-Vps75 complex at 25 °C. Reactions were analyzed by a filter binding assay, as previously described (11, 43). Concentrations of enzymes and substrates are indicated in the figure legends.
Mass Spectrometry
To identify the site and extent of Rtt109 autoacetylation, LC MS/MS (liquid chromatography mass spectrometry) was used essentially as described (18, 44). Briefly, Rtt109 or deacetylated Rtt109-Vps75 complex were resolved by SDS-PAGE, stained with Coomassie Brilliant Blue, and bands extracted. Gel bands were chemically acetylated with deuterated acetic anhydride (Acros, 99.5% atom purity) and deuterated acetic acid (Acros, 98.5% atom purity) for 6 h at 25 °C followed by in-gel trypsin digestion at 37 °C for 18 h. Extracted peptides were purified using OMIX C18 tips (Varian), dried by a speed vacuum concentrator and re-suspended in 0.1% (v/v) formic acid for analysis. LC-MS/MS conditions were as previously outlined (18) and were processed using Thermo Bioworks software. Search parameters contained Xcorr versus Charge state (1 = 1.90, 2 = 2.70, 3 = 3.50), a peptide probability of 0.01 and a mass accuracy tolerance of ≤ 0.5 amu for peptides. The percent Rtt109 autoacetylation was obtained by dividing the frequency of non-deuterated acetylation by the total acetylated species for a given peptide.
Thermal Denaturation Assays
Thermal denaturation assays were performed to determine the melting temperature (Tm) of various Rtt109 proteins. Purified proteins were diluted to 5–10 μm in buffer (50 mm Tris pH 7.5 at 25 °C, 50 mm NaCl) containing 15x Sypro Orange (Invitrogen, delivered at 5000×). 35 μl of the thermal denaturation mixture were aliquoted into PCR strip tubes and placed into a Bio-Rad CFX96 Real-Time System C1000 thermal cycler. The temperature of the thermocycler was increased over a range of 10–95 °C at a rate of 0.5 °C/min and fluorescence monitored with the FRET channels. At least three trials were performed for each Rtt109 variant. The measured fluorescence was normalized so that minimum fluorescence was set to 0 and maximum fluorescence set 1. The data were then fit to a modified Equation 1 as previously utilized by Ericsson et al. (45) to obtain the Tm,
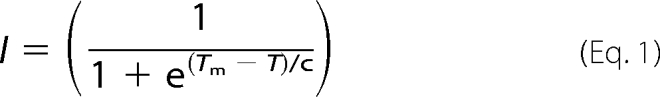 |
where I is the normalized fluorescence value at temperature T and C is a slope factor.
Intrinsic Protein Fluorescence
Prior to fluorescence measurements, 675-μl reactions containing 50 mm Tris (pH 7.5), 50 mm NaCl, 0.5 μm Rtt109-Vps75, and acetyl-CoA (varied concentrations) were equilibrated at 25 °C for 30 min. 650 μl of the reactions were excited at 295 nm (3 nm slit width) and the emission spectra recorded from 325–385 nm (3 nm slit width) in a quartz cuvette (Starna Cells, product no. NC9907274, 5 × 5 × 45 mm) on a Fluoromax-3 fluorimeter. The fraction of acetyl-CoA bound Rtt109-Vps75 was determined by Equation 2,
where Funbound is the fluorescence of Rtt109-Vps75 in the absence of acetyl-CoA, Fobserved is the observed fluorescence, and Fminimum is the lowest fluorescence observed at 340 nm. The fraction bound was multiplied by the Rtt109-Vps75 concentration to determine the concentration of acetyl-CoA bound Rtt109-Vps75 and plotted against the acetyl-CoA concentration. The data were fitted to Equation 3 to yield the dissociation constant (Kd) of acetyl-CoA to Rtt109-Vps75 complex,
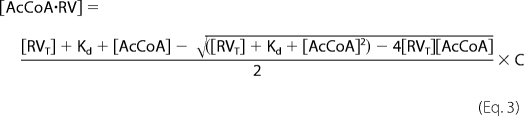 |
where [AcCoA·RV] is the concentration of acetyl-CoA-bound Rtt109-Vps75, [RVT] is the total concentration of Rtt109-Vps75, [AcCoA] is the concentration of acetyl-CoA, and C is the y axis maximum.
Acetyl-lysine Western Blotting Analysis
Rtt109 protein samples were resolved on SDS-PAGE gels and transferred to PVDF membranes. The membranes were blocked overnight in 5% (v/v) BSA-PBST (phosphate-buffered saline Tween-20; 0.05% Tween-20 v/v) at 4 °C, followed by 1 h of incubation with 2.5% (v/v) BSA-PBST containing a 1:5,000 dilution of an anti-acetyl lysine antibody (Cell Signaling, Product no. 9441L) at 25 °C. The membranes were washed three times for 5 min in PBST and incubated 1 h with 2.5% (v/v) BSA-PBST containing a 1:5,000 dilution of a horseradish peroxidase (HRP)-conjugated goat anti-rabbit (Santa Cruz Biotechnology, Product no. SC-2054) at 25 °C. Detection was performed using SuperSignal West Pico Substrate (Pierce) and exposed to film (Kodak) or analyzed by the QuantImage documentation system (GE).
RESULTS
Rtt109 Catalyzes Autoacetylation at Lys-290
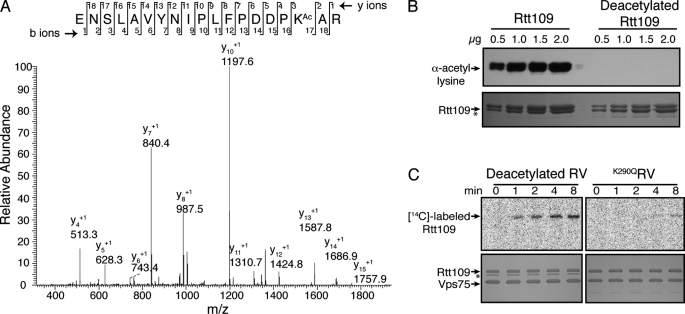
Recently, several crystal structures and mass spectrometric studies have detected nearly stoichiometric acetylation at Lys-290 from Rtt109 recombinantly expressed from Escherichia coli and purified from yeast (39–41). However, there is no direct evidence of autoacetyltransferase activity for Rtt109. Whereas this acetylation may be self-catalyzed, it was also possible that the protein acetyltransferase (Pat) in E. coli or other yeast HATs were responsible for the observed Lys-290 acetylation. Because Rtt109 recombinantly purified from E. coli was acetylated 92 ± 8% at Lys-290 (Fig. 1), generating deacetylated Rtt109 was necessary to determine whether Rtt109 is capable of autoacetylation. The NAD+-dependent deacetylase Hst2 removed >99% of the acetyl groups from Rtt109 (Fig. 1B and supplemental Fig. S1). Because Rtt109 alone is relatively unstable, Vps75 was added after the deacetylation reaction to improve the stability of the enzyme. This allowed for purification of sufficient quantities of deacetylated Rtt109-Vps75 (RV) required to measure autoacetyltransferase activity in various assays.
FIGURE 1.
Autoacetylation of Rtt109. A, Rtt109 is acetylated at Lys-290, LC MS/MS analysis of Rtt109 recombinantly expressed and purified from E. coli. A representative chromatogram depicts acetylation at Lys-290, with singly charged y-series ions labeled (inset: theoretical b and y fragment ions). Acetylated and deuterated peptides were quantified from an average of three independent purifications. B, deacetylation of Rtt109 by Hst2. Acetyl-lysine immunoblots (top) and SDS-PAGE Coomassie-stained gels (bottom) of Rtt109 and Rtt109 deacetylated by Hst2. Asterisk denotes Rtt109 doublet due to partial C-terminal truncation during the purification. Slightly truncated Rtt109 and full-length Rtt109 are equally acetylated and equally catalytically active (data not shown). C, Rtt109 catalyzes autoacetylation. Deacetylated RV (2 μm) and K290QRV (2 μm) were incubated with [14C]-acetyl-CoA (50 μm) and 20 μl of reactions were resolved on SDS-PAGE gels. Top, [14C]-acetylated Rtt109 autoradiograph image. Bottom, SDS-PAGE Coomassie-stained gel of the autoacetylation reaction. Asterisk denotes Hst2.
To determine if Rtt109 is capable of autoacetylation at Lys-290, deacetylated RV and a K290Q RV mutant (K290QRV) were incubated with radiolabeled acetyl-CoA and the acetyl transfer reaction monitored. Time points of the reaction were resolved on SDS-PAGE gels followed by autoradiography analysis. Whereas deacetylated RV demonstrated time-dependent autoacetylation, negligible acetyl transfer was observed with K290QRV (Fig. 1C). There is a small amount (<10%) of acetylation that occurs on another site(s) after extended time periods (Fig. 1C). This likely reflects non-enzymatic acetylation from excess acetyl-CoA and long reaction times. We did not observe other acetylation sites when a mass spectrometric analysis was performed using Rtt109 purified from E. coli. Furthermore, no acetylation occurred on Vps75. Thus, Rtt109 catalyzes autoacetylation, and Lys-290 is the primary site of modification.
Autoacetylation of Rtt109 at Lys-290 Occurs by an Intramolecular Mechanism
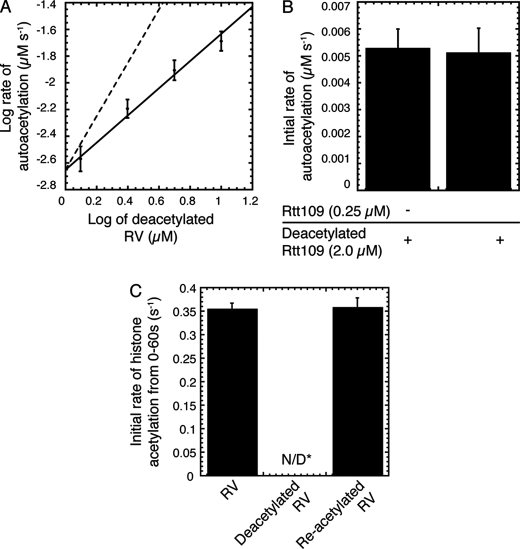
We next determined whether Rtt109 utilizes an intermolecular or intramolecular mechanism of autoacetylation. In an intramolecular mechanism, autoacetylation occurs within a single enzyme molecule and should yield a first-order rate of acetylation with respect to Rtt109 concentration. An intermolecular reaction requires an additional enzyme molecule to catalyze autoacetylation and should exhibit a second order reaction rate or higher. To determine the mechanism of acetylation, the initial rate of autoacetylation was measured as a function of deacetylated RV concentration (1.25–10 μm) at a fixed, concentration of acetyl-CoA (50 μm). The logarithmic plot of the initial rate of autoacetylation versus the deacetylated RV concentration resulted in a slope of 1.02 ± 0.05, consistent with a first-order reaction (Fig. 2A). These data provide support for an intramolecular mechanism of acetylation. Additionally the data yielded an apparent kcat of 0.0020 ± 0.0001 s−1, which approximates to one enzyme turnover every 8 min.
FIGURE 2.
Intramolecular mechanism of autoacetylation and the stimulatory role in histone acetylation. A, intramolecular mechanism of autoacetylation. Log-log plot of the initial rate versus the deacetylated RV concentration. Solid line denotes linear regression curve fit of the data. At least three trials were performed at each deacetylated RV concentration with error bars representing one standard deviation unit. Also shown is the theoretical curve fit (dashed lines) for a second order (slope = 2) reaction for comparison. B, initial rate analysis of autoacetylation in the presence or absence of Rtt109 (already acetylated), deacetylated Rtt109 and acetyl-CoA (50 μm). Experiments were performed three times with error bars representing standard error. C, autoacetylation stimulates HAT activity. The initial rate of histone acetylation from 0–60 s for acetylated RV, deacetylated RV and re-acetylated (reactivated) RV. Assays contained 50 nm RV, 10 μm H3, and 50 μm acetyl-CoA. Experiments were performed three times with error bars representing standard error. N/D*, no detectable rate of acetylation above background (no enzyme).
Rtt109 forms a high affinity complex with Vps75 such that in the RV complex, Lys-290 may not be readily accessible to other Rtt109 molecules (11,20). Therefore, we considered that Vps75 was preventing the intermolecular interaction of Rtt109 molecules. Furthermore, it was also possible that only initially acetylated Rtt109 is sufficiently active to catalyze intermolecular acetylation, which the previous experiment did not resolve because only deacetylated RV was present. To address these possibilities, rates of autoacetylation were measured in reactions containing catalytic quantities of acetylated-Rtt109 (0 or 0.25 μm), deacetylated Rtt109 (2 μm) and acetyl-CoA. It is important to note Vps75 was absent in this assay. The rates of autoacetylation were similar whether catalytic quantities of acetylated Rtt109 were present (0.0051 ± 0.0009 μm−1 s−1) or absent (0.0053 ± 0.0007 μm s−1) (Fig. 2B). Therefore, the presence of acetylated Rtt109 did not increase the rates of autoacetylation, as expected for an acetylated Rtt109 enzyme that has enhanced catalytic power and able to acetylate other Rtt109 molecules. The apparent kcat of autoacetylation of deacetylated Rtt109 without Vps75 (0.0026 ± 0.0006 s−1) were similar to deacetylated RV (0.0020 ± 0.0001 s−1), indicating Vps75 does not influence Rtt109 autoacetylation activity. Together, these data provide evidence toward an intramolecular mechanism of autoacetylation where the presence of the histone chaperone Vps75 has no apparent effect on the rate and mechanism.
Autoacetylation of Rtt109-Vps75 Stimulates Histone Acetyltransferase Activity
Crystal structures of Rtt109 revealed that acetylated Lys-290 resides near the proposed active site of Rtt109 (39–41). Moreover, previous studies reported that substituting Lys-290 to alanine results in decreased acetylation of Lys-56 on H3, while in vivo analyses demonstrated that mutation of this residue results in the loss of Rtt109-dependent functions. However, a recent study in which a thiocarbamate analog of acetyl lysine was installed at position 290 showed only ∼4-fold activity increase above a catalytically impaired K290C mutant (42). Thus, the role of acetylated Lys-290 in histone acetylation remained unclear. To investigate if Lys-290 autoacetylation regulates Rtt109 HAT activity, we compared the rates of histone H3 acetylation with acetylated and deacetylated RV (Fig. 2C). The initial rate of histone acetylation calculated from the first 60 s of the reaction was 0.36 ± 0.01 s−1 for RV, while HAT activity of the deacetylated enzyme was undetectable above background radioactivity (Fig. 2C). Although an exact number was not calculable, the rate of histone acetylation by deacetylated RV was estimated to be < 0.04 s−1. Beyond 60 s, we observed an expected exponential increase in histone acetylation, reflecting Rtt109 autoacetylation and subsequent activation of HAT activity (data not shown).
The indiscernible activity of deacetylated RV may have resulted from irreversible inactivation or denaturation of the enzyme during the preparation of deacetylated RV. To test this possibility, we evaluated the ability of deacetylated RV to be re-activated by autoacetylation. Deacetylated RV was allowed sufficient time (16 min, or ∼2 enzyme turnovers) to attain complete acetylation in the presence of acetyl-CoA, subsequently purified over cation exchange chromatography to remove excess acetyl-CoA and then assayed for HAT activity. Strikingly, re-acetylation recovered full activity, as the rate of histone acetylation (0.36 ± 0.02 s−1) of re-acetylated RV was indistinguishable from RV (0.36 ± 0.01 s−1) that had never been deacetylated (Fig. 2C). These results indicate that autoacetylation is a reversible process that functions to regulate Rtt109 HAT activity.
Autoacetylation Enhances Catalysis and Increases Acetyl-CoA Binding
To gain insight into the mechanism by which autoacetylation increases Rtt109 HAT activity, kinetic HAT analyses of various RV constructs were performed. Steady-state analysis of deacetylated RV was not possible because the kinetics are convoluted by the time-dependent autoacetylation and activation of histone acetylation. Accordingly, the analyses were performed with K290QRV and K290RRV. There are several examples in which mutation of a lysine to glutamine may function as an acetyl lysine mimetic while mutation to arginine may represent a constitutively unacetylated lysine (23, 46, 47). Contrary to these assumptions, our kinetic analyses of histone acetylation showed both K290QRV and K290RRV exhibited an ∼100-fold decrease in kcat (∼0.001 s−1) compared with RV (0.1 s−1) (Table 1). Thus, the acetyl-group at lysine 290 is indispensable for catalyzing efficient acetyl-transfer onto histone substrates and substitution with a glutamine does not mimic an acetyl-lysine.
TABLE 1.
Summary of steady-state kinetic constants for Rtt109-Vps75
| Rtt109 +Vps75 | Substrate | Kma | kcat | kcat/Km |
|---|---|---|---|---|
| μm | −1 | m−1s−1 | ||
| Wild typeb | Acetyl-CoA | 0.3 ± 0.1 | 1.1 ± 0.01 × 10−1 | 3.9 ± 0.5 × 105 |
| (Acetylated) | H3 peptide | 112 ± 11 | 1.1 ± 0.01 × 10−1 | 9.8 ± 0.9 × 102 |
| K290Q | Acetyl-CoA | 5 ± 1 | 8.2 ± 0.6 × 10−4 | 1.6 ± 0.3 × 102 |
| H3 peptide | 90 ± 20 | 1.1 ± 0.1 × 10−3 | 1.2 ± 0.3 × 101 | |
| K290R | Acetyl-CoA | 4 ± 1 | 1.1 ± 0.1 × 10−3 | 2.7 ± 0.7 × 102 |
| H3 peptide | 50 ± 9 | 7.1 ± 0.4 × 10−4 | 1.4 ± 0.3 × 101 | |
| D288Nb | Acetyl-CoA | 4.9 ± 0.9 | 2.8 ± 0.2 × 10−3 | 6 ± 1 × 102 |
| H3 peptide | 55 ± 11 | 2.1 ± 0.1 × 10−3 | 3.8 ± 0.8 × 101 |
a Standard errors are reported. All values are averages from at least three independent experiments.
b Data published previously are shown for comparison (11).
We questioned if the catalytic stimulation observed with Lys-290 acetylation was caused by increased protein stability. To determine the effect Lys-290 mutation on protein stability, thermal denaturation assays with RV, K290QRV, and K290RRV were performed. The melting temperatures of RV (Tm = 50 ± 1), K290QRV (Tm = 49 ± 1), and K290RRV (Tm = 49 ± 2) were similar, suggesting no significant differences in thermal stability between the enzyme complexes (Fig. 3A and supplemental Fig. S2). Potentially, the presence of Vps75 stabilized Rtt109 such that a measureable thermal shift was masked. To investigate this possibility and to gain insight into the stability of deacetylated enzyme, the melting temperatures of Rtt109 and deacetylated Rtt109 were measured in the absence of Vps75. As expected, because Vps75 is known to stabilize Rtt109 (11, 18–20, 34), there was a downward shift in Tm in the absence of Vps75 (Fig. 3). However, the melting temperatures of Rtt109 (Tm = 33 ± 1) and deacetylated Rtt109 (Tm = 33 ± 1) were indistinguishable, indicating acetylation does not stabilize Rtt109 (Fig. 3B and supplemental Fig. S2). These data suggest catalytic stimulation with Lys-290 acetylation is unlikely attributed to enhanced protein stability.
FIGURE 3.
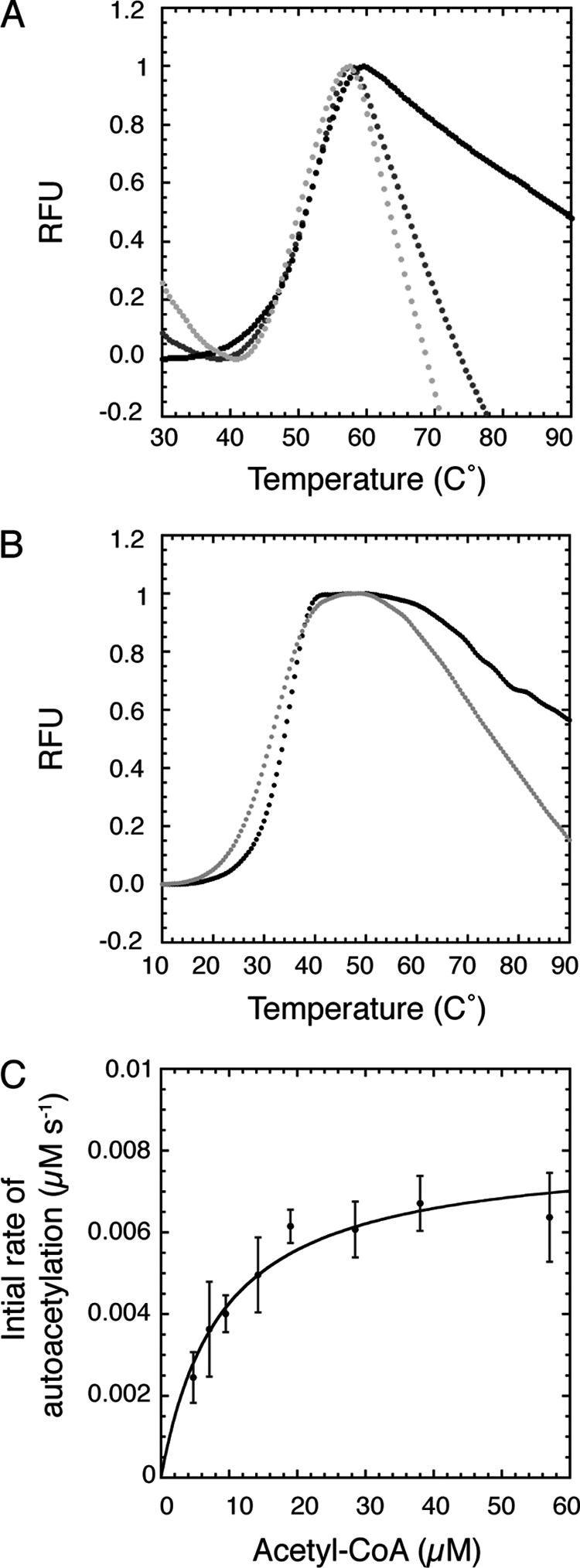
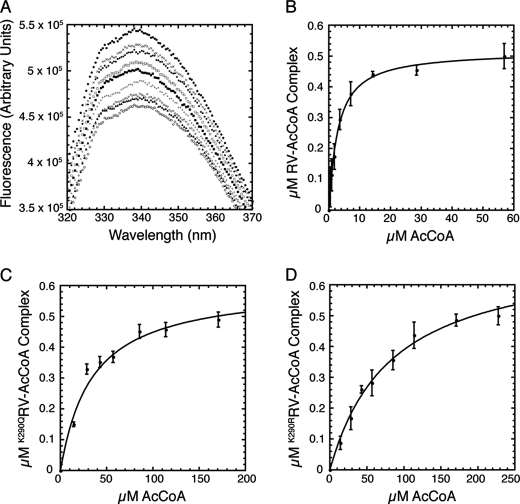
Thermal denaturation of various Rtt109 constructs and acetyl-CoA autoacetylation saturation curve of deacetylated RV. A, thermal denaturation assays of RV (black), K290QRV (dark gray), and K290R RV (light gray). A graph with representative experiments is shown. Tm = 50 ± 1 for RV, Tm = 49 ± 1 for K290QRV, and Tm = 49 ± 2 for K290RRV. B, thermal denaturation assays of Rtt109 (black) and deacetylated Rtt109 (light gray). Tm = 33 ± 1 for Rtt109 and Tm = 33 ± 1 for deacetylated Rtt109. A graph with representative experiments is shown. Error is standard error from at least three independent trials. SDS-PAGE gels of proteins used in thermal melts are in the supplemental data. C, acetyl-CoA saturation curve of the autoacetylation reaction with deacetylated RV (2.5 μm). The apparent Km = 9 ± 2 μm. Experiments were performed five times with error bars representing one standard deviation unit.
To determine if acetylated Lys-290 contributes to substrate binding, acetyl-CoA and H3 peptide saturation curves were performed with K290QRV and K290RRV. These steady-state kinetic analyses revealed ≤2-fold change in the H3 peptide Km values of the mutants compared with RV (Table 1), suggesting that Lys-290 is not critical for H3 substrate binding. However, compared with RV, an ∼10-fold increase in Km for acetyl-CoA was observed with K290QRV and K290RRV (Table 1). These kinetic results suggest acetylated Lys-290 is important for acetyl-CoA binding. If this were the case, we would predict that deacetylated RV would display a substantially perturbed Km value for the autoacetylation reaction. Indeed, there was an ∼30-fold Km difference for autoacetylation (apparent Km = 9 ± 2 μm) compared with the histone acetylation reaction (Km = 0.3 ± 0.1 μm) (Fig. 3C and Table 1). Together, these kinetic analyses strongly suggest acetylated Lys-290 functions in acetyl-CoA binding.
Because Michaelis constants reflect binding affinity but are not direct measurements, the dissociation constants of acetyl-CoA with the Lys-290 RV variants were determined by intrinsic protein fluorescence. Intrinsic protein fluorescence has been previously utilized to measure coenzyme A dissociation constants to RV(39). Because binding of acetyl-CoA to deacetylated RV would result in autoacetylation and product turnover, the Lys-290 mutants were used for the binding experiments. Acetyl-CoA binding measurements yielded a Kd value of 2.7 ± 0.3 μm for RV (Fig. 4 and supplemental Fig. S3), which is in good agreement with the previously determined Kd value of 4.4 ± 2.5 μm when acetonyl-CoA was the ligand (39). Acetyl-CoA binding measurements with K290QRV yielded a Kd value of 34 ± 5 μm and with K290RRV yielded a Kd value of 95 ± 13 μm (Fig. 4 and supplemental Fig. S3). The order-of-magnitude decrease in acetyl-CoA binding affinities of the Lys-290 mutants compared with RV provides further support for the importance of acetylated Lys-290 in binding acetyl-CoA. Together, these studies suggest that Lys-290 acetylation stimulates histone acetylation by increasing the binding affinity of Rtt109 for acetyl-CoA and enhancing catalytic transfer of acetyl group onto histone substrates.
FIGURE 4.
Acetyl-CoA binding curves with RV, K290QRV, and K290RRV. Intrinsic protein fluorescence measurements with RV constructs. Fluorescence values at 340 nm were utilized to determine Kd. Kd values are 2.7 ± 0.3 for RV, 34 ± 5 μm for K290QRV and 95 ± 13 μm for K290RRV. A, representative emission spectra of the fluorescence reduction of RV due to acetyl-CoA binding. 0 μm (filled circle), 0.44 μm (x-shape), 0.89 μm (filled diamond), 1.78 μm (open square), 3.56 μm (closed square), 7.11 μm (cross-shape), 14.22 μm (open diamond), 28.44 μm (closed triangle), and 56.90 μm (open circle) of acetyl-CoA was titrated into RV. Acetyl-CoA binding curves with RV (B) K290QRV (C), and K290RRV(D). At least four independent trials with each RV construct were performed with error bars representing one standard deviation unit.
DISCUSSION
In this study, we first sought to determine if Rtt109 catalyzes autoacetylation. Rtt109 was largely acetylated after purification from E. coli or yeast, which was an obstacle for measuring autoacetylation. Thus, we generated deacetylated Rtt109 and established that Rtt109 catalyzes intramolecular autoacetylation at Lys-290. Additionally, we found autoacetylation functions to stimulate HAT activity by increasing acetyl-CoA binding affinity and by enhancing the rate of acetyl group transfer. Moreover, HAT activity was fully reconstituted upon re-acetylation of deacetylated Rtt109, indicating this process is completely reversible and implicates autoacetylation as a regulatory mechanism.
Rtt109 and p300 Utilize Distinct Mechanisms of Autoacetylation and HAT Activation
Whereas both Rtt109 and the mammalian structural ortholog p300 catalyze autoacetylation, the mechanisms are strikingly distinct (48, 49). Consistent with an intramolecular mechanism, Rtt109 displayed first-order kinetics of autoacetylation. In contrast, p300 uses intermolecular acetylation, as demonstrated by a ∼fourth order reaction rate of autoacetylation (48). The p300 enzyme undergoes rapid multi-site acetylation (∼10 sites in 2 min), in which several acetylation sites on an autoinhibitory loop are catalytically important for histone acetylation (48, 49). Unlike p300, autoacetylation of Rtt109 occurs at the single critical site Lys-290, which is sufficient to stimulate Rtt109 HAT activity.
Acetylated Rtt109 Is Not a Catalytic Intermediate during Histone Acetylation
There are several lines of evidence that suggest acetylated Lys-290 does not represent a covalent enzyme intermediate (ping-pong mechanism) along the catalytic pathway of histone acetylation. Previously, we determined the rate-limiting step (kcat) in histone acetylation reflects the chemical attack of the substrate lysine on bound acetyl-CoA (11). Of significance, the turnover rate for autoacetylation (0.002 s−1) is 50-fold slower compared with the kcat of RV using histone H3 peptide as substrate (0.1 s−1)(Table 1). Thus, the rate of Lys-290 acetylation is not a kinetically competent rate to support the catalysis of histone acetyl transfer. Furthermore, a detailed steady-state kinetic analysis with acetylated RV provided strong support for a direct attack (sequential) mechanism of histone acetylation (11). Additionally, all other structurally and kinetically characterized HAT enzymes contain similar core catalytic domains and utilize direct attack mechanisms of catalysis, suggesting conservation of these features for Rtt109 (reviewed in Ref. 9).
The Acetyl Group at Lys-290 Is Critical for Stimulating Rtt109 HAT Activity
We found that mutation of Lys-290 to either glutamine or arginine resulted in histone acetylation rates that were equally inefficient (100-fold loss in kcat). Consistent with a critical function for acetylated Lys-290, only fully acetylated Rtt109-Vps75 exhibited robust histone acetyltransferase activity. Thus, the function of acetyl-lysine cannot be substituted with glutamine, a residue that conserves the neutral charge of the acetyl group. This is consistent with the observation that installation of a thiocarbamate analog of acetyl lysine at Lys-290 did not fully recover HAT activity (42). Moreover, introduction of alanine, arginine, glutamine, or tryptophan at residue Lys-290 into rtt109Δ yeast cells did not rescue the DNA damage sensitivity phenotype (40). Thus, in vivo and in vitro studies provide evidence for the critical requirement of acetylated Lys-290 for catalysis and cellular Rtt109-dependent functions.
We observed no significant thermal stability differences between Rtt109 and deacetylated Rtt109, therefore, it is unlikely that large conformational changes account for the increased HAT activity with autoacetylation. Instead, we suggest Lys-290 acetylation induces a local structural change of the active site that results in improved binding of acetyl-CoA and enhanced transfer of the acetyl group on the substrate lysine during chemical catalysis.
Structures of Rtt109 show the carboxylate group of conserved amino acid Asp-288 is hydrogen bonded to the amine group of the acetylated Lys-290 (39–41). Noting this interesting interaction, we had previously performed a kinetic analysis with the D288NRV mutant(11). Strikingly, D288NRV demonstrated similar kinetic parameters as K290Q RV and K290RRV, in which there were large acetyl-CoA Km defects and decreased kcat values (Table 1) (11). Together, these studies suggest that Lys-290 and Asp-288 work together to increase Rtt109 HAT activity. Potentially, Asp-288 may properly orient acetylated Lys-290, which promotes optimal positioning of acetyl-CoA for histone lysine attack.
Biological Implications of Rtt109 Autoacetylation
Compared with histone acetylation by activated RV, the Km for acetyl-CoA is ∼30-fold higher during Rtt109 autoacetylation. This relatively high Km value would be expected to sensitize Rtt109 activation to alterations in the availability of acetyl-CoA, especially at low concentrations. Several studies have shown the bulk of Rtt109 expression and activity is generally limited to yeast G1/S-phase (14, 17, 31, 50). Interestingly, acetyl-CoA levels are low during cell cycle replication (51, 52). Together, these observations suggest Rtt109 activation is regulated by the cellular acetyl-CoA concentration. Further studies will be required to determine how the metabolic status and fluctuating cellular acetyl-CoA levels influence Rtt109 autoacetylation and cell cycle replication.
In this study, we found Hst2 deacetylates Rtt109, producing intact Rtt109 enzymes that can undergo autoacetylation and activation. Although in vitro characterization has not been possible, Hst3 and Hst4 are likely the candidate deacetylases in vivo, as yeast cells deleted of Hst3 and Hst4 genes have increased H3 Lys-56 acetylation (30, 31, 53–55). Potentially, in addition to deacetylating H3 Lys-56 following S-phase, Hst3 and Hst4 may deacetylate and inactivate Rtt109, thereby lowering the overall Lys-56 acetylation levels. Deacetylation of Rtt109 by Hst2 is an intermolecular interaction, therefore, it is also possible that other HAT enzymes could acetylate and activate Rtt109. One potential candidate enzyme is Gcn5, which shares overlapping specificity with Rtt109 (e.g. H3 Lys-9 and H3 Lys-27) (37).
Importance of Autoacetylation in Regulating HAT Enzymes
This study represents the first detailed investigation of a HAT enzyme regulated by a single-site, intramolecular autoacetylation. There is increasing evidence that autoacetylation might be a common mechanism for regulating HAT activity, as Tip60, MOF, Esa1, and PCAF enzymes were also identified as acetylated proteins (48, 49, 56–58). Although acetylation of the enzymes generally stimulates HAT activity, the mechanisms of activation and regulation appear to diverge for each HAT enzyme. As discussed, p300 undergoes intermolecular autoacetylation, which controls substrate entry in the active site(48). Acetylation of Tip60 is proposed to lead to the dissociation of its oligomer status, which permits access to histone substrates (57). PCAF is suggested to be capable of both intramolecular and intermolecular acetylation, which influences its translocation to the nucleus and HAT activity(56). However, detailed analyses of these enzymes are limited and will require further investigation to determine the mechanisms of autoacetylation. Collectively, these studies suggest an emerging theme in which autoacetylation and subsequent activation by distinct mechanisms regulate HAT enzymes.
Supplementary Material
Acknowledgments
We thank all members in the research group of Dr. John Denu for helpful discussions, especially Dr. Erin Kolonko. We also thank Dr. Julie Mitchell (University of Wisconsin-Madison) for performing molecular simulations with the available Rtt109 structures, which has been useful in thinking about this study. We also appreciate the generosity of Dr. Deane Mosher (University of Wisconsin-Madison), who allowed us to use the Flouromax-3 instrument. We would also like to thank Dr. Paul Kaufman and Jessica Lopes da Rosa (University of Massachusetts Medical School). We acknowledge the University of Wisconsin-Madison Human Proteomics Program, funded by the Wisconsin Partnership Fund.
This work was supported, in whole or in part, by National Institutes of Health Grant GM059785 (to J. M. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- HAT
- histone acetyltransferase
- Rtt109
- Regulator of Ty1 Transposition 109
- Vps75
- vacuolar protein sorting 75
- Asf1
- anti-silencing function 1
- Hst2
- Hst3, and Hst 4, homolog of SIR two 2, 3, and 4
- Esa1
- essential Sas2-related acetyltransferase 1
- PCAF
- p300/CBP-associated factor
- Tip 60
- 60 kDa Tat-interactive protein
- MOF
- males-absent on the first
- LC-MS
- liquid chromatography mass spectrometry
- acetyl-CoA
- acetyl coenzyme A.
REFERENCES
- 1. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 2. Spencer V. A., Davie J. R. (1999) Gene 240, 1–12 [DOI] [PubMed] [Google Scholar]
- 3. Davie J. R. (1998) Curr. Opin. Genet. Dev. 8, 173–178 [DOI] [PubMed] [Google Scholar]
- 4. Latham J. A., Dent S. Y. (2007) Nat. Struct. Mol. Biol. 14, 1017–1024 [DOI] [PubMed] [Google Scholar]
- 5. Bhaumik S. R., Smith E., Shilatifard A. (2007) Nat. Struct. Mol. Biol. 14, 1008–1016 [DOI] [PubMed] [Google Scholar]
- 6. Li B., Carey M., Workman J. L. (2007) Cell 128, 707–719 [DOI] [PubMed] [Google Scholar]
- 7. Roth S. Y., Denu J. M., Allis C. D. (2001) Annu. Rev. Biochem. 70, 81–120 [DOI] [PubMed] [Google Scholar]
- 8. Lee K. K., Workman J. L. (2007) Nat. Rev. Mol. Cell Biol. 8, 284–295 [DOI] [PubMed] [Google Scholar]
- 9. Berndsen C. E., Denu J. M. (2008) Curr. Opin. Struct. Biol. 18, 682–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang L., Tang Y., Cole P. A., Marmorstein R. (2008) Curr. Opin. Struct. Biol. 18, 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Albaugh B. N., Kolonko E. M., Denu J. M. (2010) Biochemistry 49, 6375–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Han J., Zhou H., Horazdovsky B., Zhang K., Xu R. M., Zhang Z. (2007) Science 315, 653–655 [DOI] [PubMed] [Google Scholar]
- 13. Scholes D. T., Banerjee M., Bowen B., Curcio M. J. (2001) Genetics 159, 1449–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Driscoll R., Hudson A., Jackson S. P. (2007) Science 315, 649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han J., Zhou H., Li Z., Xu R. M., Zhang Z. (2007) J. Biol. Chem. 282, 28587–28596 [DOI] [PubMed] [Google Scholar]
- 16. Schneider J., Bajwa P., Johnson F. C., Bhaumik S. R., Shilatifard A. (2006) J. Biol. Chem. 281, 37270–37274 [DOI] [PubMed] [Google Scholar]
- 17. Masumoto H., Hawke D., Kobayashi R., Verreault A. (2005) Nature 436, 294–298 [DOI] [PubMed] [Google Scholar]
- 18. Berndsen C. E., Tsubota T., Lindner S. E., Lee S., Holton J. M., Kaufman P. D., Keck J. L., Denu J. M. (2008) Nat. Struct. Mol. Biol. 15, 948–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tsubota T., Berndsen C. E., Erkmann J. A., Smith C. L., Yang L., Freitas M. A., Denu J. M., Kaufman P. D. (2007) Mol. Cell 25, 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kolonko E. M., Albaugh B. N., Lindner S. E., Chen Y., Satyshur K. A., Arnold K. M., Kaufman P. D., Keck J. L., Denu J. M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 20275–20280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Das C., Tyler J. K., Churchill M. E. (2010) Trends Biochem. Sci. 35, 476–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park Y. J., Luger K. (2006) Biochem. Cell Biol. 84, 549–558 [DOI] [PubMed] [Google Scholar]
- 23. Chen C. C., Carson J. J., Feser J., Tamburini B., Zabaronick S., Linger J., Tyler J. K. (2008) Cell 134, 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adkins M. W., Carson J. J., English C. M., Ramey C. J., Tyler J. K. (2007) J. Biol. Chem. 282, 1334–1340 [DOI] [PubMed] [Google Scholar]
- 25. Agez M., Chen J., Guerois R., van Heijenoort C., Thuret J. Y., Mann C., Ochsenbein F. (2007) Structure 15, 191–199 [DOI] [PubMed] [Google Scholar]
- 26. Antczak A. J., Tsubota T., Kaufman P. D., Berger J. M. (2006) BMC Struct. Biol. 6, 26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Natsume R., Eitoku M., Akai Y., Sano N., Horikoshi M., Senda T. (2007) Nature 446, 338–341 [DOI] [PubMed] [Google Scholar]
- 28. Xhemalce B., Miller K. M., Driscoll R., Masumoto H., Jackson S. P., Kouzarides T., Verreault A., Arcangioli B. (2007) J. Biol. Chem. 282, 15040–15047 [DOI] [PubMed] [Google Scholar]
- 29. Värv S., Kristjuhan K., Peil K., Lõoke M., Mahlakõiv T., Paapsi K., Kristjuhan A. (2010) Mol. Cell. Biol. 30, 1467–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maas N. L., Miller K. M., DeFazio L. G., Toczyski D. P. (2006) Mol Cell 23, 109–119 [DOI] [PubMed] [Google Scholar]
- 31. Celic I., Masumoto H., Griffith W. P., Meluh P., Cotter R. J., Boeke J. D., Verreault A. (2006) Curr. Biol. 16, 1280–1289 [DOI] [PubMed] [Google Scholar]
- 32. Williams S. K., Truong D., Tyler J. K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9000–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park Y. J., Sudhoff K. B., Andrews A. J., Stargell L. A., Luger K. (2008) Nat. Struct. Mol. Biol. 15, 957–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fillingham J., Recht J., Silva A. C., Suter B., Emili A., Stagljar I., Krogan N. J., Allis C. D., Keogh M. C., Greenblatt J. F. (2008) Mol. Cell. Biol. 28, 4342–4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tang Y., Meeth K., Jiang E., Luo C., Marmorstein R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12206–12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Burgess R. J., Zhou H., Han J., Zhang Z. (2010) Mol. Cell 37, 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burgess R. J., Zhang Z. (2010) Cell Cycle 9, 2979–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tang Y., Holbert M. A., Delgoshaie N., Wurtele H., Guillemette B., Meeth K., Yuan H., Drogaris P., Lee E. H., Durette C., Thibault P., Verreault A., Cole P. A., Marmorstein R. (2011) Structure 19, 221–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tang Y., Holbert M. A., Wurtele H., Meeth K., Rocha W., Gharib M., Jiang E., Thibault P., Verreault A., Cole P. A., Marmorstein R. (2008) Nat. Struct. Mol. Biol. 15, 738–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stavropoulos P., Nagy V., Blobel G., Hoelz A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12236–12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lin C., Yuan Y. A. (2008) Structure 16, 1503–1510 [DOI] [PubMed] [Google Scholar]
- 42. Huang R., Holbert M. A., Tarrant M. K., Curtet S., Colquhoun D. R., Dancy B. M., Dancy B. C., Hwang Y., Tang Y., Meeth K., Marmorstein R., Cole R. N., Khochbin S., Cole P. A. (2010) J. Am. Chem. Soc. 132, 9986–9987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Berndsen C. E., Denu J. M. (2005) Methods 36, 321–331 [DOI] [PubMed] [Google Scholar]
- 44. Smith C. M. (2005) Methods 36, 395–403 [DOI] [PubMed] [Google Scholar]
- 45. Ericsson U. B., Hallberg B. M., Detitta G. T., Dekker N., Nordlund P. (2006) Anal. Biochem. 357, 289–298 [DOI] [PubMed] [Google Scholar]
- 46. Yu W., Lin Y., Yao J., Huang W., Lei Q., Xiong Y., Zhao S., Guan K. L. (2009) J. Biol. Chem. 284, 13669–13675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lin Y. Y., Lu J. Y., Zhang J., Walter W., Dang W., Wan J., Tao S. C., Qian J., Zhao Y., Boeke J. D., Berger S. L., Zhu H. (2009) Cell 136, 1073–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Karanam B., Jiang L., Wang L., Kelleher N. L., Cole P. A. (2006) J. Biol. Chem. 281, 40292–40301 [DOI] [PubMed] [Google Scholar]
- 49. Thompson P. R., Wang D., Wang L., Fulco M., Pediconi N., Zhang D., An W., Ge Q., Roeder R. G., Wong J., Levrero M., Sartorelli V., Cotter R. J., Cole P. A. (2004) Nat. Struct. Mol. Biol. 11, 308–315 [DOI] [PubMed] [Google Scholar]
- 50. Sundin B. A., Chiu C. H., Riffle M., Davis T. N., Muller E. G. (2004) Yeast 21, 793–800 [DOI] [PubMed] [Google Scholar]
- 51. Tu B. P., Kudlicki A., Rowicka M., McKnight S. L. (2005) Science 310, 1152–1158 [DOI] [PubMed] [Google Scholar]
- 52. Tu B. P., Mohler R. E., Liu J. C., Dombek K. M., Young E. T., Synovec R. E., McKnight S. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16886–16891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu F., Zhang Q., Zhang K., Xie W., Grunstein M. (2007) Mol. Cell 27, 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang B., Miller A., Kirchmaier A. L. (2008) Mol. Biol. Cell 19, 4993–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Haldar D., Kamakaka R. T. (2008) Eukaryot Cell 7, 800–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Santos-Rosa H., Valls E., Kouzarides T., Martínez-Balbás M. (2003) Nucleic Acids Res. 31, 4285–4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang J., Chen J. (2010) J. Biol. Chem. 285, 11458–11464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kadlec J., Hallacli E., Lipp M., Holz H., Sanchez-Weatherby J., Cusack S., Akhtar A. (2011) Nat. Struct. Mol. Biol. 18, 142–149 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.