Abstract
The vaccinia virus DNA polymerase is inherently distributive but acquires processivity by associating with a heterodimeric processivity factor comprised of the viral A20 and D4 proteins. D4 is also an enzymatically active uracil DNA glycosylase (UDG). The presence of an active repair protein as an essential component of the polymerase holoenzyme is a unique feature of the replication machinery. We have shown previously that the A20-UDG complex has a stoichiometry of ∼1:1, and our data suggest that A20 serves as a bridge between polymerase and UDG. Here we show that conserved hydrophobic residues in the N′ terminus of A20 are important for its binding to UDG. Our data argue against the assembly of D4 into higher order multimers, suggesting that the processivity factor does not form a toroidal ring around the DNA. Instead, we hypothesize that the intrinsic, processive DNA scanning activity of UDG tethers the holoenzyme to the DNA template. The inclusion of UDG as an essential holoenzyme component suggests that replication and base excision repair may be coupled. Here we show that the DNA polymerase can utilize dUTP as a substrate in vitro. Moreover, uracil moieties incorporated into the nascent strand during holoenzyme-mediated DNA synthesis can be excised by the viral UDG present within this holoenzyme, leaving abasic sites. Finally, we show that the polymerase stalls upon encountering an abasic site in the template strand, indicating that, like many replicative polymerases, the poxviral holoenzyme cannot perform translesion synthesis across an abasic site.
Keywords: DNA Polymerase, DNA Repair, DNA Synthesis, Viral Genetics, Viral Polymerase, BER, Poxvirus, Processivity Factor, Uracil DNA Glycosylase, Translesion Synthesis
Introduction
The faithful and efficient duplication of genomic DNA is one of the most conserved processes across all forms of life. Although this is a necessary and highly regulated process, it seems that each model organism has evolved unique modifications during this process. Members of the poxvirus family, of which variola virus is the most notable member and vaccinia virus is the experimental prototype, are no exception. Poxviruses are unique in that they complete the replication and maturation of their ∼200-kb double-stranded DNA genome in the cytoplasm of the infected host cell. This autonomy dictates that poxviruses encode many of the proteins necessary for nucleotide precursor synthesis and metabolism as well as the core set of enzymes and DNA binding proteins that act directly at the replication fork (1, 2). Indeed, genetic, genomic, and biochemical analysis has revealed that eight proteins are responsible for vaccinia virus DNA synthesis and maturation. This repertoire includes the catalytic DNA polymerase (E9 (3–11)), a stoichiometric component of the heterodimeric processivity factor (A20 (12–15)), a second component of the processivity factor (D4) that also possesses uracil DNA glycosylase (UDG)2 activity (16–18), a putative superfamily III helicase with known NTPase and DNA primase activity (D5 (19–23)), a serine/threonine protein kinase (B1 (24–26)), an abundant phosphoprotein with essential roles in viral replication, transcription, and morphogenesis (H5 (27)), a single-strand DNA-binding protein (I3 (28, 29)3), and a DNA ligase (A50 (30)). A Holliday-junction resolvase (A22 (31)) and a FEN-1 related endonuclease (G5 (32)) have also been shown to be important for formation of monomeric genomes. The virally encoded proteins involved in nucleotide precursor synthesis and metabolism have been reviewed elsewhere (33, 34).
We have for some time been interested in the catalytic polymerase and its unusual heterodimeric processivity factor. The polymerase contains proofreading exonuclease activity as well as polymerase activity (3, 4). Forward genetic studies have identified alleles encoding enzymes with altered fidelity (9) as well those with resistance or hypersensitivity to inhibitors such as aphidicolin, cytosine arabinoside, phosphonoacetic acid, and cidofivir (9–11, 35–37). The enzyme is intrinsically distributive, adding ∼10 nt per template binding event in the presence of moderate concentrations of NaCl or MgCl2+ (5). A processive form of the enzyme exists within infected cells; however, we have previously identified the A20 protein and the virally encoded UDG protein, which is traditionally a DNA repair enzyme, as the two components necessary for processivity (13, 15). A20 appears to bind directly to Pol as well as to other components of the replication machinery such as the D5 NTPase/primase and the abundant H5 phosphoprotein (38). The N′-terminal 25 residues of A20 have been shown to be necessary and sufficient for interaction with UDG (39). A20 and UDG interact tightly in vivo and in vitro, and we assume that A20 serves to bridge Pol and UDG and that the intrinsic DNA scanning activity of UDG prolongs the association of Pol with the template and renders it processive. This type of processivity factor would be quite distinct from the toroidal sliding clamps associated with prokaryotic (Escherichia coli β complex (40, 41)) and eukaryotic (e.g. mammalian proliferating cell nuclear antigen (42, 43)) replication machinery and much more like the herpes simplex virus (HSV UL42 monomer (44)) and cytomegalovirus (UL44 dimer (45)) processivity factors.
Although UDG proteins from other model systems have been found to associate with the DNA replication complex via protein-protein interactions (46–51), the essential nature of the vaccinia virus UDG and its integration into the polymerase complex itself is unique. In other model systems, deletion mutants lacking the UDG are viable (52, 53). The importance of controlling the amount of uracil in the vaccinia virus genome is underscored by the observation that vaccinia also encodes a dUTPase (F2 (54)). Although the vaccinia virus UDG is an essential protein (16, 55), its glycosylase activity is not mandatory for productive infection in actively dividing tissue culture cells (56). However, the combination of a UDG protein that is catalytically inert with the deletion of the viral dUTPase is detrimental to replication in quiescent cells (57). Furthermore, this double mutant strain is attenuated in a murine model (57). Together, these observations suggest that monitoring the levels of uracil in the vaccinia genome is of utmost importance. Because dTTP → dUTP substitutions do not alter the primary sequence of the encoded protein, the presence of dUMP residues within the genome must have an impact on as yet unidentified properties such as genome stability or protein-protein interactions.
The current study addresses several questions of interest regarding the processivity factor encoded by vaccinia virus, including the association between A20 and UDG and the possible self-association of UDG. Most importantly, we address whether the moving polymerase holoenzyme can both incorporate UTP and excise the uracil moiety and whether the polymerase can perform translesion synthesis when it encounters either a dUMP residue or abasic site in the template strand. These studies have implications for the coupling of synthesis and repair during the replication of the poxvirus genome.
EXPERIMENTAL PROCEDURES
Reagents
Restriction endonucleases, E. coli polymerase I, Klenow fragment of E. coli DNA polymerase, T4 DNA ligase, T4 polynucleotide kinase, calf intestinal phosphatase, pancreatic RNase, deoxynucleoside triphosphates (dNTP) PCR grade, Expand High Fidelity Taq polymerase, Taq polymerase, and DNA molecular weight standards were purchased from Roche Diagnostics and were used per the manufacturer specifications. 32P- and 3H-labeled nucleoside triphosphates were purchased from PerkinElmer Life Sciences. 3X-FLAG peptide and EZview Red ANTI-FLAG M2 Affinity gel beads were obtained from Sigma. Lipofectamine 2000 was acquired from Invitrogen. E. coli single-stranded DNA-binding protein was purchased from Agilent Technologies (Cedar Creek, TX). Ribonucleoside triphosphates (NTPs) were purchased from GE Healthcare. T7-Coupled Reticulocyte Lysate System for in vitro coupled transcription/translation (IVTT) was purchased from Promega (Madison, WI). Glycogen was purchased from Fermentas (Glen Burnie, MD). Oligonucleotide primers were purchased from Integrated DNA Technologies (Coralville, IA).
Cells and Virus
Monolayer cultures of African green monkey BSC40 cells and human thymidine kinase-negative (TK−) 143B cells were maintained in Dulbecco modified Eagle's medium (Invitrogen) containing 5% fetal calf serum. Wild-type (WT) vaccinia virus (WR strain, except as noted to be IHD-W strain) was grown in BSC40 cells. Dts48 (generously provided by Richard Condit, University of Florida, Gainsville, FL) was grown in BSC40 cells. The recombinant vaccinia virus vTF7.3 (58) was a gift from Bernard Moss (NIH, Bethesda, MD). The recombinant vaccinia viruses enabling T7-mediated overexpression of E9 (Pol), A20, 3XFLAG-UDG, or 3XFLAG-UDG⦶ (6, 13, 15) have been previously described. Viral stocks were prepared from cytoplasmic lysates of infected cells by ultracentrifugation through 36% sucrose; titers were determined on BSC40 cells. For virological studies using Dts48, 31.5 and 39.7 °C were used as the permissive and nonpermissive temperatures, respectively.
Site-directed Mutagenesis of the A20 Protein
A20 alleles with amino acid substitutions in the 5′-region of the ORF were generated by PCR using genomic viral DNA as the template. For each mutant generated, the A20 5′ primer introduced a BspHI site that overlapped the initiating ATG codon, whereas the A20 3′ primer introduced a BamHI restriction enzyme site downstream of the termination codon (oligonucleotide sequences can be found in Table 1). Glass-purified PCR products were digested with BspHI and BamHI, glass-purified, and ligated to pTM1 DNA (59) that had been digested with NcoI and BamHI and treated with calf intestinal phosphatase. All plasmids were verified by restriction enzyme digestion and DNA sequencing.
TABLE 1.
Oligonucleotides used in this study
For cloning oligonucleotides, restriction enzyme sites are represented by bold or italic type. The initiation or termination codons are underlined. Nucleotides chosen for mutagenesis are denoted by the double underline. For oligonucleotides used in the generation of DMCs, the nucleotides that are single- or double-underlined are complementary to each other and are the sequences that facilitate circularization of the linear DNA template.
| Cloning oligonucleotides | |
| A20 5′ Leu7,10 → Ala | 5′-GCGGATCCTCATGACTTCTAGCGCTGATGCAACTAACGCAAAAGAATTAC-3′ |
| A20 5′ Leu13,14,16 → Ala | 5′-GCGGATCCTCATGACTTCTAGCGCTGATTTAACTAACTTAAAAGAAGCAGCTAGTGCGTACAAAAG-3′ |
| A20 5′ Asp6-Glu12 → Ala | 5′-GCGGATCCTCATGACTTCTAGCGCTGCTTTAACTAACTTAAAAGCATTACTTAG-3′ |
| pTM1-A20 5′ | 5′-CCCGGATCCCTCATGACTTCTAGCGCT-3′ |
| A20 3′ | 5′-GCGGATCCTCACTCGAATAATCTT3′ |
| Oligonucleotides used in the generation of DMCs | |
| Bridging oligo | 5′-GGTTATGGTGGAGTGGTATA-3′ |
| 70-Mer for DMC | 5′-CACCATAACCTCCACCCTCCCCAATATTCACCATCAACCCTTCACCTCACTTCACTCCACTATACCACTC-3′ |
| 70-Mer for U-DMC | 5′-CACCATAACCTCCACCCTCCCCAATATTCACCATCAACCC/U/TTCACCTCACTTCAC/U/TCCACTATACCACTC-3′ |
| DMC-1 | 5′-GGTGAATATTGGGGAGGGTGGAGG-3′ |
| DMC-2 | 5′-GGGTGGAGGTTATGGTGGAGTGG-3′ |
Isolation of Genomic Dts48 Viral DNA and Mapping of the A20 Allele
One confluent 15-cm dish of BSC40 cells was infected with Dts48 at an multiplicity of infection of 0.2 and maintained at 31.5 °C for 48 h. The viral genomic DNA was purified from cytoplasmic extracts and used as the template for two independent PCRs using the A20 5′ and A20 3′ primers (Table 1). The products from each independent PCR reaction were subjected to DNA sequencing in duplicate and compared with the sequence from the allele encoded by the WT IHD-W strain.
In Vitro-coupled Transcription/Translation of 3XFLAG-UDG and A20 Variants
In vitro coupled transcription/translation reactions (IVTT) were carried out employing the TnT T7-Coupled Reticulocyte Lysate System from Promega. 50-μl reactions were programmed with pTM1 plasmids that allowed the co-expression of 3XFLAG-UDG (15) and either WT A20, Dts48A20, or A20 site-directed mutants prepared for this study. 1/10th of each reaction was removed for analysis as the “input” fraction. The remainder of the reaction was processed for retrieval using ANTI-FLAG M2 affinity gel beads as previously described (15). Input and eluate samples were resolved by 12% SDS-PAGE and visualized by autoradiography.
Gel Filtration
Purification was performed on an Amersham Biosciences ATKA FPLC at 4 °C. Approximately 40 μg (800 μl) of affinity-purified 3XFLAG-UDG was combined with 230 μg of gel filtration chromatography standards (Bio-Rad, supplemented with 5 μg of BSA) and loaded onto a HiPrep 16/60 Sephacryl S-100 HR column (Amersham Biosciences). The column was developed with gel filtration buffer (50 mm sodium phosphate (pH 7.4), 150 mm NaCl) at a flow rate of 0.5 ml/min. One-ml fractions were collected, resolved by 12% SDS-PAGE, and visualized by silver staining.
Construction of a Recombinant Virus Enabling T7 Polymerase-mediated Overexpression of 3XFLAG-UDG-⊗
Viral recombinants were generated and screened as previously described (6, 15).
Expression and Purification of the 3XFLAG Epitope-tagged Complexes
Overexpression of fUDG, A20, and E9 (Pol), singly and in combination, was achieved by co-infecting BSC40 cells with vTF7.3 and vTM-3XFLAG-UDG, vTM-3XFLAG-UDG-⦶, vTM-3XFLAG-UDG-⊗, vTM-A20, and/or vTM-Pol (each at a multiplicity of infection of 2). Complexes were recovered on α-FLAG resin and further purified by Mono Q chromatography as previously described (15).
Incorporation of 3H-dUTP by the Vaccinia Virus DNA Polymerase
An artificially nicked dsDNA template (activated salmon sperm) was used to monitor radiolabeled nucleotide incorporation by the vaccinia DNA polymerase. The catalytic viral DNA polymerase was expressed and purified as previously described (6). Reactions (100 μl) (6, 60) contained 3.45 nm vaccinia DNA polymerase in the presence of 100 μm each of dGTP, dCTP, and dATP and increasing concentrations of [3H]dUTP (0–19 μm). Reactions were terminated after 10 min at 37 °C by the addition of 20% TCA, 0.2 m NaPPi and precipitated on ice. TCA-precipitable material was collected on GF-C fiber filters (Whatman) and counted in Opti-Fluor liquid scintillation mixture (PerkinElmer). Replicate values were averaged and plotted. The Km of the viral polymerase for dUTP was determined by nonlinear regression analysis using GraphPad Prism 5.01 (GraphPad Software, La Jolla, CA).
Single-primed M13 DNA Replication Assay
RFII reactions were performed as previously detailed (13, 15). For experiments involving Dts48, the source of processive polymerase holoenzyme was the post-nuclear supernatant isolated from virally infected cells (Dts48 experiments). For other studies, purified fUDG-A20-Pol holoenzyme was prepared as described above.
Preparation of the DNA Minicircle (DMC) Templates
DNA minicircles were generated as previously described (61). Two 70-mer oligos, which comprised the same core sequence with the exception that one contained two internal dUMP residues, were converted to a single-stranded circle as previously described (62). The circularized single-stranded DNA product was purified by denaturing gel electrophoresis on a 6 m urea, 10% acrylamide, 0.5× TBE gel. After elution of the DNA from the acrylamide, the single-stranded circularized DNA was annealed to one of two oligos (see Table 1 for sequence) to generate the various primed templates (DMC-1, DMC-2, U-DMC-1, U-DMC-2), which were used as templates for in vitro minicircle replication assays.
Minicircle Replication Assays
Reactions (25 μl) contained 13 nm DNA polymerase holoenzyme and 14 nm DMC (or U-DMC) template in 10 mm Tris-HCl (pH 7.4), 40 ng/μl bovine serum albumin, 8% glycerol, 0.1 mm EDTA, 5 mm DTT, 8 mm MgCl2, 60 μm each of dGTP and TTP, and 20 μm α-[32P]dATP. Reactions were primed by incubation at 30 °C for 3 min in the presence of TTP. Reactions were initiated by the addition of dGTP and α-[32P]dATP and quenched at 5, 10, 20, 60, and 180 s by the addition of EDTA to 50 mm. Samples were resolved by denaturing gel electrophoresis on a 6 m urea, 10% acrylamide, 0.5× TBE gel. Reaction products were visualized by autoradiography and quantitated on a PhosphorImager Storm Scanner using ImageQuant software. Sizing standards were generated by radiolabeling oligonucleotide primers of various sizes on their 5′ termini using T4 polynucleotide kinase and γ-[32P]dATP.
DNA Glycosylase Assay
The 24-mer oligomers were radiolabeled on their 5′ terminus using T4 polynucleotide kinase and γ-[32P]dATP. The uracil-containing oligo (ssU) (5′-CGACTTGAAGCUACGTATCGATTA-3′) has the same core sequence as the control oligo (ssG), which contains a G at position 12 instead of the dUMP residue. Standard reactions (20 μl) contained 0.2 pmol of labeled oligo, 20 mm Tris-HCl (pH 7.4), 60 mm NaCl, 1 mm EDTA, 1 mm DTT, 100 μg of bovine serum albumin/ml, and 11 or 27 ng of fUDG (either free or complexed within the holoenzyme). Reactions were incubated at 37 °C for 5 min and quenched by the addition of 1.5% SDS, 0.3 mg/ml proteinase K at 37 °C for 15 min. Reactions were treated with 10% v/v piperidine (37 °C, 15 min). Reaction products were lyophilized with heat and washed twice with water before being resolved on a denaturing 20% acrylamide, 7 m urea, 0.5× TBE gel. Data were visualized by autoradiography and quantitated on a PhosphorImager Storm Scanner using ImageQuant software.
Preparation of Digital Figures
Original data were scanned on a SAPHIR scanner (Linotype-Hell Co., Hauppauge, NY) and were adjusted with Adobe Photoshop software (Adobe Systems Inc., San Jose, CA). Data from processivity assays were acquired on a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, CA) and quantitated using ImageQuant software (Molecular Dynamics). Images from immunoblot analysis were acquired using the AlphaImager documentation system (Alpha Innotech, San Leandro, CA). The sequence alignments were performed by using the ClustalW method and Lasergene software (DNASTAR, Inc., Madison, WI) using sequences retrieved from the Poxvirus Bioinformatics Resource Center. Final figures were assembled and labeled with Canvas software (Deneba Systems, Miami, FL). Dashed lines separating lanes on Figs. 1–4 represent the juxtaposition of samples that were analyzed on the same gel but were not originally adjacent to each other.
FIGURE 1.
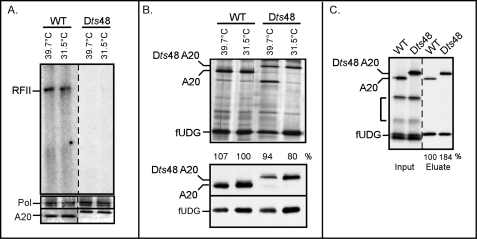
Characterization of a temperature-sensitive virus with a lesion in the A20 ORF. A, extracts from cells infected with Dts48 under non-permissive conditions lack processive polymerase activity. BSC40 cells were infected with WT virus (IHD-W strain) or Dts48 (multiplicity of infection of 2) and incubated at non-permissive (39.7 °C) or permissive (31.5 °C) temperature for 24 h. Post-nuclear supernatants were prepared and assayed for processive polymerase activity using a single-primed M13 DNA template in the presence of [32P]dATP. Reactions were resolved on a 0.8% agarose gel and visualized by autoradiography. RFII products, representing synthesis of the complete 7.2-kb daughter strand in a single binding event, are marked. The bottom panels represent immunoblot analyses of the same lysates using α-Pol and α-A20 antibodies. B, the Dts48-A20 protein retains the ability to interact with fUDG in vivo. BSC40 cells were infected and transfected so as to express 3XFLAG-UDG (fUDG) and either WT A20 or Dts48 A20 and incubated at 39.7 or 31.5 °C. At 24 h post-infection, cells were harvested, and clarified lysates were subjected to affinity purification using α-FLAG beads. FUDG and any associated proteins were visualized by silver staining (note the different electrophoretic mobilities of the two A20 proteins). The numbers below the lanes indicate the relative amount of WT or Dts48 A20 that was retrieved with fUDG; the amount of WT A20 retrieved was set at 100%. The bottom two panels represent immunoblot analyses of the lysates using α-A20 or α-FLAG antibodies. C, the Dts48-A20-fUDG association is conserved in vitro. IVTT reactions were programmed to synthesize fUDG and either WT or Dts48 A20; fUDG and any associated proteins were retrieved on α-FLAG beads. The input and eluate fractions were analyzed by SDS-PAGE and visualized by autoradiography. The bracket indicates A20 fragments produced from internal initiation events. The numbers below the eluate lanes indicate the relative amount of WT or Dts48 A20 that was retrieved with fUDG; the level of WT A20 retrieved was set at 100%.
FIGURE 2.
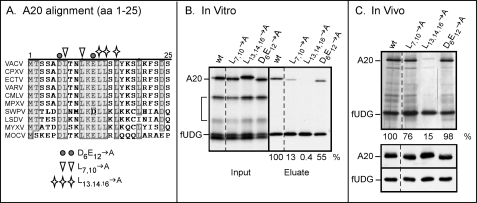
Conserved nonpolar residues within the first 25 amino acids of the A20 protein are required for association with fUDG. A, alignment of the first 25 amino acids of the A20 proteins from diverse poxviruses is shown. The A20 protein sequences from diverse poxviruses were aligned using the ClustalW program; only the alignment of the first 25 amino acids is shown. Residues boxed and shaded in gray are conserved in 9 of the 10 aligned protein sequences. Residues chosen for mutagenesis are noted. The representative viruses are vaccinia virus (VACV; western reserve-WR strain), cowpox virus (CPXV), ectromelia virus (ECTV), variola virus (VARV), camelpox virus (CMLV), monkeypox virus (MPXV), swinepox virus (SWPV), lumpy skin disease virus (LSDV), myxoma virus (MYXV), and molluscum contagiosum virus (MOCV). B, in vitro analysis of the protein-protein interactions between fUDG and the various A20 mutants is shown. IVTT reactions were programmed to express fUDG and either WT or mutant A20 proteins. FUDG and associated proteins were retrieved by affinity purification on α-FLAG beads. The input and eluate fractions were analyzed by SDS-PAGE, visualized by autoradiography, and quantitated by PhosphorImager analysis. The bracket indicates A20 fragments produced from internal initiation events. The numbers below the eluate lanes indicate the relative amounts of WT and mutant A20 proteins that were retrieved with fUDG; the amount of WT A20 retrieved was set at 100%. C, in vivo analysis of the protein-protein interactions between fUDG and the various A20 mutants is shown. BSC40 cells were infected and transfected so as to express fUDG and either WT or mutant A20. The fUDG and associated proteins within the clarified lysates were subjected to affinity purification on α-FLAG beads and analyzed by SDS-PAGE and silver staining. The numbers below the lanes indicate the relative amounts of WT and mutant A20 that were retrieved with fUDG; the amount of WT A20 retrieved was set at 100%. The two bottom panels represent immunoblot analyses of the lysates with α-A20 and α-FLAG antibodies.
FIGURE 3.
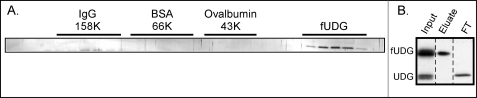
fUDG purified from infected cells and synthesized in vitro is monomeric. A, use of gel filtration to assess the native molecular weight of purified fUDG is shown. FUDG synthesized in virally infected cells was purified from clarified lysates using affinity chromatography; ∼40 μg (in combination with gel filtration standards) was applied to a HiPrep 16/60 Sephacryl S-100 HR column. Fractions were resolved on SDS-PAGE and visualized by silver staining; only the relevant portion of the gel is shown. Fractions containing peak amounts of the gel filtration standards are noted above the gel. B, fUDG synthesized in vitro does not self-associate. IVTTs were programmed to synthesize fUDG and UDG lacking an epitope tag. FUDG and any associated proteins were retrieved on α-FLAG beads. A fraction of the input, elute, and flow-through (FT) fractions was analyzed by SDS-PAGE and visualized by autoradiography.
FIGURE 4.
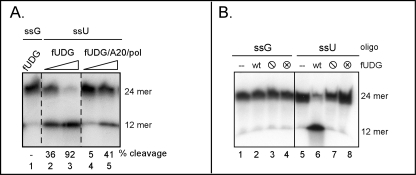
Characterization of the glycosylase activity of the vaccinia virus UDG. A, UDG within the polymerase holoenzyme is enzymatically active. A 5′-radiolabeled 24-mer oligo containing a single uracil residue at position 12 (ssU) was incubated with increasing concentrations of either fUDG (lanes 2 and 3) or a preparation of trimeric holoenzyme (fUDG-A20-Pol) that was free of any uncomplexed fUDG (lanes 4 and 5). A control oligonucleotide containing a G residue (ssG) at position 12 was also incubated with fUDG (lane 1). Reactions were treated with piperidine before separation by denaturing gel electrophoresis and visualization by autoradiography. The migration of the radiolabeled substrate (24-mer) and resultant UDG-dependent cleavage product (12-mer) are noted. B, characterization of UDG variants is shown. The control or uracil-containing oligonucleotide were left untreated (−) (lanes 1 and 5) or incubated with equivalent amounts of WT fUDG (lanes 2 and 6), fUDG-⦶ (lacks glycosylase activity) (lanes 3 and 7), or fUDG-⊗ (lacks uracil recognition) (lanes 4 and 8). Reaction products were treated with piperidine and separated on a denaturing gel.
RESULTS
The vaccinia virus DNA polymerase holoenzyme is known to be a trimeric complex of Pol-A20-fUDG (15, 63), but little is known about how the three proteins interact or mediate processivity. No information is available on the regions of A20 or Pol that mediate the interaction between these two proteins. The A20 interacting domain on the D4 protein has not yet been identified, and even small deletions to either terminus of the D4 protein appears to disrupt its expression and/or function (39).4 An internal amino acid substitution within D4 that impairs the interaction with A20 has been identified within the Dts30 mutant (Gly179 → Arg) (15). The N′-terminal 25 amino acids of A20 are necessary and sufficient to interact with D4 on their own (39), but internal amino acid substitutions elsewhere in the A20 protein (185ERSFDDK → AASFAAA) can also impair this interaction (15). Thus, much remains to be learned about the structure and function of these three proteins.
Analysis of the A20-fUDG and fUDG-UDG Interactions
Genotypic and Preliminary Phenotypic Analyses of a Temperature-sensitive Mutant with a Lesion in the A20 ORF
The re-evaluation of the Dales collection of ts-vaccinia virus (64–66) revealed a mutant with a lesion in the A20 gene (Dts48). We chose to investigate this mutant further to compare it with others that have been generated by targeted mutagenesis (12, 14). DNA sequencing analysis of the endogenous A20 gene within Dts48 indicated that the allele contained a single Gly252 → Ala transition that would be responsible for a Gly84 → Glu amino acid substitution.
Preliminary characterization of Dts48 had revealed a defect in DNA replication at the restrictive temperature (39.7 °C) (65). Our goals were to assess the impact of the ts mutation on both distributive and processive polymerase activity and on the A20-UDG interaction. Cells were infected with either WT vaccinia virus (IHD-W strain) or Dts48 at the permissive (31.5 °C) or nonpermissive (39.7 °C) temperature for 8 h, and post nuclear supernatants were prepared. The level of distributive polymerase activity within the lysates was assessed by monitoring the incorporation of [α-32P]dNTPs into an activated salmon sperm DNA template (60); no differences were seen between the WT and Dts48 extracts (not shown). Processive polymerase activity was assessed by assaying the same lysates for their ability to convert a single-primed M13 DNA template to the double-stranded 7-kb RFII product under conditions in which the free DNA polymerase (in the absence of UDG-A20) is highly distributive. Although extracts prepared from WT infections had processive polymerase activity (Fig. 1A, RFII), extracts prepared from cells infected with Dts48 at either temperature were unable to direct RFII formation. These in vitro data suggest that the replication defect of Dts48 is due to impaired processive polymerase activity. We were not surprised to observe that extracts prepared from 31.5 °C Dts48 infections showed a defect in vitro even though the virus can replicate adequately under these conditions in vivo. Other ts mutants with lesions in Pol, A20, and D4 exhibit this same discrepancy (7, 14, 15). We presume that, during infection, the replication complex is stabilized by factors within the cytoplasmic milieu that are lacking in our in vitro reactions.
Immunoblot analysis of the various lysates indicated that the steady state levels of Pol (116 kDa) and A20 (48 kDa) were not reduced during non-permissive Dts48 infections (Fig. 1A, bottom panels). Thus, the mutant A20 protein is temperature-sensitive for function rather than stability. The A20 protein encoded by Dts48 exhibits an altered electrophoretic mobility, which we predict is due to the substitution of a large, charged amino acid (glutamate) in place of a glycine.
To assess whether the Dts48 A20 protein was defective in its interaction with D4, protein-protein interactions were monitored both in vivo and in vitro. Cells were infected with a virus encoding the T7 RNA polymerase at either 31.5 or 39.7 °C and transfected with plasmids encoding 3XFLAG-UDG (fUDG; 29 kDa) and either WT or Dts48 A20 under the control the T7 promoter (58). Clarified cell lysates were prepared, and fUDG, along with any associated proteins, were retrieved by affinity purification on α-FLAG affinity resin. As evidenced by both silver-staining and immunoblot analyses (Fig. 1B), both WT A20 and Dts48 A20 co-purified with fUDG under all conditions. It can be noted that the expression of both A20 and fUDG was somewhat reduced at 39.7 °C, but this minor reduction did not alter the profile of protein-protein interaction. The same plasmids were used to express fUDG, WT A20, and Dts48 A20 in vitro using IVTT (Fig. 1C), and again α-FLAG resin was used to purify fUDG and associated proteins. These data confirmed that the Dts48 A20 protein retains the ability to interact with fUDG.
Conserved Hydrophobic Residues within the First 25 Amino Acids of A20 Are Required for Its Association with D4
The N′-terminal 25 amino acids of the A20 protein have been previously shown to be necessary and sufficient for binding to D4 (39). The ability of this interaction to persist in the presence of 750 mm NaCl suggests that hydrophobic, rather than ionic, interactions might be involved (15). The N′-terminal 25 amino acids of the A20 homologs encoded by diverse poxviruses, whose overall sequence identity with the vaccinia A20 protein ranged from >95% (CPXV, ECTV, VARV, CMLV, and MPX) to <50% (SWPV, LSDV, MYXV) or <30% (MOCV) were aligned and compared (Fig. 2A). We identified three clusters of highly conserved non-polar (Leu7,10 and Leu13,14,16) or charged (Asp6-Glu12) residues and constructed mutant alleles in which these clusters were changed to alanine residues. These alleles were placed in the pTM1 (15, 58) vector to facilitate analysis of their expression and protein-protein interactions in vitro and in vivo. IVTT reactions were employed to assess whether the amino acid substitutions affected the direct interaction between A20 and fUDG. IVTTs were programmed to co-express fUDG and WT or mutant A20. fUDG, along with any associated proteins, was retrieved by affinity purification and visualized by autoradiography (Fig. 2B). We observed that converting the conserved nonpolar residues to alanine reduced the association of A20 with fUDG by 7 to 250-fold (Leu7,10 → Ala and Leu13,14,16 → Ala, respectively), whereas the association of the Asp6-Glu12 → Ala variant with fUDG was reduced by less than 2-fold. These data strongly suggest that the A20-fUDG interaction is hydrophobic in nature.
To assess the impact of the mutations on the interaction of A20 and UDG in vivo, the same plasmids, which encode fUDG and the A20 alleles under the control of the T7 promoter, were transfected into cells infected with the T7 polymerase-expressing virus. At 24 h post-infection, clarified lysates were prepared. Immunoblot analysis confirmed that all of the A20 variants were expressed to equivalent levels (Fig. 2C, bottom). The lysates were incubated with α-FLAG resin to purify fUDG along with any associated proteins, and the eluates were analyzed by silver staining (Fig. 2C, top). Although the Asp6-Glu12 → Ala A20 protein was retrieved as well as WT A20, the association of both the Leu7,10 → Ala and Leu13,14,16 → Ala A20 variants with fUDG was reduced, with the latter protein showing the greatest deficit (Fig. 2C). We hypothesize that the cellular milieu may be responsible for the milder defect seen in vivo than we had observed in vitro (compare Figs. 2,B and C). Cumulatively, these data provide evidence that conserved nonpolar residues within the N terminus of A20 may facilitate its association with fUDG.
UDG Is a Monomer in Solution
The mechanism by which UDG contributes to polymerase processivity remains unknown, but insight into whether it functions as a monomer or as a higher order multimer might shed light on this question. For example, if UDG and/or the UDG-A20 complex were found predominantly as multimers, it might suggest that the processivity factor wraps at least partway around the DNA helix. The crystal structure of UDG found evidence for UDG dimers (67), and the authors reported (data not shown) that recombinant UDG was dimeric in solution at the concentration studied. To assess the native molecular weight of our affinity-purified fUDG, gel filtration standards were mixed with 40 μg of fUDG (in 800 μl) and resolved on a HiPrep 16/60 Sephacryl S-100 HR column using an Amersham Biosciences ATKA FPLC. fUDG was found in fractions consistent with a native molecular weight significantly below that of ovalbumin (Mr 43,000) (Fig. 3A). Given the fact that fUDG has a predicted Mr of 27,900, these data indicate that our preparation of fUDG purified from infected cells is monomeric.
As an alternative approach to assessing fUDG multimerization, we programmed IVTT reactions to express both fUDG (Fig. 3B) and untagged UDG and applied the reactions to α-FLAG resin. fUDG was retrieved on the resin, but the untagged UDG protein remained unbound (Fig. 3B, FT). Because we know that the fUDG protein expressed in IVTT is competent to bind to A20, its inability to self-interact with untagged UDG is meaningful. In sum, the data obtained from these two experimental approaches suggest that fUDG is in fact monomeric.
Can the UDG-A20-Pol Holoenzyme Initiate Base Excision Repair as Well as Perform Processive DNA Synthesis?
The Viral UDG Is Enzymatically Active within the DNA Polymerase Holoenyzme
To test the hypothesis that the vaccinia holoenzyme might perform both DNA synthesis and repair, we first needed to determine whether UDG retained its enzymatic activity within the context of the trimeric fUDG-A20-Pol complex. To ensure that the holoenzyme preparation did not contain any excess free (uncomplexed) fUDG, the affinity-purified complex was passaged over a Mono Q ion exchange column to resolve the free fUDG from the trimeric complex (15). Increasing amounts of the trimeric complex were then incubated with a 5′-radiolabeled 24-mer containing an internal dUMP residue at position 12 (ssU). Reactions containing free fUDG were performed in parallel. After incubation, reactions were treated with piperidine to cleave the DNA at any abasic sites that had been generated by the glycosylase, and the intact and cleaved oligonucleotides were resolved electrophoretically and visualized by autoradiography. As evidenced by the piperidine-generated cleavage products seen in Fig. 4A (12-mer), abasic sites were generated after incubation of the substrate with either free fUDG (lanes 2 and 3) or the trimeric holoenzyme (lanes 4 and 5). As expected, abasic sites were not generated using a control substrate that lacks any dUMP residues (lane 1).
For subsequent use in these studies, we also generated two variants of UDG predicted to lack catalytic activity (15, 56, 57). The fUDG-⦶ protein contains substitutions in two key amino acid residues (Asp68 and His181) that are required for cleavage of the glycosidic bond (56, 68). The fUDG-⊗ protein contains substitutions in three key residues (Tyr70, Phe79, Asn120) that are crucial for creating the uracil recognition pocket (69, 70). Our laboratory has previously reported that these catalytically impaired variants of fUDG retain the ability to impart processivity within the context of the trimeric complex (15). Here we verify that, unlike WT fUDG (lane 6), neither fUDG-⦶ nor fUDG-⊗ exhibited any enzymatic activity in our DNA glycosylase assay (Fig. 4B, lanes 7 and 8).
Effect of dUTP on DNA Synthesis in Vitro
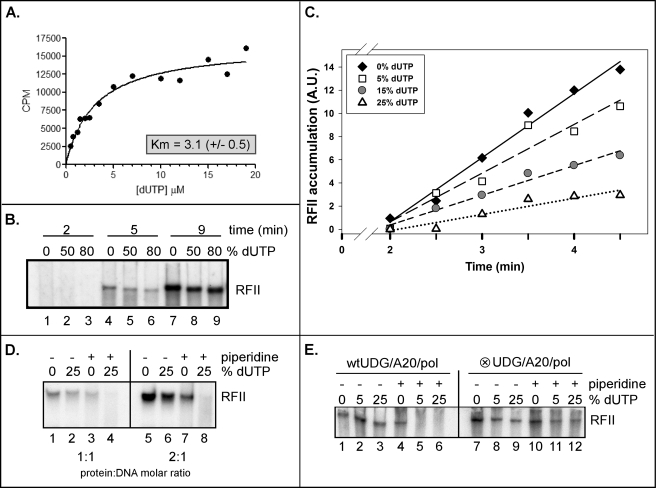
The inclusion of an enzymatically active UDG within the DNA holoenzyme complex (Fig. 4A) suggested to us that base excision repair of misincorporated dUTP residues might be a feature of vaccinia virus DNA replication. We were, therefore, interested in assessing if the viral DNA polymerase could directly incorporate dUTP. To this end, purified DNA polymerase (6) was incubated with activated salmon-sperm DNA in the presence of increasing concentrations of [3H]dUTP (0–19 μm) and a constant concentration (100 μm) of dGTP, dCTP, and dATP. We observed a nonlinear (hyperbolic) relationship between the initial enzyme velocity and dUTP concentration (Fig. 5, panel A). Data collected in duplicate and analyzed by nonlinear fit regression analysis (GraphPad Prism software) yielded a Km value for dUTP of 3.1 ± 0.5 μm. Our laboratory has previously published a kinetic analysis of the viral DNA polymerase (6) and obtained a similar Km value for dTTP of 4.0 μm. Our findings are in good agreement with studies analyzing the ability of other polymerases (Pol β or the HSV DNA polymerase) to utilize dUTP (71–73) and clearly demonstrate that the vaccinia polymerase can incorporate dUTP during DNA synthesis.
FIGURE 5.
The DNA polymerase holoenzyme is able to incorporate, recognize, and remove dUMP residues in a growing nascent DNA strand. A, the catalytic DNA polymerase can incorporate dUTP into an activated dsDNA template. DNA synthesis assays containing activated salmon-sperm DNA and purified Pol were performed in the presence of increasing concentrations of dUTP (0–19 μm) and 100 μm dGTP, dCTP, and dATP. The incorporation of 3H-dUTP into acid-precipitable material was quantified (CPM) and plotted against the dUTP concentration (μm) (the data represent the average of two experiments). The data were then subjected to nonlinear regression analysis, and the Km value was determined. B, RFII products synthesized in the presence of high concentrations of dUTP have an altered electrophoretic migration. An RFII time course (2, 5, and 9 min) was performed in the presence of 0, 50, or 80% dUTP (percentage of the TTP pool replaced by dUTP) using the viral DNA polymerase holoenzyme (Pol-A20-fUDG) that had been freed of uncomplexed fUDG. Products were resolved on an agarose/TBE gel and visualized by autoradiography. C, shown is a graphic representation of an RFII time course synthesis in the presence of increasing concentrations of dUTP. An RFII time course (0–4.5 min) was performed under conditions in which 0, 5, 15, or 25% of the TTP had been replaced by dUTP. Reactions were resolved on an agarose/TBE gel and visualization by autoradiography. The accumulation of the RFII products was quantified; data points and linear regression lines are shown. A.U., arbitrary units. D, vaccinia virus DNA replication and repair may be coupled. RFII reactions were performed using equimolar (1:1 protein:DNA molar ratio; lanes 1–4) or near equimolar (2:1; lanes 5–8) amounts of affinity-purified, Mono Q-purified DNA polymerase holoenzyme in the absence (0%) or presence (25% of the TTP pool) of dUTP. Reactions were allowed to proceed for 5 min after which time they were left untreated (−) or treated (+) with piperidine to cleave the DNA at any abasic sites that had been generated. E, dUMP excision required an enzymatically active complexed UDG. RFII reactions were performed using purified trimeric holoenzyme containing either WT fUDG or fUDG-⊗, which fails to recognize uracil.
To determine what effect the inclusion of dUTP in the nucleotide pool would have on processive polymerase activity, we performed RFII analysis using the affinity-purified DNA polymerase holoenzyme in the presence of increasing amounts of dUTP. A single-primed M13 template was incubated with the polymerase complex for increasing amounts of time under conditions in which 0, 50, or 80% of the TTP pool was replaced with dUTP. Reaction products were analyzed by agarose gel electrophoresis, and we observed that although RFII products were readily synthesized, there was a minor but reproducible shift in the electrophoretic mobility of the RFII product (Fig. 5B; compare lanes 5, 6, 8, and 9 to lanes 4 and 7). We also noted that there was a slight reduction in the amount of RFII product formed in the presence of higher concentrations of dUTP (Fig. 5B; compare lanes 4 to 6 and 7 to 9). To investigate this finding further, we performed a RFII time course focusing on the earliest time points (0–4.5 min) combined with conditions in which 0, 5, 15, or 25% of the TTP pool was replaced with dUTP. Quantification of RFII formation (Fig. 5C) confirmed that the rate of RFII synthesis diminished as the fraction of the TTP pool that was replaced with dUTP was increased. Although we demonstrated that the Km value for dUTP of the polymerase is comparable with the Km value for dTTP, the data presented in Fig. 5C suggest that the rate of synthesis is nevertheless affected by the presence of dUTP in the reaction mixture.
DNA Replication and Repair May Be Coupled in Vitro
The data shown above clearly indicate that the polymerase can incorporate dUTP during DNA synthesis. The next question we addressed was whether the UDG within the holoenzyme complex was excising the uracil moieties introduced into the nascent strands during RFII synthesis. RFII reactions were performed using near-equimolar amounts of the polymerase complex and the single-primed M13 template (1:1 or 2:1 molar ratio of protein-DNA) (Fig. 5D). Reactions were carried out in the absence or presence of dUTP (0 or 25% of the TTP pool), terminated by the addition of proteinase K, and either left untreated or treated with piperidine to cleave any abasic sites that had been generated by UDG. As seen in Fig. 5D, RFII products were generated in both the presence or absence of dUTP (lanes 1 and 2 and lanes 5 and 6), but the products synthesized in the presence of dUTP were sensitive to piperidine cleavage. Quantitation of the data indicated that at either enzyme concentration, RFII products generated in the presence of 25% dUTP were 10-fold more sensitive to piperidine treatment than those synthesized in the absence of dUTP (Fig. 5D, compare lane 4 to 3 and lane 8 to 7). Because the reactions were performed using equimolar ratios of enzyme-DNA, these data suggest that DNA synthesis and the removal of uracil residues may be mediated by the same holoenzyme, as it translocates along the single-primed DNA template.
An additional set of experiments was performed using a holoenzyme containing the enzymatically inactive fUDG-⊗ variant (Fig. 5E, lanes 7–12); WT holoenzyme was assayed in parallel (lanes 1–6). The data generated with the WT holoenzyme were similar to those described for Fig. 5D; note the loss of RFII product in lanes 5 and 6. However, the RFII products generated by the fUDG-⊗-containing holoenzyme in the presence of dUTP were piperidine-resistant. These data confirm that the abasic sites generated by the WT holoenzyme were excised by the WT fUDG within that holoenzyme and not by a contaminant in the preparation.
How Does the Holoenzyme Function upon Encountering a Uracil Moiety in the Template Strand?
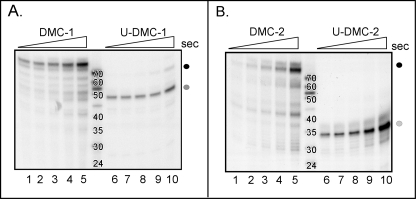
To assess the behavior of the holoenzyme when it encountered uracil moieties in the template strand, we generated minicircle templates modeled on those used in studies of HSV and bacteriophages T7 and T4 (61, 74–77). Two 70-mer oligonucleotides were prepared, one containing two internal dUMP residues (U-DMC) and one lacking dUMP residues (DMC). These oligonucleotides were converted to covalently closed minicircles (62) and annealed to one of two primers (generating DMC-1 and U-DMC-1 and generating DMC-2 and U-DMC-2). The primers were positioned such that the distance between their 3′ termini and the first dUMP residue within the minicircle were 23 or 9 nt, respectively.
When DMC-1 was incubated with a WT trimeric DNA polymerase holoenzyme in the presence of 32P-dNTPs, we observed the conversion of the 24-mer primer to a full-length 70-nt product within 5 s of incubation (Fig. 6A, lane 1, black circle). Full-length product continued to accumulate throughout the 3-min time course (Fig. 6A, lanes 1–5). The smaller reaction products that were also seen may reflect nonspecific polymerase stuttering/idling. In contrast, when comparable reactions were performed using the U-DMC-1 primer/template, the major product that accumulated was 47 nt in length (Fig. 6A, lanes 6–10, gray circle), which corresponds to the extension of the 24-nt primer to, but not beyond, the first dUMP residue encountered in the template.
FIGURE 6.
Stalling of the DNA polymerase holoenzyme complex upon encountering a uracil residue in the template DNA strand. A, the polymerase holoenzyme stalls when it reaches a dUMP residue in the template strand. A single-primed, ssDNA minicircle template (DMC-1) was incubated with purified polymerase holoenzyme (free of uncomplexed fUDG) for increasing amounts of time (5, 10, 20, 60, and 180 s, lanes 1–5, respectively). Parallel reactions were performed with a second template containing two internal dUMP residues in the template strand (U-DMC-1) (lanes 6–10). Reactions were quenched, resolved by denaturing gel electrophoresis, and visualized by autoradiography. The black circle represents the full-length replication product (∼70 nt) generated with DMC-1, whereas the gray circle represents the stalled replication product generated with U-DMC-1 (∼47 nt). Size standards were generated by radiolabeling the 5′ end of DNA oligonucleotides of the sizes shown. B, shown is confirmation that polymerase stalling was due to encountering a dUMP residue within the template strand. The same minicircle templates were annealed to a different primer, generating the primer/templates DMC-2 and U-DMC-2. Reactions were performed as described in A. The size of the stalled replication product is now ∼34 nt (gray circle), which is the distance from the new primer terminus to the first dUMP encountered by the polymerase.
To confirm that the stalling we observed with the U-DMC-1 template was indeed due to the polymerase encountering a uracil moiety in the template strand, we performed comparable reactions using the U-DMC-2/DMC-2 minicircle templates. With these templates, the predicted size of the product that would be generated if the holoenzyme stalled at the first dUMP residue is 34 nt. Indeed, whereas the major product formed using the DMC-2 template was the full-length 70 nt (Fig. 6B, lanes 1–5, black circle), the major product generated using the U-DMC-2 template was ∼34 nt (Fig. 6B, lanes 6–10, gray circle). Together, these data provide convincing evidence that the vaccinia virus DNA polymerase holoenzyme stalls upon encountering a dUMP residue within the template strand.
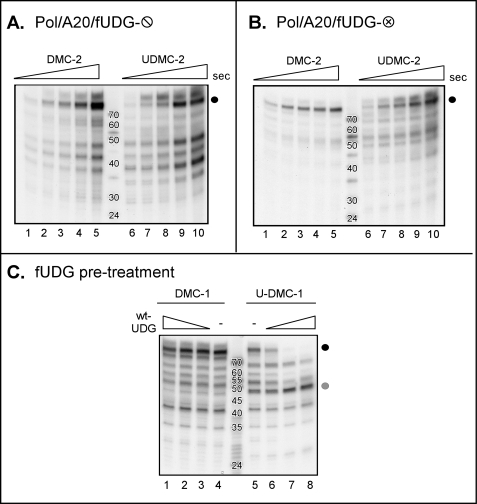
It was important for us to distinguish whether the barrier to further primer extension on the U-DMC-1 or -2 templates was the dUMP moiety itself or an abasic site generated by the fUDG during the incubation. Therefore, we performed a comparable set of experiments using preparations of trimeric holoenzyme containing glycosylase-deficient fUDG-⦶ or fUDG-⊗ (Fig. 7, A and B). With these preparations of holoenzyme that lack an active UDG, the full-length 70-nt product was readily detected using either the DMC-2 or U-DMC-2 template, even after only 5 s of incubation (Fig. 7, A and B, lanes 6–10, black circle). These results demonstrate that the enzymatic activity of the UDG protein within the holoenzyme, which can convert dUMP residues to abasic sites, is required for the stalling of the enzyme when a dUMP residue is encountered on the template.
FIGURE 7.
The DNA polymerase holoenzyme stalls when it encounters an abasic site on the template strand. A and B, stalling of the polymerase on templates containing dUMP residues is dependent on the presence of active UDG in the holoenzyme. Minicircle replication time course assays were performed using either DMC-2 (lanes 1–5) or U-DMC-2 (lanes 6–10) and DNA polymerase holoenzymes containing inactive forms of UDG (Pol-A20-fUDG-⦶; panel A) (Pol-A20-fUDG-⊗; panel B). Reactions were performed and analyzed as described in Fig. 6. C, pretreatment of dUMP-containing templates with active fUDG is sufficient to induce stalling by a polymerase holoenzyme containing inactive UDG. DMC-1 and U-DMC-1 templates were left untreated (lanes 4 and 5) or treated with increasing concentrations of WT-fUDG (lanes 1–3 and 6–8). DNA synthesis was then initiated by the addition of dNTPs and Pol-A20-fUDG-⦶. Full-length(∼70 nt) products are marked by the black circle, whereas the gray circle marks the stalled replication product (∼47 nt).
The Trimeric DNA Polymerase Holoenzyme Cannot Perform Translesion Synthesis When It Encounters an Abasic Site
To obtain definitive proof for the conclusion that the polymerase stalls when it encounters an abasic site in the template rather than a dUMP residue, we preincubated the DMC-1 and U-DMC-1 templates with a WT preparation of free fUDG. This pretreatment should generate two abasic sites within the U-DMC-1 template but none in the DMC-1 template. Primer extension was then initiated by the addition of dNTPs and a purified preparation of holoenzyme containing the glycosylase-deficient fUDG-⦶ variant (Pol-A20-fUDG). As expected, pretreatment of the DMC-1 template with WT-fUDG had no effect on the subsequent formation of full-length 70-nt product (Fig. 7C, black circle, compare lanes 1–3 with lane 4 (untreated)). However, the ability of Pol-A20-fUDG-⦶ holoenzyme to convert the U-DMC-1 template to the full-length 70-nt product (Fig. 7C, lane 5, black circle) was progressively impaired by pretreatment of the template with increasing amounts of enzymatically active fUDG (lanes 6–8, see the loss of the 70-nt product) (a corresponding increase in the ∼47-nt product that accumulates as a result of polymerase stalling at the position of the dUMP residue is seen (gray circle)). These data strongly support the conclusion that it is the presence of an abasic site within the template strand that is responsible for the stalling of the holoenzyme complex, indicating that the vaccinia virus holoenzyme cannot perform this type of translesion synthesis.
DISCUSSION
In this report we have extended our genetic and biochemical characterization of the vaccinia virus DNA polymerase holoenzyme. We have previously shown that three viral proteins (Pol-A20-D4) are necessary and sufficient for assembly of the processive vaccinia virus DNA polymerase holoenzyme (15). Others have recently confirmed our observation (63). Our working model is that the A20 protein serves as a bridge within the complex, binding to both Pol and D4 (15). No data have been obtained to date on how the Pol and A20 proteins interact, and the primary sequence of A20 has not provided any clues as to its structure. We have been unable to express and purify recombinant A20 on its own and believe that in vivo A20 may always exist as part of the A20-UDG heterodimer. The N′-terminal 25 amino acids of the A20 have previously been shown to be necessary and sufficient for association with D4 (39), and we have demonstrated here that alterations to highly conserved Leu residues within this region disturb the interaction. In contrast, mutation of highly conserved charged residues within this region did not disturb the A20-UDG interaction (Fig. 2). These data strengthen the hypothesis that the A20-UDG interaction is mediated by hydrophobic interactions.
No insights into which regions of D4 interact with A20 have been reported, and in fact a recent targeted mutational study of UDG failed to identify mutants with defects in this property (63). The crystal structure of the vaccinia UDG has been solved; UDG was found to crystallize as a dimer, and highly concentrated preparations of recombinant UDG were also found to be dimeric in solution (67). This observation was provocative as several prokaryotic and eukaryotic processivity factors multimerize and function as toroidal sliding clamps (40–43). These toroidal rings are loaded onto the DNA in an ATP-dependent manner and encircle the DNA without directly binding to it (for review, see Ref. 78). We have not favored this model for the viral processivity factor, in part because there is no evidence for an ATP-dependent clamp loader among the viral replication proteins. Herein, we show that the native molecular weight of the D4 protein purified from virally infected cells is consistent with it being in a monomeric form (Fig. 3A). Additionally, epitope-tagged D4 synthesized in in vitro IVTT reactions failed to retrieve an untagged version of D4, providing further evidence that D4 does not assemble into higher order structures (Fig. 3B). We suggest that the dimeric form observed previously (63) may have reflected the very high protein concentrations of recombinant preparations of D4. Our current working model is that, rather than a multimeric ring of D4 serving to tether the polymerase to the DNA topologically, the D4 protein is likely to confer processivity though its intrinsic DNA scanning activity. UDGs are predicted to scan along a DNA template in a processive manner, binding, kinking, and compressing the DNA backbone as they probe for dUMP residues using a “pinch-push-pull” mechanism (69, 70). This processive scanning mechanism, which ultimately allows the UDG protein to probe for uracil in the DNA, is most likely mediated by interactions with the phosphate backbone and independent of uracil recognition (79).
The constituents of the DNA polymerase holoenzyme may effectively allow vaccinia virus to couple ongoing DNA replication with the active repair of misincorporated dUMP residues. This hypothesis requires that the UDG complexed within the holoenzyme retains enzymatic activity, which we have clearly demonstrated herein (Fig. 4A). When in vitro replication assays were performed using highly purified holoenzyme at nearly equimolar concentrations of template and enzyme, DNA products synthesized in the presence of dUTP were sensitive to piperidine treatment (Fig. 5D). Thus, the DNA polymerase holoenzyme was able to incorporate dUMP residues into the growing nascent DNA strand, and in turn, the UDG within the complex was able to recognize the uracil moieties and excise them, leaving abasic sites. Although UDGs in several model systems have been shown to associate with polymerases or polymerase-associated proteins (46–51), this is the first example of a traditional DNA repair protein (UDG) being an intrinsic and active component of the core replication machinery.
If base excision repair (BER) does occur during vaccinia DNA synthesis in vivo, there needs to be a means by which the abasic sites generated by UDG are repaired. We are currently investigating whether components of the viral replication machinery have the activities required for the completion of BER or whether cellular repair proteins with AP endonuclease or lyase activities are recruited to viral replication sites for this purpose. This topic has been addressed for HSV, and it was reported that BER could be reconstituted in vitro using the HSV UDG (UL2), human AP endonuclease, the HSV DNA polymerase (UL30, which has intrinsic lyase activity (80)), and human ligase IIIα-XRCC1 (81).
We were also interested in determining the impact of the holoenzyme encountering a dUMP residue within the template strand. Replication reactions were, therefore, carried out using synthetic single-primed DNA minicircles that lacked (61, 62) or contained two internal dUMP residues. We observed that the purified DNA polymerase holoenzyme complex stalled at the position of the first dUMP residue (Fig. 6). Holoenzyme stalling was dependent upon the presence of an active UDG, as full-length products were readily accumulated when the holoenzyme contained an inactive variant of UDG (Fig. 7, A and B). The conclusion that it was not the uracil moiety that caused polymerase stalling but an abasic site generated by the viral UDG was verified by demonstrating that conversion of the dUMP residues in the template to abasic sites caused stalling by a glycosylase-deficient holoenzyme (Fig. 7C). These data are the first demonstration that the vaccinia virus DNA polymerase is unable to perform translesion synthesis when encountering an abasic site.
It will be of interest to determine what happens when the vaccinia holoenzyme encounters other types of DNA lesions. For the well studied bacteriophage polymerases encoded by T4 and T7 and for the HSV polymerase, the inability to perform translesion synthesis across from an abasic site is correlated with their proofreading exonuclease activity. Exonuclease-deficient variants of the various polymerases are able to more effectively synthesize across an abasic site than their exo-proficient counterparts (82–85). Further studies will be required to determine whether the vaccinia polymerase follows this same paradigm. Whereas inactivation of the exonuclease domain of the T4 and HSV polymerases is not deleterious to viral infectivity (86, 87), there are data to suggest that the exonuclease activity is essential in poxviruses (29). Although it has been hypothesized that the essentiality of the exonuclease is associated with its contribution to DNA recombination (29), its role during BER and translesion synthesis also merits study.
In sum, these studies provide support for the hypothesis that the inclusion of an active UDG in the polymerase holoenzyme may permit the coupling of DNA synthesis and base excision repair during the replication of the poxvirus genome. Further analysis of how viral or cellular proteins complete BER during poxvirus infection will be of significant interest. In addition, we hope to gain a more detailed understanding of the interactions between A20 and UDG and A20 and Pol and in so doing understand how A20-UDG confers processivity on the catalytic subunit of the viral polymerase.
Acknowledgments
We thank past and present members of the Traktman laboratory for critical input and discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant 2 R01 AI21758. This work was also supported by developmental grants from the Great Lakes Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (1-U54-AI057153; to P. T.).
P. Traktman, manuscript in preparation.
E. S. Stanitsa and P. Traktman, unpublished data.
- UDG
- uracil DNA glycosylase
- fUDG
- 3XFLAG-UDG
- IVTT
- in vitro coupled transcription/translation
- DMC
- DNA minicircle
- TBE
- Tris borate-EDTA
- BER
- base excision repair
- Pol
- polymerase
- nt
- nucleotides.
REFERENCES
- 1. Boyle K., Traktman P. (2009) in Viral Genome Replication (Cameron C. E., Gotte M., Raney K. D. eds) pp. 225–248, Springer Science + Business Media LLC, New York [Google Scholar]
- 2. Moss B., De Silva F. S. (2006) in DNA Replication and Human Disease (DePamphilis M. ed) pp. 707–728, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 3. Challberg M. D., Englund P. T. (1979) J. Biol. Chem. 254, 7820–7826 [PubMed] [Google Scholar]
- 4. Challberg M. D., Englund P. T. (1979) J. Biol. Chem. 254, 7812–7819 [PubMed] [Google Scholar]
- 5. McDonald W. F., Traktman P. (1994) J. Biol. Chem. 269, 31190–31197 [PubMed] [Google Scholar]
- 6. McDonald W. F., Traktman P. (1994) Protein Expr. Purif. 5, 409–421 [DOI] [PubMed] [Google Scholar]
- 7. McDonald W. F., Klemperer N., Traktman P. (1997) Virology 234, 168–175 [DOI] [PubMed] [Google Scholar]
- 8. Sridhar P., Condit R. C. (1983) Virology 128, 444–457 [DOI] [PubMed] [Google Scholar]
- 9. Taddie J. A., Traktman P. (1991) J. Virol. 65, 869–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taddie J. A., Traktman P. (1993) J. Virol. 67, 4323–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Traktman P., Kelvin M., Pacheco S. (1989) J. Virol. 63, 841–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ishii K., Moss B. (2001) J. Virol. 75, 1656–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Klemperer N., McDonald W., Boyle K., Unger B., Traktman P. (2001) J. Virol. 75, 12298–12307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Punjabi A., Boyle K., DeMasi J., Grubisha O., Unger B., Khanna M., Traktman P. (2001) J. Virol. 75, 12308–12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stanitsa E. S., Arps L., Traktman P. (2006) J. Biol. Chem. 281, 3439–3451 [DOI] [PubMed] [Google Scholar]
- 16. Millns A. K., Carpenter M. S., DeLange A. M. (1994) Virology 198, 504–513 [DOI] [PubMed] [Google Scholar]
- 17. Stuart D. T., Upton C., Higman M. A., Niles E. G., McFadden G. (1993) J. Virol. 67, 2503–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Upton C., Stuart D. T., McFadden G. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4518–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boyle K. A., Arps L., Traktman P. (2007) J. Virol. 81, 844–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Silva F. S., Moss B. (2005) Virol. J. 2, 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Silva F. S., Lewis W., Berglund P., Koonin E. V., Moss B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18724–18729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Evans E., Traktman P. (1992) Chromosoma 102, S72–S82 [DOI] [PubMed] [Google Scholar]
- 23. Evans E., Klemperer N., Ghosh R., Traktman P. (1995) J. Virol. 69, 5353–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rempel R. E., Anderson M. K., Evans E., Traktman P. (1990) J. Virol. 64, 574–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rempel R. E., Traktman P. (1992) J. Virol. 66, 4413–4426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Traktman P., Anderson M. K., Rempel R. E. (1989) J. Biol. Chem. 264, 21458–21461 [PubMed] [Google Scholar]
- 27. D'Costa S. M., Bainbridge T. W., Kato S. E., Prins C., Kelley K., Condit R. C. (2010) Virology 401, 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rochester S. C., Traktman P. (1998) J. Virol. 72, 2917–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gammon D. B., Evans D. H. (2009) J. Virol. 83, 4236–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paran N., De Silva F. S., Senkevich T. G., Moss B. (2009) Cell Host Microbe 6, 563–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garcia A. D., Moss B. (2001) J. Virol. 75, 6460–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Senkevich T. G., Koonin E. V., Moss B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17921–17926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beaud G. (1995) Biochimie 77, 774–779 [DOI] [PubMed] [Google Scholar]
- 34. Traktman P. (1996) DNA Replication in Eukaryotic Cells (DePomphilis M. L. ed) pp. 775–798, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 35. Andrei G., Gammon D. B., Fiten P., De, Clercq E., Opdenakker G., Snoeck R., Evans D. H. (2006) J. Virol. 80, 9391–9401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. DeFilippes F. M. (1984) J. Virol. 52, 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. DeFilippes F. M. (1989) J. Virol. 63, 4060–4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McCraith S., Holtzman T., Moss B., Fields S. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4879–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ishii K., Moss B. (2002) Virology 303, 232–239 [DOI] [PubMed] [Google Scholar]
- 40. Kong X. P., Onrust R., O'Donnell M., Kuriyan J. (1992) Cell 69, 425–437 [DOI] [PubMed] [Google Scholar]
- 41. Stukenberg P. T., Studwell-Vaughan P. S., O'Donnell M. (1991) J. Biol. Chem. 266, 11328–11334 [PubMed] [Google Scholar]
- 42. Krishna T. S., Kong X. P., Gary S., Burgers P. M., Kuriyan J. (1994) Cell 79, 1233–1243 [DOI] [PubMed] [Google Scholar]
- 43. McConnell M., Miller H., Mozzherin D. J., Quamina A., Tan C. K., Downey K. M., Fisher P. A. (1996) Biochemistry 35, 8268–8274 [DOI] [PubMed] [Google Scholar]
- 44. Randell J. C., Coen D. M. (2004) J. Mol. Biol. 335, 409–413 [DOI] [PubMed] [Google Scholar]
- 45. Appleton B. A., Loregian A., Filman D. J., Coen D. M., Hogle J. M. (2004) Mol. Cell 15, 233–244 [DOI] [PubMed] [Google Scholar]
- 46. Bogani F., Corredeira I., Fernandez V., Sattler U., Rutvisuttinunt W., Defais M., Boehmer P. E. (2010) J. Biol. Chem. 285, 27664–27672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Parlanti E., Locatelli G., Maga G., Dogliotti E. (2007) Nucleic Acids Res. 35, 1569–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Prichard M. N., Lawlor H., Duke G. M., Mo C., Wang Z., Dixon M., Kemble G., Kern E. R. (2005) Virol. J. 2, 55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ranneberg-Nilsen T., Dale H. A., Luna L., Slettebakk R., Sundheim O., Rollag H., Bjørås M. (2008) J. Mol. Biol. 381, 276–288 [DOI] [PubMed] [Google Scholar]
- 50. Strang B. L., Coen D. M. (2010) J. Gen. Virol. 91, 2029–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Uetz P., Dong Y. A., Zeretzke C., Atzler C., Baiker A., Berger B., Rajagopala S. V., Roupelieva M., Rose D., Fossum E., Haas J. (2006) Science 311, 239–242 [DOI] [PubMed] [Google Scholar]
- 52. Mullaney J., Moss H. W., McGeoch D. J. (1989) J. Gen. Virol. 70, 449–454 [DOI] [PubMed] [Google Scholar]
- 53. Reddy S. M., Williams M., Cohen J. I. (1998) Virology 251, 393–401 [DOI] [PubMed] [Google Scholar]
- 54. Broyles S. S. (1993) Virology 195, 863–865 [DOI] [PubMed] [Google Scholar]
- 55. Holzer G. W., Falkner F. G. (1997) J. Virol. 71, 4997–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. De Silva F. S., Moss B. (2003) J. Virol. 77, 159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. De Silva F. S., Moss B. (2008) Virol. J. 5, 145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fuerst T. R., Niles E. G., Studier F. W., Moss B. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 8122–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Elroy-Stein O., Fuerst T. R., Moss B. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 6126–6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Traktman P., Boyle K. (2004) Methods Mol. Biol. 269, 169–186 [DOI] [PubMed] [Google Scholar]
- 61. Zhu Y., Trego K. S., Song L., Parris D. S. (2003) J. Virol. 77, 10147–10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fire A., Xu S. Q. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 4641–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Druck Shudofsky A. M., Silverman J. E., Chattopadhyay D., Ricciardi R. P. (2010) J. Virol. 84, 12325–12335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dales S., Milovanovitch V., Pogo B. G., Weintraub S. B., Huima T., Wilton S., McFadden G. (1978) Virology 84, 403–428 [DOI] [PubMed] [Google Scholar]
- 65. Kato S. E., Moussatche N., D'Costa S. M., Bainbridge T. W., Prins C., Strahl A. L., Shatzer A. N., Brinker A. J., Kay N. E., Condit R. C. (2008) Virology 375, 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lackner C. A., D'Costa S. M., Buck C., Condit R. C. (2003) Virology 305, 240–259 [DOI] [PubMed] [Google Scholar]
- 67. Schormann N., Grigorian A., Samal A., Krishnan R., DeLucas L., Chattopadhyay D. (2007) BMC Struct. Biol. 7, 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ellison K. S., Peng W., McFadden G. (1996) J. Virol. 70, 7965–7973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Parikh S. S., Mol C. D., Slupphaug G., Bharati S., Krokan H. E., Tainer J. A. (1998) EMBO J. 17, 5214–5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Parikh S. S., Putnam C. D., Tainer J. A. (2000) Mutat. Res. 460, 183–199 [DOI] [PubMed] [Google Scholar]
- 71. Focher F., Mazzarello P., Verri A., Hübscher U., Spadari S. (1990) Mutat. Res. 237, 65–73 [DOI] [PubMed] [Google Scholar]
- 72. Focher F., Verri A., Verzeletti S., Mazzarello P., Spadari S. (1992) Chromosoma 102, S67–S71 [DOI] [PubMed] [Google Scholar]
- 73. Neumüller M., Karlström A. R., Källander C. F., Gronowitz J. S. (1990) Biotechnol. Appl. Biochem. 12, 34–56 [PubMed] [Google Scholar]
- 74. Falkenberg M., Lehman I. R., Elias P. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3896–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lee J., Chastain P. D., 2nd, Kusakabe T., Griffith J. D., Richardson C. C. (1998) Mol. Cell 1, 1001–1010 [DOI] [PubMed] [Google Scholar]
- 76. Salinas F., Benkovic S. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7196–7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yang J., Trakselis M. A., Roccasecca R. M., Benkovic S. J. (2003) J. Biol. Chem. 278, 49828–49838 [DOI] [PubMed] [Google Scholar]
- 78. Bruck I., O'Donnell M. (2001) Genome Biol. 2, REVIEWS3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zharkov D. O., Mechetin G. V., Nevinsky G. A. (2010) Mutat. Res. 685, 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bogani F., Boehmer P. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11709–11714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bogani F., Chua C. N., Boehmer P. E. (2009) J. Biol. Chem. 284, 16784–16790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. McCulloch S. D., Kunkel T. A. (2006) DNA Repair 5, 1373–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Reineks E. Z., Berdis A. J. (2003) J. Mol. Biol. 328, 1027–1045 [DOI] [PubMed] [Google Scholar]
- 84. Tanguy, Le Gac N., Delagoutte E., Germain M., Villani G. (2004) J. Mol. Biol. 336, 1023–1034 [DOI] [PubMed] [Google Scholar]
- 85. Zhu Y., Song L., Stroud J., Parris D. S. (2008) DNA Repair 7, 95–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Frey M. W., Nossal N. G., Capson T. L., Benkovic S. J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 2579–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hwang Y. T., Liu B. Y., Coen D. M., Hwang C. B. (1997) J. Virol. 71, 7791–7798 [DOI] [PMC free article] [PubMed] [Google Scholar]