Abstract
The dihydroceramide desaturase (DES) enzyme is responsible for inserting the 4,5-trans-double bond to the sphingolipid backbone of dihydroceramide. We previously demonstrated that fenretinide (4-HPR) inhibited DES activity in SMS-KCNR neuroblastoma cells. In this study, we investigated whether 4-HPR acted directly on the enzyme in vitro. N-C8:0-d-erythro-dihydroceramide (C8-dhCer) was used as a substrate to study the conversion of dihydroceramide into ceramide in vitro using rat liver microsomes, and the formation of tritiated water after the addition of the tritiated substrate was detected and used to measure DES activity. NADH served as a cofactor. The apparent Km for C8-dhCer and NADH were 1.92 ± 0.36 μm and 43.4 ± 6.47 μm, respectively; and the Vmax was 3.16 ± 0.24 and 4.11 ± 0.18 nmol/min/g protein. Next, the effects of 4-HPR and its metabolites on DES activity were investigated. 4-HPR was found to inhibit DES in a dose-dependent manner. At 20 min, the inhibition was competitive; however, longer incubation times demonstrated the inhibition to be irreversible. Among the major metabolites of 4-HPR, 4-oxo-N-(4-hydroxyphenyl)retinamide (4-oxo-4-HPR) showed the highest inhibitory effect with substrate concentration of 0.5 μm, with an IC50 of 1.68 μm as compared with an IC50 of 2.32 μm for 4-HPR. N-(4-Methoxyphenyl)retinamide (4-MPR) and 4-Oxo-N-(4-methoxyphenyl)retinamide (4-oxo-4-MPR) had minimal effects on DES activity. A known competitive inhibitor of DES, C8-cyclopropenylceramide was used as a positive control. These studies define for the first time a direct in vitro target for 4-HPR and suggest that inhibitors of DES may be used as therapeutic interventions to regulate ceramide desaturation and consequent function.
Keywords: Enzyme Inhibitors, Lipid, Neuroblastoma, Retinoid, Sphingolipid, Dihydroceramide Desaturase, Fenretinide
Introduction
Sphingolipids are known to be modulators of various cell functions. They are not only components of cell membranes but also play a role in cell survival, apoptosis, senescence, and differentiation (1, 2). Ceramide, a central molecule in the metabolism of sphingolipids and glycosphingolipids, is involved in these regulatory cellular events. Intracellulary, ceramide is generated by different pathways. De novo synthesis of ceramide starts with condensation of l-serine with palmitoyl-CoA. Further reduction and subsequent N-acylation generates dihydroceramide. Ceramide is finally generated by introduction of the 4,5-double bond into dihydroceramide by dihydroceramide desaturase (DES)2 (3).
The DES enzyme was characterized previously, and an in vitro assay was developed to determine its activity (4). In subsequent studies, a family of sphingolipid Δ4-desaturases (homologs of the Drosophila melanogaster degenerative spermatocyte gene 1 (des-1)) were identified via a bioinformatics approach (5). These proteins contain three His-containing consensus motifs that are characteristic of a group of membrane fatty acid desaturases. The human homolog of des-1 is now referred to as DEGS-1, although it was first cloned in 1997 and named as membrane lipid desaturase because its physiologic substrate was not determined at the time (6). DEGS-1 is the only dihydroceramide desaturase reported to be present in human cells, and its mouse homolog (mDES1) was shown to have desaturase activity (7). hDES2, the human homolog of the mouse DES2 (mDes2) gene, like mDES2 has dihydroceramide hydroxylase activity (8). Although mDES2 has been reported to have both desaturase and hydroxylase activity, no desaturase activity was detected in HEK 293 human embryonic kidney cells overexpressing hDES2 (8). In this work, we refer to enzyme as DES in experiments with rat liver microsomes and as DEGS-1 in experiments with human SMS-KCNR cells.
We previously developed an assay to evaluate the in situ activity of DEGS-1 using cell-permeable dihydroceramidoids (dhCCPS analogs) (9). We showed in these studies that the synthetic retinoid N-(4-hydroxyphenyl)retinamide (4-HPR or fenretinide) is an inhibitor of DEGS-1. 4-HPR is currently in clinical trials for neuroblastoma, leukemia, lymphoma, lung, breast, head and neck, prostate, and ovarian cancer. Functionally, inhibition of DEGS-1 by 4-HPR led to the accumulation of endogenous dihydroceramides and cell cycle arrest.
In the present study, a truncated dihydroceramide analog, N-C8:0-d-erythro-dihydroceramide (C8-dhCer), was used to further characterize this enzyme, by establishing the conditions for the in vitro assay, as well as determination of kinetic parameters. Importantly, we investigated whether the enzyme served as a direct target for 4-HPR and its metabolites: 4-oxo-N-(4-hydroxyphenyl)retinamide (4-oxo-4-HPR), N-(4-methoxyphenyl)retinamide (4-MPR), and 4-oxo-N-(4-methoxyphenyl)retinamide (4-oxo-4-MPR). The results show that 4-HPR indeed inhibits the enzyme in vitro in clinically relevant concentrations. The implications of these results for therapeutic applications of 4-HPR and for DES as a candidate therapeutic target are discussed.
EXPERIMENTAL PROCEDURES
Materials
C8-dhCer was purchased from American Radiolabeled Chemicals (St. Louis, MO). N-Octanoyl-d-erythro-dihydrosphingosine and C8-cyclopropenylceramide (C8-CPPC) were purchased from Matreya, LLC (Pleasant Gap, PA). Bond Elut® C18 columns were purchased from Varian (Palo Alto, CA). EcoLumeTM Liquid Scintillation Fluid was purchased from MP Biomedicals (Cleveland, OH). Fenretinide (N-(4-hydroxyphenyl)retinamide), bicine, and trichloroacetic acid were obtained from Sigma. CHAPS was purchased from Thermo Fisher Scientific. The ceramidoids (CCPS) d-erythro-2-N-[12′-(1″-pyridinium)dodecanoyl]sphingosine bromide (C12-CCPS) and d-erythro-2-N-[12′-(1″-pyridinium)dodecanoyl]-4,5-dihydrosphingosine bromide (C12-dhCCPS; d-erythro-C12-dihydroceramide) were synthesized by the Lipidomics Core Facility at the Medical University of South Carolina (10).
The 4-HPR metabolites 4-oxo-4-HPR and 4-MPR were prepared as described previously (11, 12). The yellow solid 4-oxo-4-MPR was synthesized analogously to 4-oxo-4-HPR by reaction of 4-methoxyaniline with 4-oxoretinoic acid activated as its acid chloride: UV (methanol) λmax 376 nm (ϵ 60,000); 1H NMR (acetone-d6) * 1.29 (s, 6, (CH3)2), 1.79 (s, 3, 5-CH3), 1.85 (t, 2, 2-CH2), 2.07 (s, 3, 9-CH3), 2.42 (s, 3, 13-CH3), 2.45 (t, 2, 3-CH2), 3.76 (s, 3, OCH3), 6.04 (s, 1, 14-H), 6.34–6.44 (m, 4, vinyls), 6.97 (d, 2, Ar, J = 9 Hz), 7.09 (dd, 1, 11-H), 7.63 (d, 2, Ar, J = 9 Hz), 9.09 (s, 1, NH); MS (ESI) for C27H33NO3+Na, calculated 442.2358, observed 442.2351. HPLC analysis on a Beckman Instruments (San Ramon, CA) model 127 pump with model 166 UV detector (set at 350 nm) and a 250 × 4.6 mm Ultrasphere ODS column with 85% methanol/water at 1 ml/min showed a retention time of 7 min with a minor amount (6%) of the 13-cis isomer at 5.8 min and no other impurities. The UV spectrum was recorded on a Beckman Instruments DU-40 spectrophotometer, and the 1H NMR spectrum was recorded at 400 MHz on a Bruker Instruments DRX400 instrument (Billerica, MA). The MS was recorded at the Campus Chemical Instrument Center of The Ohio State University on a Micromass Q-Tof II spectrometer (Milford, MA).
Cell Lines and Culture Conditions
The SMS-KCNR cell line was obtained from Dr. C. Pat Reynolds (Texas Tech University). Cells were maintained in growth medium (RPMI 1640) containing 10% FCS (Invitrogen) at 37 °C in 5% CO2. C8-CPPC was dissolved in methanol at a stock concentration of 10 mm, and 4-HPR, 4-oxo-4-HPR, 4-MPR, and 4-oxo-4-MPR were dissolved in 95% ethanol at a stock concentration of 100 mm. Stock solutions were diluted to the required concentrations (0.01–2.5 μm for C8-CPPC, and 0.5–10 μm for 4-HPR and its metabolites) just prior to use, then directly added to the cells in growth medium. The final volume of methanol, ethanol, or DMSO in the medium was <0.02%, which had no effect on cell growth or survival.
Preparation of Rat Liver Microsomes
To prevent enzyme inactivation, all of the following procedures were carried out at 4 °C. Microsomes were prepared as described (13). Briefly, livers from male Sprague-Dawley rats were rinsed twice in ice-cold normal saline and homogenized in buffer (0.25 m sucrose, 10 mm HEPES, 1 mm EDTA, pH 7.4) with a Polytron® PT 1200 E (Kinematica, Inc., Bohemia, NY) homogenizer. The homogenate was centrifuged at 10,000 × g for 15 min followed by ultracentrifugation of the resulting supernatant at 104,000 × g for 60 min. The microsomal pellet was resuspended in potassium phosphate buffer (50 mm, pH 7.4). Aliquots were stored at −80 °C until use. Protein concentration was determined by the Bradford method (Bio-Rad) according to the manufacturer's directions.
Solubilization of Lipid Substrates and Inhibitors
A solution containing labeled and unlabeled substrates as well as inhibitors was dried under a stream of nitrogen in a 1.5-ml Eppendorf tube. 1.1 mg of CHAPS dissolved in 10 μl of water was added to each tube, mixed thoroughly, and sonicated for 3 min in a bath sonicator. This step was repeated three times, and the tubes were vortexed for 30 s each time.
In Vitro Dihydroceramide Desaturase Assay
The assay was performed as described previously (14) with some modifications. Heat-inactivated microsomes were used as negative controls. All reactions were performed with 100 μg of protein and 20 min incubation time at 37 °C unless otherwise mentioned. (Data from optimization studies showed that 100 μg of protein and 20 min of incubation time were both within the linear range of the assay; data not shown; see supplemental data). Labeled substrate (see Fig. 1A, 2 nm equal to 0.125 μCi, and ∼100,000 dpm) and 500 nm unlabeled substrate were used in all reactions unless otherwise specified. Briefly, the reaction mixture contained 2 mm of NADH (dissolved in double distilled H2O), 20 mm bicine, pH 8.5, 50 mm NaCl, and 50 mm sucrose. The substrates as well as the inhibitors dissolved in CHAPS were pipetted into glass tubes. The mixture was incubated for 20 min at 37 °C with shaking. The reaction was terminated by the addition of 100 μl of 8% BSA (w/v), immediately followed by 100 μl of 72% trichloroacetic acid (w/v). To remove denatured protein, the reaction mixture was centrifuged at 1100 × g for 20 min at 4 °C. The supernatant was transferred to a 13 × 100 mm glass tube containing 350 μl of 1 m Na2HPO4 to bring the pH to 5.5. The Varian Bond Elute® C18 columns were washed with 1 ml of methanol followed by a wash of 1 ml of water right before use, and then the supernatant was passed over the columns. The flow-through fraction and a wash fraction of 2 ml of water were collected. A 900 μl fraction of each eluate was transferred to counting tubes, and tubes were filled with Eco-LumeTM solvent, and the radioactivity was determined using a liquid scintillation counter. Released radioactivity was determined, and the formation of product was calculated based on the specific activity of the substrate. In calculating the results, the amount of radioactivity was halved to take into account only the amount of radioactivity that was relevant to the enzyme activity. This is important because the action of the enzyme abstracts only one of the two hydrogen atoms, and only one is “randomly” labeled with 3H.
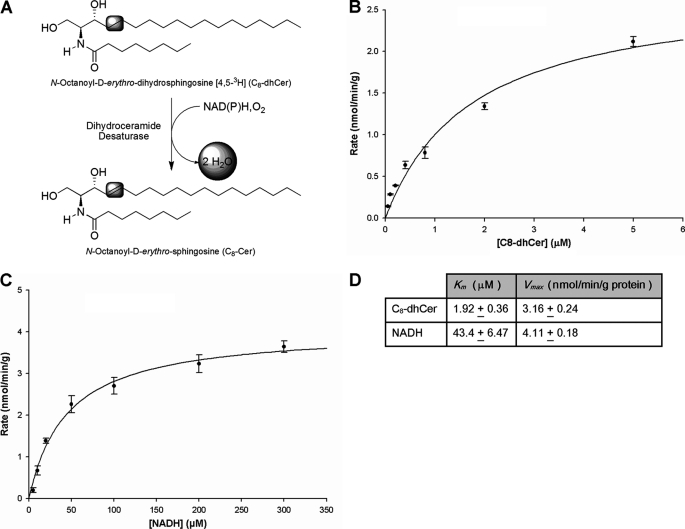
FIGURE 1.
Kinetic parameters for in vitro DES assay. A, conversion of C8-dhCer into C8-Cer. This scheme depicts the oxidation of C8-dhCer into C8-Cer by the enzyme dihydroceramide desaturase. The formed water is radiolabeled, and it was the target for measurements in our assays. B and C, Michaelis-Menten plots of DES activity for determination of Km and Vmax for C8-dhCer and NADH. B, determination of Km and Vmax for C8-dhCer. A fixed amount of labeled C8-dhCer and increasing amounts of unlabeled C8-dhCer starting from 0.05 μm to 10 μm were used. C, determination of Km and Vmax for NADH. Increasing amounts of NADH were used and [C8-dhCer] was 0.25 Km. Assays were performed in vitro using rat liver microsomes with an incubation time of 20 min. Values represent the mean ± S.D. of at least two separate determinations performed in triplicate. Km and Vmax values are shown in Table 1. D, table showing Km and Vmax values for NADH and C8-dhCer.
Note that for assays using SMS-KCNR total cell homogenates instead of rat liver microsomes, cells were grown in T-150 (Corning®) flasks at a density of 3 × 106 cells per flask. The confluency on the day of treatment was ∼35–40%. Cells were treated with 5 μm of 4-HPR for 2 h. Cells were then washed with PBS, and the medium was replaced by fresh medium. Control flasks contained 0.1% ethanol, which did not affect DES activity (data not shown). Cells were incubated with fresh medium for 24, 48, or 72 h. After each time point, flasks were washed twice with ice-cold PBS after removing the medium. Cells were scraped and centrifuged at 1000 × g for 5 min at 4 °C. Total cell homogenates were prepared from the cell pellets as described previously (15). Briefly, pellets were resuspended in homogenization buffer (5 mm HEPES, pH 7.4, containing 50 mm sucrose) and kept on ice for 10 min. Cell suspensions were homogenized employing a 1-ml insulin syringe using 10 strokes. Homogenates were centrifuged at 250 × g for 5 min at 4 °C to remove unbroken cells. The assays were performed as described above, substituting 400 μg of total cell homogenate for rat liver microsomes at 37 °C for 20 min. (Data from previous optimization studies using total cell homogenates demonstrated that 400 μg of protein and 20 min of incubation time were both within the linear range of the assay (16).)
Statistical Analysis and Determination of Kinetic Constants
Data were analyzed using SigmaPlot® Enzyme Kinetics Module (Systat Software, Inc. San Jose, CA) software to determine the IC50, Ki, Km constants, and Vmax value.
In Situ Dihydroceramide Desaturase Assay
C12-dhCCPS was used as a cellular substrate for this enzyme. C12-dhCCPS was dissolved in 100% ethanol at concentrations of 100 mm, and this stock solution was diluted immediately prior to use and directly added to the cells in medium containing 10% fetal calf serum to obtain a final concentration of 0.5 μm. Cells were incubated with C12-dhCCPS for 6 h. The cells were collected at this time points, and levels of C12-dhCCPS and its product C12-CCPS were detected by LC/MS (9, 10). The percentage of the conversion C12-CCPS was then calculated.
LC-MS Analysis of Endogenous Ceramides and Cellular Level of CCPS and dhCCPS Analogs
Advanced analyses were performed by the Lipidomics Core Facility at the Medical University of South Carolina on a ThermoFinningan TSQ 7000, triple-stage quadrupole mass spectrometer operating in a multiple reaction monitoring positive ionization mode as described (17).
RESULTS
Determination of Dihydroceramide Desaturase Activity in Vitro by Measurement of Formed Water
This method was first described by Geeraert et al. (14). In this procedure, the enzyme activity is determined by formation of tritiated water that accompanies the 4,5-double bond formation if the substrate is labeled appropriately (Fig. 1A). C8-dhCer was employed as the substrate, and NADH served as a cofactor. After 20 min of incubation, the reaction mixture, and a wash fraction of 2 ml with water were loaded onto C18 columns, and the eluate was collected. A fraction was taken, and the radioactivity was determined.
Determination of Kinetic Parameters for C8-dhCer and NADH
For enzyme kinetic determinations, a fixed amount of labeled C8-dhCer of 100,000 dpm (0.125 μCi = 2 nm), and increasing amounts of unlabeled C8-dhCer starting from 0.05 μm to 10 μm were used. The Km and Vmax for C8-dhCer (Fig. 1B) as well as for NADH (Fig. 1C) were determined, and the Km for C8-dhCer was 1.92 ± 0.36 μm, and Vmax was 3.16 ± 0.24 nmol/min/g protein. To determine the kinetic parameters for NADH, the concentration of substrate used was three times the Km of C8-dhCer. The NADH concentration was varied from 5 μm up to 300 μm. The Km for NADH was 43.4 ± 6.47 μm, and the Vmax was 4.11 ± 0.18 nmol/min/g protein, respectively.
Determination of Ki for 4-HPR
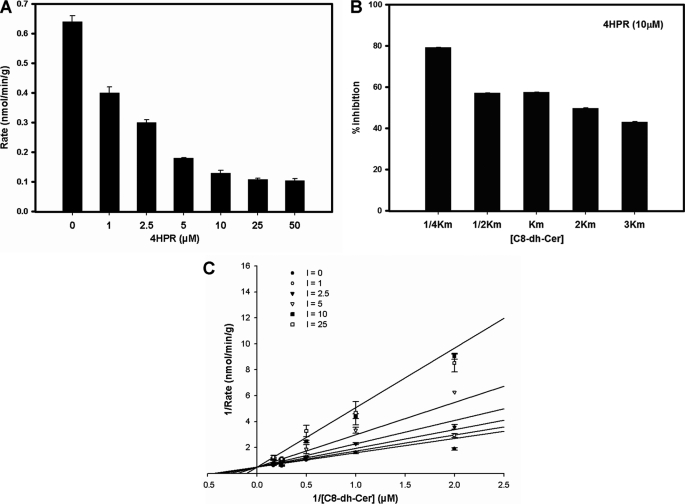
4-HPR is a synthetic analog of retinoic acid that inhibits DES (9, 18, 19). We have previously demonstrated the inhibitory effects of 4-HPR on DEGS-1 in situ in intact cells using the SMS-KCNR neuroblastoma cell line. Inhibition of DEGS-1 resulted in increased levels of endogenous dihydroceramides leading to cell growth inhibition. In this study, a dose response experiment was first conducted using the in vitro enzyme assay at 20 min and varying the concentrations of 4-HPR with a fixed amount of substrate (0.25 Km, equal to 0.5 μm) (Fig. 2A). Next, to demonstrate that with increasing substrate concentrations the percentage of inhibition would decrease, assays were conducted with a fixed amount of 4-HPR (10 μm) and increasing amounts of substrate (between 0.25 Km to 3 Km) (Fig. 2B). To determine the Ki (the dissociation constant for binding of inhibitor to enzyme) for 4-HPR and whether it inhibited in a competitive manner, experiments were performed with a fixed amount of substrate and increasing amounts of inhibitor. The Km substrate concentrations utilized were 0.25, 0.5, 1, 2, and 3. The amount of 4-HPR ranged from 0, 1, 2.5, 5, 10, 25, and 50 μm. The results showed that at 20 min 4-HPR interacted with the dihydroceramide substrate in a competitive manner (Fig. 2C). The Ki value was determined using SigmaPlot® and its enzyme kinetics module. The Ki for 4-HPR was 8.28 ± 1.25 μm.
FIGURE 2.
The effects of 4-HPR on DES activity and determination of Ki for 4-HPR. A, the effects of increasing concentrations of 4-HPR on DES activity in rat liver microsomes. DES activity was measured using a fixed amount of substrate (2 nm labeled and 500 nm unlabeled substrate) and increasing concentrations of 4-HPR (0, 1, 2.5, 5, 10, 25, and 50 μm). Values represent the mean ± S.D. of three independent experiments performed in triplicate. B, the effects of increasing amounts of substrate on 4-HPR inhibition of DES activity in rat liver microsomes. DES activity was measured using a fixed concentration of 4-HPR (10 μm) and increasing amounts of substrate (between 0.25 and 3 Km). Values represent % inhibition as compared with controls and the mean ± S.D. of two independent experiments performed in triplicate. C, Lineweaver-Burke plot for Ki demonstrating DES activity with increasing concentrations of 4-HPR. For these experiments, the [substrate] was fixed and ranged from 0.25, 0.5, 1, 2, to 3 Km, and the [4-HPR] used ranged from 0, 1, 2.5, 5, 10, 25, and 50 μm. Values represent the mean ± S.D. of five independent experiments performed twice in triplicate.
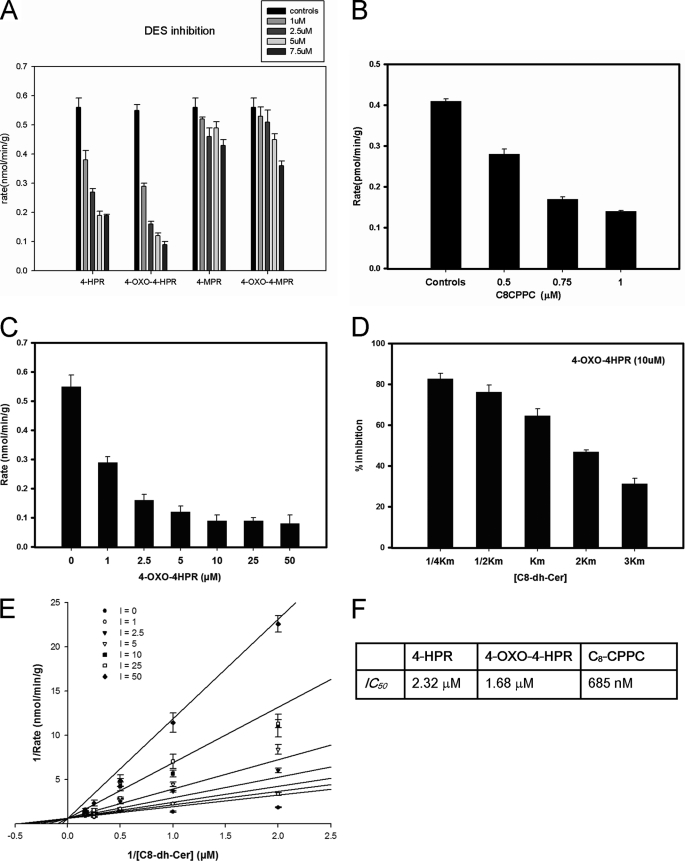
Effects of 4-HPR and Its Metabolites on DES Activity
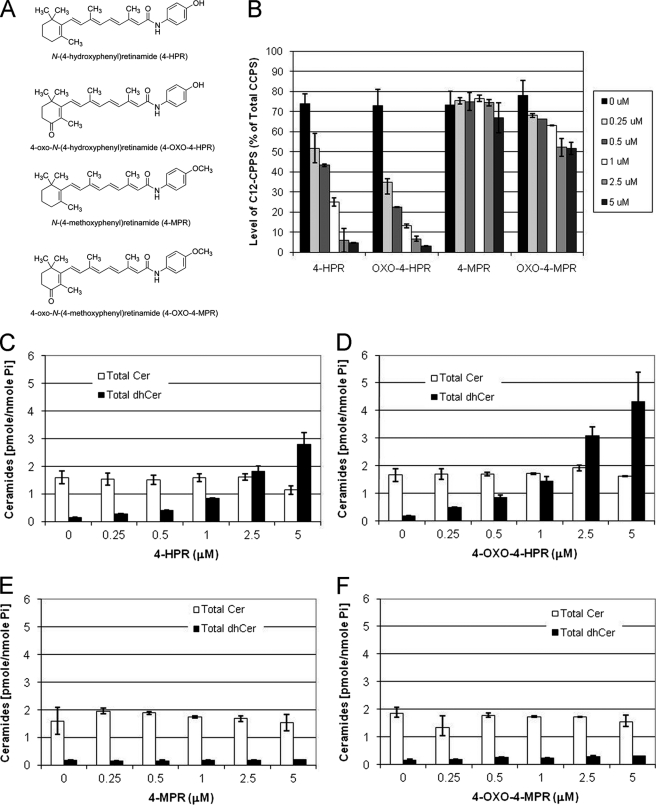
Two metabolites of 4-HPR have been identified in human plasma, 4-MPR and 4-oxo-4-HPR (20). 4-MPR is the most abundant metabolite in human plasma. It has been found to be ineffective in inducing cell growth inhibition and does not generate reactive oxygen species. 4-oxo-4-HPR is an oxidized form of 4-HPR with modification in position 4 of the cyclohexene ring. Similar to 4-HPR, it induces reactive oxygen species generation and apoptosis (11). 4-oxo-4-MPR is a putative metabolite of 4-HPR. The structures of the metabolites are depicted in Fig. 4.
FIGURE 4.
The effects of 4-HPR, 4-HPR metabolites, and C8-CPPC on desaturase activity in vitro. A, the effects of increasing concentrations of 4-HPR and 4-HPR metabolites on DES activity in rat liver microsomes. In these experiments, [C8-dhCer] was 0.25 Km, and the inhibitor concentrations were between 1 and 7.5 μm. The values represent the mean ± S.D. of two independent experiments performed in triplicate. B, the effects of increasing concentrations of C8-CPPC on DES activity in rat liver microsomes. In these experiments, [C8-dhCer] was 0.25 Km, and C8-CPPC concentrations were 0.5, 0.75, and 1 μm. The values represent the mean ± S.D. of two independent experiments performed in triplicate. C, the effects of increasing concentrations of 4-oxo-HPR on DES activity in rat liver microsomes. DES activity was measured using a fixed amount of substrate (2 nm labeled and 500 nm unlabeled substrate) and increasing concentrations of 4-oxo-HPR (0, 1, 2.5, 5, 10, 25, and 50 μm). The values represent the mean ± S.D. of two independent experiments performed in triplicate, D, the effects of increasing amounts of substrate on 4-oxo-HPR inhibition of DES activity in rat liver microsomes. DES activity was measured using a fixed concentration of 4-oxo-HPR (10 μm) and increasing amounts of substrate (from 0.25 to 3 Km). The values represent % inhibition as compared with controls and the mean ± S.D. of two independent experiments performed in triplicate. E, Lineweaver-Burke plot for Ki demonstrating DES activity with increasing concentrations of 4-oxo-HPR. For these experiments, the [substrate] was fixed and ranged from 0.25, 0.5, 1, 2, to 3 Km, and the [4-oxo-HPR] used ranged from 0, 1, 2.5, 5, 10, 25, and 50 μm. Values represent the mean ± S.D. of five independent experiments performed twice in triplicate. F, table of IC50 values for 4-HPR, 4-oxo-4HPR, and C8-CPPC. Values represent the mean ± S.D. of three independent experiments performed in triplicate.
First, we tested the ability of 4-HPR to inhibit desaturase activity using our in situ assay with C12-dhCCPS (Fig. 3A). SMS-KCNR human neuroblastoma cells were treated with increasing concentrations (0.25, 0.5, 1, 2.5, and 5 μm) of 4-HPR, 4-MPR, 4-oxo-4-HPR, or 4-oxo-4-MPR for 6 h. The substrate C12-dhCCPS (0.5 μm) was added at the same time as 4-HPR and its metabolites, and the conversion to C12-CCPS was measured by LC/MS. 4-HPR and 4-oxo-4-HPR inhibited desaturase activity in a dose-dependent manner (Fig. 3B). Inhibition was observed even at the lowest dose. In untreated (control) cells, ∼74% of measured CCPS were converted to C12-CCPS at 6 h. The conversion was decreased in cells treated with 0.25, 0.5, 1, 2.5, and 5 μm 4-HPR and 4-oxo-4-HPR starting at 0.25 μm, decreasing conversion levels to ∼52 and ∼35%, respectively (Fig. 3B). The percentage of the conversion to C12-CCPS in cells treated with 0.5, 1, 2.5, and 5 μm 4-HPR was ∼43, ∼25, ∼6, and ∼5% respectively. The percentage of the conversion to C12-CCPS in cells treated with 0.5, 1, 2.5, and 5 μm 4-oxo-4-HPR was ∼22, ∼13, ∼6, and ∼3%, respectively. Measurement of total endogenous ceramides (Cer), dihydroceramides (dhCer), and sphingosine levels was also performed (Fig. 3, C–F) by LC/MS. Approximately 1.8-, 2.7-, 5.5-, 11.7-, and 18-fold increases in total endogenous dihydroceramides were observed in cells treated with 0.25, 0.5, 1, and 2.5 μm 4-HPR, respectively, correlating with the inhibition of DEGS-1 (Fig. 5B). Approximately 2.7-, 4.8-, 8-, 17.3-, and 24.3-fold increases in total endogenous dihydroceramides were observed in cells treated with 0.25, 0.5, 1, and 2.5 μm 4-oxo-HPR, respectively (Fig. 3D). 4-oxo-4-HPR was a slightly more potent inhibitor of DEGS-1 than 4-HPR. There were no significant changes seen in endogenous sphingosine, dihydrosphingosine, or sphinogosine-1-phosphate levels (data not shown). The other metabolites (4-MPR and 4-oxo-MPR) had minimal effects on DEGS-1 activity or endogenous ceramide levels (Fig. 3, B, E, and F).
FIGURE 3.
Effects of 4-HPR and its metabolites on in situ desaturase activity and endogenous sphingolipids. A, structures of 4-HPR and its metabolites. B, desaturase activity was measured using the in situ assay. SMS-KCNR cells were treated with increasing concentrations of 4-HPR, 4-oxo-4-HPR, 4-MPR, or 4-oxo-4-MPR for 6 h. C12-dhCCPS was added at the same time as 4-HPR or its metabolites. Cells were collected after 6 h, and the conversion to C12-CCPS was determined by LC/MS. Total endogenous levels of ceramides (Cer) and dihydroceramides (dhCer) in SMS-KCNR cells treated with 4-HPR (C), 4-oxo-4-HPR (D), 4-MPR (E), or 4-oxo-4-MPR (F) were measured by LC/MS as described under “Experimental Procedures.” Sphingolipid levels were normalized to total lipid phosphate. The data presented are representative of the mean of two independent experiments ± S.D. The error bars represent the S.D., and when not seen, they are smaller than the thickness of the lines on the graphs.
FIGURE 5.
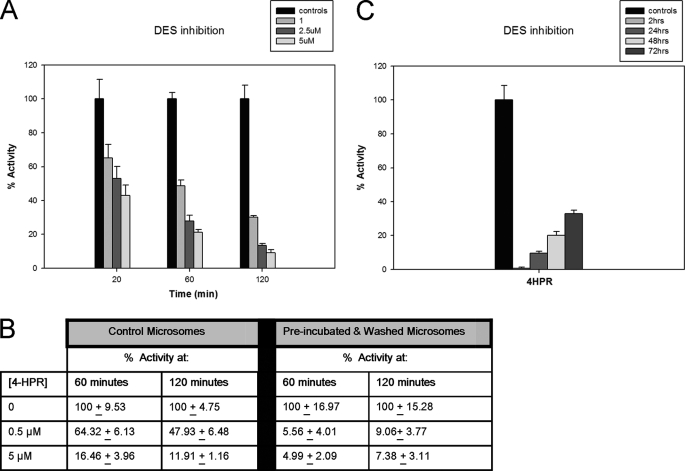
The effects of 4-HPR on the reversibility of inhibition of desaturase activity in rat liver microsomes or SMS-KCNR cells. A, the effects of inhibition by 4-HPR over time. Rat liver microsomes were treated with 1, 2.5, and 5 μm of 4-HPR, and the in vitro assay was performed for either 20, 60, or 120 min along with controls. Values represent the remaining activity (% to controls), and they represent the mean ± S.D. of two independent experiments performed in quadruplicate. B, the effects of dilution of 4-HPR on DES activity. Rat liver microsomes were preincubated with either 0.5 or 5 μm 4-HPR for 60 min at 37 °C. After preincubation, the microsomes were centrifuged and washed twice. The in vitro enzyme assay was performed with 100 μg of either “control microsome” or “preincubated and washed microsomes” for 60 or 120 min. Control microsomes were not washed or preincubated with 4-HPR. The values represent the remaining activity (% to controls), and they represent the mean ± S.D. of two independent experiments performed in quadruplicate. C, the effect of 4-HPR inhibition on SMS-KCNR cells. Cells were treated with 5 μm of 4-HPR for 2 h. After 2 h, cells were washed with PBS, and the medium was replaced by fresh medium. Total cell homogenates were prepared from SMS-KCNR cells after 2 h of 4-HPR treatment, and at 24, 48, and 72 h after 4-HPR treatment. Determination of DES activity was performed at these time points, using the in vitro assay with 400 μg of total cell homogenate for 20 min. Values represent the remaining activity (% to controls), and they represent the mean ± S.D. of two independent experiments performed in quadruplicate.
Determination of IC50 for 4HPR, Its Metabolites, and C8CPPC
We then tested the effects of 4HPR and its metabolites using the in vitro assay with rat liver microsomes at 20 min. For determination of IC50 (the concentration of inhibitor at which there was 50% inhibition of enzyme activity at 20 min), the substrate concentration utilized was 0.25 Km (0.5 μm). The inhibitor concentration ranged between 1 and 7.5 μm (Fig. 4A). The IC50 was determined using SigmaPlot® and its enzyme kinetics module by plotting the log of inhibitor concentration against percentage of inhibition. The IC50 for 4-HPR was 2.32 μm. Among 4-HPR metabolites, 4-oxo-4-HPR was the most potent inhibitor with an IC50 of 1.68 μm. The inhibition was slightly more than 4-HPR, an effect consistent with the previous experiment on in situ DEGS-1 inhibition, where 4-oxo-4-HPR showed more inhibition than 4-HPR. 4-MPR and 4-oxo-4-MPR had minimal effects on DES activity. C8-cyclopropenylceramide, a known competitive inhibitor of the DES enzyme (21–23) was used as a positive control in these assays. The IC50 for C8CPPC was 685 nm (Fig. 4B). Fig. 4F summarizes the IC50 values for inhibitors studied in these experiments.
Effects of 4-Oxo-HPR and Its Metabolites on DES Activity
As for 4-HPR, a dose response experiment was conducted using the in vitro enzyme assay at 20 min and varying the concentrations of 4-oxo-HPR with a fixed amount of substrate (0.25 Km, equal to 0.5 μm) (Fig. 2A). Assays were also conducted with a fixed amount of 4-oxo-HPR (10 μm) and increasing amounts of substrate (between 0.25 to 3 Km) (Fig. 2B). We also determined the Ki for 4-oxo-4-HPR at 20 min of incubation in the same manner as we did for 4-HPR. In brief, the substrate concentration in each experiment was constant (ranging from 0.25 to 3× Km), and the inhibitor concentration ranged from 1 to 50 μm. As with 4-HPR, the results showed that at 20 min, 4-oxo-HPR interacted with the dihydroceramide substrate in a competitive manner (Fig. 4C). The Ki value was determined using SigmaPlot® and its enzyme kinetics module. The Ki for 4-oxo-4-HPR was 6.61 ± 1.25 μm.
Determination of Reversibility of Enzyme Inhibition by 4-HPR
We examined whether 4-HPR could be an irreversible inhibitor because an irreversible inhibitor would appear competitive until a significant amount of the enzyme had been inactivated. To test the veracity of this, the in vitro assay with rat liver microsomes was performed were with 1, 2.5, and 5 μm of 4-HPR for either 20, 60, and 120 min along with controls for each time point (Fig. 5A). The percent inhibition for microsomes treated with 1 μm 4-HPR for 20, 60, or 120 min was ∼35, ∼52, and ∼69%, respectively. The percent inhibition for microsomes treated with 2.5 μm 4-HPR for 20, 60, or 120 min was ∼46, ∼73, and ∼84%, respectively. The percent inhibition for microsomes treated with 5 μm 4-HPR for 20, 60, or 120 min was ∼57, ∼79, and ∼89%, respectively. In these experiments, inhibition increased with time suggesting time-dependent irreversible inhibition.
To further determine whether 4-HPR was an irreversible inhibitor, rat liver microsomes were preincubated with concentrations of 4-HPR that would inhibit DES by ∼30% (0.5 μm) or ∼90% (5 μm) for 60 min at 37 °C. After preincubation, the microsomes were centrifuged and washed twice with 5 ml of 50 mm potassium phosphate buffer, pH 7.4. The assay was performed with 100 μg of microsomes that had not been preincubated or washed or with 100 μg of preincubated and washed microsomes for 60 or 120 min. This method of washing has been reported to give a dilution of inhibitor of >800,000:1 (24, 25). If an enzyme inhibits irreversibly, then little recovery of enzyme activity should be noted. There was very little recovery of enzyme activity in the preincubated and washed (diluted) samples (Fig. 5B). Enzyme activity was ∼6 and ∼9% of control for microsomes preincubated with 0.5 μm 4-HPR and assayed for 60 or 120 min and ∼5 and ∼7% of control for microsomes preincubated with 5 μm 4-HPR and assayed for 60 or 120 min, respectively.
We then examined the reversibility of DEGS-1 inhibition in SMS-KCNR cells (Fig. 5C). Cells were treated with 5 μm of 4-HPR for 2 h. After 2 h, cells were washed with PBS, and the medium was replaced by fresh medium. Total cell homogenates were prepared from SMS-KCNR cells after 2 h of 4-HPR treatment, and at 24, 48, and 72 h after 4-HPR treatment. Determination of DES activity was performed at these time points, using the in vitro assay with total cell homogenates for 20 min. After 2 h of 5 μm 4-HPR treatment, there was almost no activity detected. After replacement of medium and time, there was some recovery of enzyme activity; there was ∼90, ∼80, and ∼67% inhibition of DES activity after 24, 48, and 72 h, respectively.
DISCUSSION
In this study, we established the conditions for an in vitro assay for DES using rat liver microsomes and a tritium-labeled dihydroceramide analog, and evidence is provided for potent direct inhibition of DES by 4-HPR and its active metabolites. These results have implications for the mechanism of action of 4-HPR and for a potential role for DES as a target for chemotherapy.
The in vitro assay optimized in this study was accurate and very reproducible. As solubilizing agents, we tested BSA, CHAPS, and ethanol/dodecane, as the substrate was difficult to get into solution without solubilizing agents. The best solubilization was achieved using CHAPS (data not shown). Under these conditions (with NADH as a co-factor), reactions proceeded according to classical Michaelis-Menten kinetics and were linear with time and protein concentration. The Km value for the dihydroceramide substrate was in the low μm range (about 2 μm) whereas the Km for the NADPH substrate was determined at 43 μm.
The development of the assay allowed us to investigate whether 4-HPR functions as a direct inhibitor of DES. 4-HPR is employed as a chemotherapeutic agent especially in neuroblastoma as well as leukemia and various solid tumors, including lung and prostate cancer. In addition to being employed as a cytotoxic drug in clinical trials, it has also been reported to act as a chemopreventive agent in breast cancer and oral leukoplakia (26–28).
Our previous results showed that at clinically achievable μm concentrations, 4-HPR inhibits DEGS-1 activity in cells (using an in situ assay) in SMS-KCNR human neuroblastoma cells, causing marked increases in the levels of endogenous dihydroceramides, leading to cell cycle arrest by hypophosphorylation of retinoblastoma protein (9). Initial investigation of the mechanism failed to reveal effects on protein or message levels for DEGS-1. These results suggested the possibility that 4-HPR may act as a direct inhibitor of the enzyme, especially because it is a lipid amide with some resemblance to the dihydroceramide substrate of the enzyme.
4-HPR has a very broad range (0.7–10 μm) of cytotoxicity and may have different effects dependent on the concentration and type of cancer cell used (29). In neuroblastoma, high concentrations of 4-HPR (>5 μm) have been shown to induce apoptosis and necrosis (3), whereas lower concentrations (< 3 μm) of 4-HPR have been reported to induce G1-S phase arrest and hypophosphorylation of retinoblastoma (9, 30, 31). For neuroblastoma patients enrolled on a phase I pediatric trial, the mean plasma steady-state concentration on day 7 was as 9.9 μmol/liter with the maximal tolerated dosage of 2475 mg/m2 per day (32). Another more recent pediatric phase I study using a new formulation of 4-HPR to improve bioavailability (4HPR/Lym-X-Sorb oral powder), reported the day 7 mean peak plasma concentrations to be 19.7 μm at the recommended dose of 1700 mg/m2 per day (33). These doses safely achieved levels active against neuroblastoma in vivo with minimal toxicity. Although our in vitro study employed rat liver microsomes, we may infer that the levels of 4-HPR used for the in vitro assays would correspond with levels that are clinically achievable in patients.
Indeed, the results from this study show that 4-HPR functions as a potent and direct inhibitor of DES in rat liver microsomes with a Ki of 8 μm at 20 min. The in vitro assay initially showed a competitive pattern of inhibition with respect to dihydroceramide. Further studies with longer assay times and preincubation with 4-HPR strongly support that the inhibition is irreversible. As there was little recovery of enzyme activity in samples preincubated with 4-HPR and assayed for 2 h, the inhibition is unlikely to be slow reversible inhibition. Further studies are underway to characterize the inhibition. Our experiments using SMS-KCNR cell homogenates demonstrated some 1recovery of enzyme function over time. Enzyme activity was likely restored by de novo enzyme synthesis in these cells. These results demonstrate for the first time a direct clinically relevant target for the action of 4-HPR. As such, DES may emerge as a novel target for chemotherapeutic development.
DES is an important enzyme that has the potential to modulate the dihydroceramide/ceramide ratio in the cell; however, the regulation of DES activity has not been well characterized. Moreover, although the cell biology of ceramide has been well studied with roles in regulation of cell apoptosis, growth, and differentiation, the functions of dihydroceramides have been largely obscure. Consequently, dihydroceramide has been considered primarily as an inactive precursor of ceramide (34). However, recent studies have shown dihydroceramides to be involved in important cellular responses such as cell cycle arrest, apoptosis, ceramide channel formation, autophagy, and oxidative stress (9, 16, 18, 35–37). These studies indicate the significance of endogenous levels of dihydroceramides in cell regulatory processes, and they suggest that DES has the potential to regulate cell function through both ceramide and dihydroceramide (and their respective sphingolipid metabolites).
In addition to 4-HPR and C8-CPPC, other cellular inhibitors of DES such as γ-tocopherol, celecoxib, resveratrol, XM462, photodynamic therapy, and hydrogen peroxide have been identified recently (16, 35, 38–41); however, these are unlikely to function as direct inhibitors and probably function by modulating the redox status of the cell that has been shown to modulate DES activity.
The current results also implicate key metabolites of 4-HPR as inhibitors of DES activity in situ in SMS-KCNR neuroblastoma cells and in vitro in rat liver microsomes. The 4-oxo-4-HPR metabolite has been detected in human plasma and can also be formed through induction of CYP26A1 (20). It was the most potent inhibitor among the metabolites tested. 4-MPR and 4-oxo-4-MPR showed minimal inhibition. 4-oxo-4-HPR has also been reported to induce apoptosis in both 4-HPR-sensitive and -resistant cells, and it has been demonstrated to have more potent cytotoxicity than 4-HPR or 4-MPR alone (11). Furthermore, it has also been reported that clinically achievable levels of 4-oxo-4HPR are possible with daily oral dosing of 4-HPR (29).
Exploring and studying inhibitors of ceramide metabolism pathways is a valuable approach in modulating ceramide-mediated biological processes. As such, targeting ceramide metabolism has been shown to be an effective method of cancer cell destruction (2). Drugs that augment ceramide and dihydroceramide levels induce cell toxicity and apoptosis in cancer cells. The current results and previous results suggest that inhibition of DES is a novel target for cancer therapy. The balance of ceramides and dihydroceramides within a cell may be key to cancer cell proliferation.
Supplementary Material
Acknowledgments
We thank Dr. Leslie Wooten-Blanks and Dr. Li Li for helpful discussions. We thank Dr. Jacek Bielawski, Dr. Alicja Bielawska, Barbara Rembiesa, and Jason Pierce from the Lipidomics Core Facility in the Department of Biochemistry and Molecular Biology at the Medical University of South Carolina.
This work was supported, in whole or in part, by National Institutes of Health Grants K01-CA100767 (to J. M. K.), P01-CA97132 (to Y. A. H. and L. M. O.), R01-AG016583 (to L. M. O.), R01-CA49837 (to R. W. C.), C06-RR018823 (Lipidomics Core), and by a MERIT award by the Office of Research and Development (to L. M. O.). This work was also supported by the Department of Veterans Affairs, Ralph H. Johnson VA Medical Center (Charleston, SC) by Grant P20-RR17677 from the National Center for Research Resources and by grants from the Rally Foundation for Childhood Cancer Research, the Monica Kreber Golf Tournament, the Chase After a Cure Foundation, and Hyundai Hope on Wheels (to J. M. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- DES
- dihydroceramide desaturase
- 4-HPR
- fenretinide and N-(4-hydroxyphenyl)retinamide
- C8-dhCer
- N-C8:0-d-erythro-dihydroceramide
- 4-oxo-4-HPR
- 4-oxo-N-(4-hydroxyphenyl)retinamide
- 4-MPR
- N-(4-methoxyphenyl)retinamide
- 4-oxo-4-MPR
- 4-oxo-N-(4-methoxyphenyl)retinamide
- C8-CPPC
- C8-cyclopropenylceramide
- DEGS-1
- human dihydroceramide desaturase
- dhCCPS
- dihydroceramidoids
- CCPS
- ceramidoids
- C12-dhCCPS
- d-erythro-2-N-[12′-(1″-pyridinium)dodecanoyl]-4,5-dihydrosphingosine bromide
- C12-CCPS
- d-erythro-2-N-[12′-(1″-pyridinium)dodecanoyl]sphingosine bromide
- dhCer
- dihydroceramide.
REFERENCES
- 1. Hannun Y. A., Obeid L. M. (2008) Nat. Rev. Mol. Cell. Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 2. Ogretmen B., Hannun Y. A. (2004) Nat. Rev. Cancer 4, 604–616 [DOI] [PubMed] [Google Scholar]
- 3. Wang H., Maurer B. J., Reynolds C. P., Cabot M. C. (2001) Cancer research 61, 5102–5105 [PubMed] [Google Scholar]
- 4. Michel C., van Echten-Deckert G., Rother J., Sandhoff K., Wang E., Merrill A. H., Jr. (1997) J. Biol. Chem. 272, 22432–22437 [DOI] [PubMed] [Google Scholar]
- 5. Ternes P., Franke S., Zähringer U., Sperling P., Heinz E. (2002) J. Biol. Chem. 277, 25512–25518 [DOI] [PubMed] [Google Scholar]
- 6. Cadena D. L., Kurten R. C., Gill G. N. (1997) Biochemistry 36, 6960–6967 [DOI] [PubMed] [Google Scholar]
- 7. Omae F., Miyazaki M., Enomoto A., Suzuki M., Suzuki Y., Suzuki A. (2004) Biochem. J. 379, 687–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mizutani Y., Kihara A., Igarashi Y. (2004) FEBS. Lett. 563, 93–97 [DOI] [PubMed] [Google Scholar]
- 9. Kraveka J. M., Li L., Szulc Z. M., Bielawski J., Ogretmen B., Hannun Y. A., Obeid L. M., Bielawska A. (2007) J. Biol. Chem. 282, 16718–16728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Szulc Z. M., Bielawski J., Gracz H., Gustilo M., Mayroo N., Hannun Y. A., Obeid L. M., Bielawska A. (2006) Bioorg Med. Chem. 14, 7083–7104 [DOI] [PubMed] [Google Scholar]
- 11. Villani M. G., Appierto V., Cavadini E., Bettiga A., Prinetti A., Clagett-Dame M., Curley R. W., Formelli F. (2006) Cancer Res. 66, 3238–3247 [DOI] [PubMed] [Google Scholar]
- 12. Mershon S. M., Anding A. L., Chapman J. S., Clagett-Dame M., Stonerock L. A., Curley R. W., Jr. (2007) Bioorg Med. Chem. Lett. 17, 836–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schulze H., Michel C., van Echten-Deckert G. (2000) Methods Enzymol. 311, 22–30 [DOI] [PubMed] [Google Scholar]
- 14. Geeraert L., Mannaerts G. P., van Veldhoven P. P. (1997) Biochem. J. 327, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sukhodub A. L., Burchell A. (2005) J. Pharmacol. Toxicol. Methods 52, 330–334 [DOI] [PubMed] [Google Scholar]
- 16. Idkowiak-Baldys J., Apraiz A., Li L., Rahmaniyan M., Clarke C. J., Kraveka J. M., Asumendi A., Hannun Y. A. (2010) Biochem. J. 427, 265–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bielawski J., Szulc Z. M., Hannun Y. A., Bielawska A. (2006) Methods 39, 82–91 [DOI] [PubMed] [Google Scholar]
- 18. Zheng W., Kollmeyer J., Symolon H., Momin A., Munter E., Wang E., Kelly S., Allegood J. C., Liu Y., Peng Q., Ramaraju H., Sullards M. C., Cabot M., Merrill A. H., Jr. (2006) Biochim. Biophys. Acta 1758, 1864–1884 [DOI] [PubMed] [Google Scholar]
- 19. Schulz A., Mousallem T., Venkataramani M., Persaud-Sawin D. A., Zucker A., Luberto C., Bielawska A., Bielawski J., Holthuis J. C., Jazwinski S. M., Kozhaya L., Dbaibo G. S., Boustany R. M. (2006) J. Biol. Chem. 281, 2784–2794 [DOI] [PubMed] [Google Scholar]
- 20. Villani M. G., Appierto V., Cavadini E., Valsecchi M., Sonnino S., Curley R. W., Formelli F. (2004) Clin. Cancer Res. 10, 6265–6275 [DOI] [PubMed] [Google Scholar]
- 21. Triola G., Fabriàs G., Llebaria A. (2001) Angew Chem. Int. Ed. Engl. 40, 1960–1962 [PubMed] [Google Scholar]
- 22. Triola G., Fabrias G., Dragusin M., Niederhausen L., Broere R., Llebaria A., van Echten-Deckert G. (2004) Mol. Pharmacol. 66, 1671–1678 [DOI] [PubMed] [Google Scholar]
- 23. Triola G., Fabriàs G., Casas J., Llebaria A. (2003) J. Org. Chem. 68, 9924–9932 [DOI] [PubMed] [Google Scholar]
- 24. Rege B., Carter K. M., Sarkar M. A., Kellogg G. E., Soine W. H. (2002) Drug Metab. Dispos. 30, 1337–1343 [DOI] [PubMed] [Google Scholar]
- 25. Tipton K. F., Fowler C. J., McCrodden J. M., Strolin Benedetti M. (1983) Biochem. J. 209, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bonanni B., Lazzeroni M., Veronesi U. (2007) Expert Rev. Anticancer Ther. 7, 423–432 [DOI] [PubMed] [Google Scholar]
- 27. Torrisi R., Decensi A., Formelli F., Camerini T., De Palo G. (2001) Drugs 61, 909–918 [DOI] [PubMed] [Google Scholar]
- 28. Chiesa F., Tradati N., Marazza M., Rossi N., Boracchi P., Mariani L., Formelli F., Giardini R., Costa A., De Palo G., et al. (1993) J. Cell. Biochem. Suppl. 17F, 255–261 [DOI] [PubMed] [Google Scholar]
- 29. Formelli F., Cavadini E., Luksch R., Garaventa A., Villani M. G., Appierto V., Persiani S. (2008) Cancer Chemother. Pharmacol. 62, 655–665 [DOI] [PubMed] [Google Scholar]
- 30. DiPietrantonio A. M., Hsieh T. C., Olson S. C., Wu J. M. (1998) Int. J. Cancer 78, 53–61 [DOI] [PubMed] [Google Scholar]
- 31. Wu J. M., DiPietrantonio A. M., Hsieh T. C. (2001) Apoptosis 6, 377–388 [DOI] [PubMed] [Google Scholar]
- 32. Villablanca J. G., Krailo M. D., Ames M. M., Reid J. M., Reaman G. H., Reynolds C. P., Reynolds P. C. (2006) J. Clin. Oncol. 24, 3423–3430 [DOI] [PubMed] [Google Scholar]
- 33. Marachelian A., Kang M. H., Hwang K., Villablanca J. G., Groshen S., Matthay K. K., Maris J., DeSantes K. B., Reynolds C. P., Maurer B. J. (2009) J. Clin. Oncol. 27, 15s (Abstr. 10009) [Google Scholar]
- 34. Bielawska A., Crane H. M., Liotta D., Obeid L. M., Hannun Y. A. (1993) J. Biol. Chem. 268, 26226–26232 [PubMed] [Google Scholar]
- 35. Signorelli P., Munoz-Olaya J. M., Gagliostro V., Casas J., Ghidoni R., Fabriàs G. (2009) Cancer Lett. 282, 238–243 [DOI] [PubMed] [Google Scholar]
- 36. Wang H., Maurer B. J., Liu Y. Y., Wang E., Allegood J. C., Kelly S., Symolon H., Liu Y., Merrill A. H., Jr., Gouazé-Andersson V., Yu J. Y., Giuliano A. E., Cabot M. C. (2008) Mol. Cancer Ther. 7, 2967–2976 [DOI] [PubMed] [Google Scholar]
- 37. Stiban J., Fistere D., Colombini M. (2006) Apoptosis 11, 773–780 [DOI] [PubMed] [Google Scholar]
- 38. Munoz-Olaya J. M., Matabosch X., Bedia C., Egido-Gabás M., Casas J., Llebaria A., Delgado A., Fabriàs G. (2008) Chem. Med. Chem. 3, 946–953 [DOI] [PubMed] [Google Scholar]
- 39. Jiang Q., Wong J., Fyrst H., Saba J. D., Ames B. N. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17825–17830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schiffmann S., Sandner J., Schmidt R., Birod K., Wobst I., Schmidt H., Angioni C., Geisslinger G., Grösch S. (2009) J. Lipid Res. 50, 32–40 [DOI] [PubMed] [Google Scholar]
- 41. Separovic D., Bielawski J., Pierce J. S., Merchant S., Tarca A. L., Ogretmen B., Korbelik M. (2009) Br. J. Cancer 100, 626–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.