Abstract
Pancreatic cancer remains a devastating malignancy with a poor prognosis and is largely resistant to current therapies. To understand the resistance of pancreatic tumors to Fas death receptor-induced apoptosis, we investigated the molecular mechanisms of Fas-activated survival signaling in pancreatic cancer cells. We found that knockdown of the Fas-associated protein with death domain (FADD), the adaptor that mediates downstream signaling upon Fas activation, rendered Fas-sensitive MiaPaCa-2 and BxPC-3 pancreatic cells resistant to Fas-induced apoptosis. By contrast, Fas activation promoted the survival of the FADD knockdown MiaPaCa-2 and BxPC-3 cells in a concentration-dependent manner. The pharmacological inhibitor of ERK, PD98059, abrogated Fas-promoted cell survival in FADD knockdown MiaPaCa-2 and BxPC-3 cells. Furthermore, increased phosphorylation of Src was demonstrated to mediate Fas-induced ERK activation and cell survival. Immunoprecipitation of Fas in the FADD knockdown cells identified the presence of increased calmodulin, Src, and phosphorylated Src in the Fas-associated protein complex upon Fas activation. Trifluoperazine, a calmodulin antagonist, inhibited Fas-induced recruitment of calmodulin, Src, and phosphorylated Src. Consistently, trifluoperazine blocked Fas-promoted cell survival. A direct interaction of calmodulin and Src and their binding site were identified with recombinant proteins. These results support an essential role of calmodulin in mediating Fas-induced FADD-independent activation of Src-ERK signaling pathways, which promote survival signaling in pancreatic cancer cells. Understanding the molecular mechanisms responsible for the resistance of pancreatic cells to apoptosis induced by Fas-death receptor signaling may provide molecular insights into designing novel therapies to treat pancreatic tumors.
Keywords: Apoptosis, Calmodulin, Death Domain, Src, Tumor, Fas, Pancreatic Tumor
Introduction
Pancreatic cancer is an extraordinarily lethal solid malignancy. The American Cancer Society estimates 43,140 new cases and 36,800 deaths from pancreatic cancer in 2010, making this cancer the fourth leading cause of cancer deaths (1). With current therapy, the 5-year survival rate of pancreatic cancer is only 4%. Therefore, understanding the pathogenesis of pancreatic cancer and developing more effective therapies for pancreatic cancer is very important.
Activation of the Fas death receptor-initiated apoptotic pathway is considered to be a promising approach to treat human pancreatic cancer (2–4). The Fas death receptor is a prototypical member of the tumor necrosis factor receptor superfamily that mediates apoptosis induced by extrinsic signals, such as Fas ligands or pharmacological reagents (5, 6). Upon activation, the Fas receptors are multimerized and form a platform for binding additional components that form the death-inducing signaling complex (DISC)2 (7, 8). The DISC consists of Fas, the adaptor protein Fas-associated death domain (FADD), caspase-8, as well as caspase-10 (9–11). Caspase-8 and -10 undergo cleavage and subsequent activation within the DISC to initiate the cascade of caspase reactions, which ultimately activate caspase-3, -6, and -7, which are responsible for apoptotic cell death.
We have previously shown that calmodulin (CaM) binds to Fas and regulates the Fas-mediated apoptosis signaling pathway. CaM is a 17-kDa calcium-binding protein that is ubiquitously expressed in all types of cells. CaM acts as a major calcium sensor and regulator through its interaction with a diverse group of cellular proteins, including enzymes such as kinases, phosphatases, and nitric-oxide synthase, as well as receptors, ion channels, G-proteins, and transcription factors in calcium-dependent and -independent manners (12, 13). CaM binds directly to Fas, and this binding is increased upon Fas stimulation. The CaM antagonist trifluoperazine (TFP) decreases CaM/Fas binding (14, 15). We have reported that TFP induces apoptosis in cholangiocarcinoma cells, Jurkat cells, and osteoclasts. However, the definitive role of CaM in regulating Fas-induced signaling pathways remains unclear.
In addition to its role as a death-inducing receptor, Fas activation has also been reported to accelerate liver regeneration in mice subjected to partial hepatectomy (16, 17) and to promote proliferation of human T lymphocytes as well as growth factor-deprived fibroblasts and maturation of dendritic cells in culture (18, 24). Recently, activation of Fas was found to enhance tumorigenesis such as colon, ovarian, renal, breast, and liver cancer (19, 20). Consistently, some pancreatic cancer cells are resistant to Fas-mediated cell death, despite their expression of the Fas death receptor (21–23).
The molecular mechanisms underlying Fas-activated survival/proliferative signals involve Fas-induced activation of nuclear factor κB (NFκB) and extracellular signal-regulated kinase (ERK) signaling pathways, which have been shown to promote cell survival and proliferation (16, 24, 25). The Fas-activated recruitment of cellular FLICE-inhibitory protein (c-FLIP) into the DISC via FADD is an important step in activating the downstream NFκB and ERK signaling pathways (26–31). FLIP is the enzymatically inactive homologue of caspase-8, which is expressed at high levels in cancer cells and proliferating cells. When recruited into the DISC, FLIP blocks the recruitment and thus activation of caspase-8 and converts the death signal into a cell survival/proliferation signal. We have previously shown that CaM can also bind to FLIP in cholangiocarcinoma cells, which is important for the function of FLIP in regulating Fas-mediated signaling pathways (35). Additional mechanisms have been reported to regulate Fas-activated survival and proliferation independent of recruitment of FLIP into the DISC. In hepatocytes and thymocytes, Fas induces cell survival/proliferation independent of the adaptor protein FADD (17, 32).
In this study, we investigated the molecular mechanisms of Fas-activated survival signals in pancreatic cancer cells. We demonstrated that Fas activated pancreatic cancer cell survival via ERK activation in a FADD-independent manner. Further characterization demonstrated Fas-activated recruitment of Src into the DISC, and its activation was responsible for the activation of ERK and cell survival. A direct interaction of CaM and Src was identified, which mediated Src activation in the DISC. These results demonstrate Fas activates the Src/ERK signaling pathway and promotes cancer cell survival in a calmodulin-dependent manner.
EXPERIMENTAL PROCEDURES
Cell Culture, Antibodies, and Reagents
The human pancreatic cancer cell lines MiaPaCa-2 and BxPC-3 were from the American Type Culture Collection (ATCC, Manassas, VA). Cells were grown in RPMI 1640 medium (Invitrogen) supplemented with penicillin (5 units/ml), streptomycin (5 μg/ml), and 10% heat-inactivated fetal bovine serum. Fas agonist antibody CH-11 and antibodies to FADD and CaM were purchased from Millipore (Billerica, MA). Human GST-Src recombinant protein and antibodies to c-FLIP and caspase-3 were from Enzo Life (Plymouth Meeting, PA). Antibody to caspase-8 was purchased from BD Biosciences. Antibodies to Fas, phospho-IκB, IκB, and GAPDH were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies to phospho-ERK, ERK, phospho-Src, and Src were purchased from Cell Signaling Technology (Danvers, MA). Protein G-agarose was from Invitrogen. Anti-mouse IgM-conjugated agarose, trifluoperazine, PD98059, and PP2 were obtained from Sigma.
Knockdown of FADD with Lentivirus-delivered shRNA
Lentiviral constructs expressing a 21-nucleotide FADD short hairpin RNA (shRNA) targeting the human FADD gene (GenBankTM accession number NM_003824.3) were purchased from Open Biosystems (Huntsville, AL). The construct was packed into lentivirus-like particles pseudotyped with the vesicular stomatitis virus glycoprotein as we described previously (33). Transduction was performed by incubating cancer cells with recombinant lentivirus, and stably transduced cells were selected with puromycin (2 μg/ml).
Cell Survival Assay
Cells were grown in 96-well plates at 4 × 103 cells per well overnight and treated for 48 h. After incubation, 20 μl of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium solution (Promega, Madison, WI) were added into each well. The plate was incubated for 2 h at 37 °C. The absorbance at 490 nm was recorded with a microplate reader (BioTek, Winooski, VT).
Assessment of Apoptosis
Cells were exposed to CH-11 for the time indicated in the figure legends. Apoptosis was determined by annexin V-FITC and propidium iodide staining (BD Biosciences) and analyzed by flow cytometry.
Western Blot Analysis
Protein extracts from cells were prepared as described previously (34). Concentrations of protein were determined with a BCA protein assay kit (Thermo Scientific, Waltham, MA). Proteins were separated by SDS-PAGE and transferred to Immobilon P membranes (Millipore) as described previously (34). Membranes were blocked in 5% nonfat milk and incubated with primary antibodies overnight at 4 °C. Horseradish peroxidase-conjugated secondary antibodies in the blocking buffer were incubated for 1 h at room temperature. Signals were detected using Immobilon Western chemiluminescent horseradish peroxidase substrate detection kit (Millipore).
Immunoprecipitation
Five hundred micrograms of extracted proteins were incubated with 1 μg of anti-CaM or anti-Src antibody for 2 h. Immune complexes were recovered from the supernatant by incubation with 60 μl of 1:1 slurry of protein G-agarose beads overnight at 4 °C. Beads were then washed with lysis buffer, and 20 ml of 2× Laemmli sample buffer was added to the beads followed by heating at 95 °C for 5 min and chilling on ice. After brief centrifugation, proteins in the supernatant were analyzed by Western blot with specific antibodies.
DISC Analysis
Immunoprecipitation for DISC analysis was performed by a modification of a method described previously (14). Cells were trypsinized, and 5 × 107 suspended cells were incubated with 1 μg/ml Fas activating antibody (CH-11) for various times at 37 °C and then washed with PBS and lysed in lysis buffer (20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% Triton X-100, 10% glycerol) for 30 min on ice. In control cells, Fas activating antibody (CH-11) was added to cell lysates at a final concentration of 1 μg/ml to immunoprecipitate nonstimulated Fas receptors. After centrifugation at 15,000 × g for 15 min at 4 °C, the supernatant was immunoprecipitated with 40 μl of goat anti-mouse IgM-agarose (Sigma) overnight at 4 °C and analyzed by Western blotting.
Expression and Purification of Fusion Proteins in Escherichia coli
The human Src cDNA from pDONR223-Src (Addgene, Cambridge, MA) was cloned into pcDNA3.1 (Invitrogen) or pCMV-Tag2A (Stratagene, La Jolla, CA) and confirmed by sequencing. The QuikChange site-directed mutagenesis kit (Stratagene) was used to make Src mutations. The primers for making mutation are as follows: SRC mutation forward, GGGCCTCAACGTGGCGGCTGCAGCGGCTGCCGCAGCGGCTGCCGGCGGCTTCTACATCACCTCC, and SRC mutation reverse, GGTGATGTAGAAGCCGCCGGCAGCGCTGCGGCAGCCGCTGCAGCCGCCACGTTGAGGCCCTTGGCGTTG. Expression and purification of GST proteins were performed as described previously (15). GST proteins were expressed in E. coli, strain DH-5α, which were transformed with the empty pGEX-5X-3 (Amersham Biosciences) construct containing the GST gene. After inducing with isopropyl β-d-thiogalactoside at 30 °C, the bacteria were lysed in GST lysis buffer (PBS, 50 mm EDTA, 10% glycerol, 0.5% aprotinin, 1 mm dithiothreitol, and 1 mm PMSF) with lysozyme and purified with a GST expression and purification kit (Amersham Biosciences). For expression and purification of His-SUMO fusion protein, the cDNA fragment corresponding to human Fas cytoplasmic domain (amino acids 191–335) was amplified from a human umbilical vein endothelial cell library by PCR, or cDNA of Src from pDONR223-Src (Addgene, Cambridge, MA) was cloned into a pET28A-His-SUMO construct (provided by Dr. Jinbiao Ma, University of Alabama at Birmingham) and confirmed by sequencing. His fusion proteins were expressed in E. coli, strain BL-21(DE3), by inducing with isopropyl β-d-thiogalactoside at 30 °C. The bacteria were lysed in lysis buffer with lysozyme and purified with His expression and purification kit (Qiagen, Valencia, CA).
Protein Pulldown
Protein pulldown with CaM-Sepharose 4B (GE Healthcare) or Control Sepharose CL-4B beads (Sigma) was performed as described previously (35). Briefly, proteins were added in binding buffer (20 mm Tris, 150 mm NaCl, EDTA-free protease inhibitor mixture) and incubated with 60 μl (1:1) slurry of beads overnight at 4 °C. The beads were washed five times with lysis buffer, and proteins were eluted in 2× SDS buffer.
Transient Transfection
Cells at 80% confluence in 6-well plates were transfected with 2 g of plasmids. Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. 24 h post-transfection, the medium was changed, and cells were lysed or treated with CH-11 as indicated.
Statistical Analysis
Results are expressed as means ± S.D. Differences between two groups were identified with Student's t test. Significance was defined as p < 0.05.
RESULTS
FADD Knockdown Attenuates Fas-induced Apoptosis in Pancreatic Cancer Cells
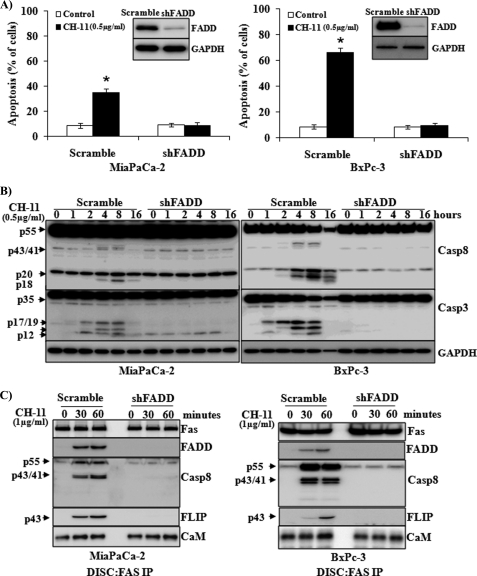
We analyzed the expression of Fas receptor in several pancreatic cells and identified that the expression of Fas is higher in pancreatic cancer cell lines MiaPaCa-2 and BxPC-3 compared with that in ASPC-1 and PANC-1 cells. Consistently, low expression of Fas by ASPC-1 and PANC-1 cells renders them resistant to Fas-induced apoptosis (data not shown). Therefore, to further understand Fas-activated signaling pathways in pancreatic cells, we utilized the pancreatic cancer cells expressing higher levels of Fas, MiaPaCa-2 and BxPC-3 cells. The expression of the Fas receptor was similar in MiaPaCa-2 and BxPC-3 cells. Upon stimulation, the death receptor Fas recruits adaptor protein FADD, which binds to caspase-8 or FLIP to activate apoptotic or survival signaling pathways. In addition, Fas has been shown to induce cell survival/proliferation independent of FADD (17, 32). To determine whether FADD is required for Fas-activated apoptotic or proliferative signals in pancreatic cancer cells, we generated MiaPaCa-2 and BxPC-3 cells with FADD knockdown using lentivirus-delivered shRNA that specifically targets FADD. Western blot analysis confirmed the knockdown of FADD in these cells (Fig. 1A, insets). FADD knockdown in MiaPaCa-2 and BxPC-3 cells dramatically decreased Fas-induced apoptosis, compared with that in control cells (Fig. 1A). Consistently, Fas-induced activation of caspase-8 and caspase-3 was inhibited in the FADD knockdown cells compared with those in control cells (Fig. 1B). Fas-activated recruitment of FADD and caspase-8 into the DISC was demonstrated in the control cells after exposure to CH-11 for 30 and 60 min (Fig. 1C). In addition, increased recruitment of FLIP, the enzymatically inactive homologue of caspase-8, was also demonstrated in response to CH-11 treatment. By contrast, recruitment of caspase-8 and FLIP into Fas-activated DISC was blocked in FADD knockdown cells (Fig. 1C). We have previously demonstrated the recruitment of CaM into the Fas-induced DISC in cholangiocarcinoma cells (14). Increased interaction of CaM with Fas was also demonstrated in MiaPaCa-2 and BxPC-3 pancreatic cells upon Fas stimulation (Fig. 1C). FADD knockdown did not abrogate CaM binding to Fas under basal conditions, which was increased upon CH-11 stimulation (Fig. 1C).
FIGURE 1.
FADD knockdown attenuates Fas-induced apoptosis in pancreatic cancer cells. MiaPaCa-2 and BxPC-3 cells with FADD knockdown were generated (see “Experimental Procedures”). A lentiviral vector carrying a scrambled shRNA was used as control. A, stably infected cells were exposed to Fas agonistic antibody CH-11 (0.5 μg/ml) or control condition for 24 h, and apoptosis was analyzed by annexin V/propidium iodide staining. The percentage of apoptotic cells in MiaPaCa-2 (left panel) or BxPC-3 (right panel) with scramble or FADD shRNA is shown. The insets shown are Western blot analyses of the expression of FADD in these cells. The expression of GAPDH was used as a loading control. The results represent means ± S.D. from three independent experiments. B, cells were exposed to CH-11 for the indicated times, and Western blot analysis was performed to determine the expression of caspase (Casp)-8 and caspase-3. The expression of GAPDH was used as a loading control. Representative blots from three independent experiments are shown. C, cells were exposed to CH-11 for 0, 30, and 60 min. Fas-induced DISC complex was immunoprecipitated. The recruitment of FADD, caspase-8, c-FLIPL, and CaM into DISC was analyzed by Western blot. Representative blots from three independent experiments are shown. Statistical significance is as follows: *, p ≤ 0.05 compared with control group.
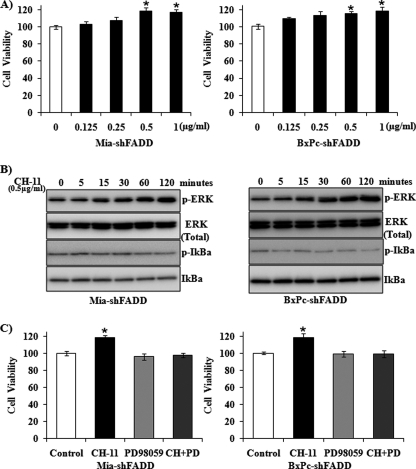
FADD Knockdown Renders Increased Cell Survival via ERK Activation in Response to Fas Stimulation
To determine whether FADD is required for Fas-induced survival signaling pathways, we determined the effect of Fas stimulation on the survival of FADD knockdown cells. The Fas agonist antibody CH-11 was found to increase accumulation of FADD knockdown cells in a concentration-dependent manner (Fig. 2A). To identify the downstream signaling pathways that mediate Fas-induced survival signals, we determined the activation of ERK and NFκB, which have been shown previously to be activated by Fas to promote cell proliferation and survival (16, 18, 25). Activation of ERK, but not NFκB, was demonstrated in FADD knockdown cells in response to Fas stimulation (Fig. 2B). Furthermore, the pharmacological inhibitor of ERK, PD98059, abrogated the CH-11-promoted cell survival in both MiaPaCa-2 and BxPC-3 cells with FADD knockdown, whereas PD98059 alone did not affect cell viability (Fig. 2C).
FIGURE 2.
Fas promotes survival of FADD knockdown cells through ERK activation. A, MiaPaCa-2 and BxPC-3 cells with FADD knockdown were exposed to increasing concentrations of CH-11 for 48 h. The accumulation of cells in each condition, determined by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium assay, was compared with that in control condition (1st column, defined as 100%). Results shown are means ± S.D. from three independent experiments. B, MiaPaCa-2 and BxPC-3 cells with FADD knockdown were treated with CH-11 for the indicated times. The expression of phospho-ERK, ERK, phospho-IκB, and IκB was determined by Western blot analysis. Representative blots from three independent experiments are shown. C, MiaPaCa-2 and BxPC-3 cells with FADD knockdown were treated with CH-11 (0.5 μg/ml), with or without PD98059 (an ERK inhibitor, 5 μm), for 48 h. The accumulation of cells in each condition was compared with that in control condition (1st column, defined as 100%). Results shown are from three independent experiments. Statistical significance is as follows: *, p ≤ 0.05 compared with control group.
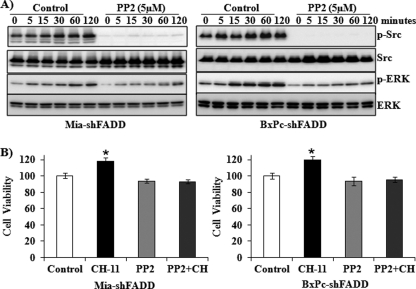
Fas-activated Src Phosphorylation Regulates ERK Activation in FADD Knockdown Cells
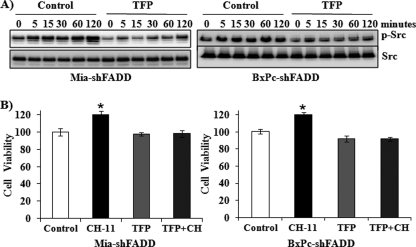
To investigate the molecular mechanisms of ERK activation upon Fas stimulation, we determined the effect of CH-11 on the activation of Src kinase, which has been demonstrated to regulate Fas death receptor signaling in colon cancer and glioblastoma tumorigenesis and metastasis experiments (36, 37). Increased activation of Src, as demonstrated by its phosphorylation, was demonstrated in FADD knockdown cells in response to CH-11 in a time-dependent manner. The Fas-induced activation of Src activation was blocked by the Src inhibitor, PP2, which also decreased the Fas-induced activation of ERK (Fig. 3A). Furthermore, the Src inhibitor PP2 abrogated CH-11-promoted cell survival of the FADD knockdown cells (Fig. 3B).
FIGURE 3.
Fas-induced Src activation regulates Fas-activated ERK and cell survival. A, MiaPaCa-2 and BxPC-3 cells with FADD knockdown were pretreated with or without 5 μm PP2, a Src inhibitor, for 30 min and subsequently exposed to CH-11 (0.5 μg/ml) for 0, 5, 15, 30, 60, and 120 min. Western blot analysis was performed to determine the expression of phospho-Src, Src, phospho-ERK, and ERK. Representative blots from three independent experiments are shown. B, MiaPaCa-2 and BxPC-3 cells with FADD knockdown were treated with CH-11 (0.5 μg/ml), with or without PP2, for 48 h. The accumulation of cells in each condition was compared with that in control condition (1st column, defined as 100%). Results shown are from three independent experiments. Statistical significance is as follows: *, p ≤ 0.05 compared with control group.
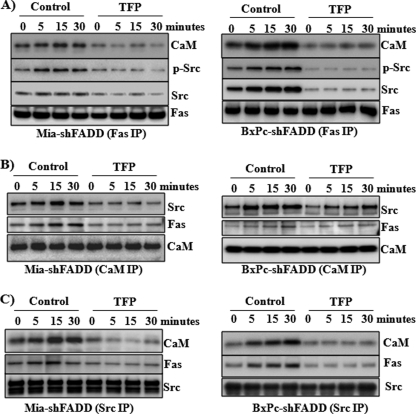
Src Association with Fas Is CaM-dependent
To understand whether Fas-induced activation of Src was through a direct interaction of Fas and Src, we analyzed the Fas-recruited protein complex upon Fas activation of the FADD knockdown cells. Similar to that seen in control pancreatic cells, increased recruitment of CaM was also found in the Fas-activated protein complex in the FADD knockdown cells (Fig. 4A). The presence of Src and phosphorylated Src was increased in the Fas-recruited protein complex in the FADD knockdown cells exposed to CH-11. To determine whether the binding of CaM to Fas was required for the presence of Src in the Fas-recruited protein complex, we treated the cells with a CaM antagonist, TFP. TFP inhibited the Fas-induced recruitment of CaM as well as Src and phosphorylated Src into the Fas-activated DISC (Fig. 4A). Further analysis by immunoprecipitation with the use of antibodies for CaM (Fig. 4B) or Src (Fig. 4C) confirmed the Fas-induced increased interaction among Fas, CaM, and Src, which was inhibited by the CaM antagonist TFP.
FIGURE 4.
TFP decreases the interaction of Fas, Src, and CaM. MiaPaCa-2 and BxPC-3 cells with FADD knockdown were pretreated for 30 min with a CaM inhibitor, TFP (10 μm), and sequentially exposed to CH-11 (1 μg/ml) for 0, 5, 15, and 30 min. Immunoprecipitation (IP) was performed with antibodies against Fas (A), Src (B), and CaM (C). Western blot analysis was performed to analyze the presence of Fas, Src, and CaM in the immunoprecipitated protein complexes. Representative blots from three independent experiments are shown.
CaM Mediates Fas-induced Src Activation and Cell Survival
The role of CaM in Fas-induced Src activation and cell survival was further investigated. Fas activation induced a time-dependent activation of Src in the FADD knockdown cells, which was seen at 5 min and sustained at 120 min after Fas stimulation (Fig. 5A). TFP, however, inhibited Fas-induced activation of Src (Fig. 5A). Consistently, TFP was found to block Fas-promoted cell survival in the FADD knockdown MiaPaCa-2 and BxPC-3 cells, whereas TFP alone did not affect cell viability (Fig. 5B).
FIGURE 5.
TFP blocks Fas-induced Src activation and cell survival. A, MiaPaCa-2 and BxPC-3 cells with FADD knockdown were pretreated with TFP (10 μm) or control condition for 30 min and susequently exposed to CH-11 (0.5 μg/ml) for 0, 5, 15, 30, 60, and 120 min. Western blot analysis was performed to determine the expression of phospho-Src and Src. Representative blots from three independent experiments are shown. B, MiaPaCa-2 and BxPC-3 cells with FADD knockdown were treated with CH-11 (0.5 μg/ml), with or without TFP, for 48 h. The accumulation of cells in each condition was compared with that in control condition (1st column, defined as 100%). Results are from three independent experiments. Statistical significance is as follows: *, p ≤ 0.05 compared with control group.
Direct Interaction of CaM and Src
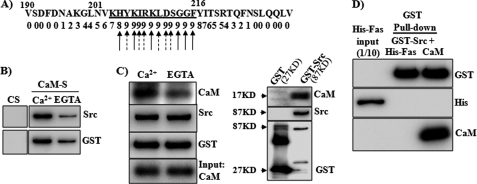
To further characterize the association of CaM with Src in the Fas-recruited complex, we determined whether Src directly interacts with Fas or CaM. Analysis of the Src protein sequence identified a potential CaM-binding site on the Src protein between amino acids 204 and 214. This sequence fits the basic 1-5-10 CaM binding motif (RK)(RK)(RK)(FILVW)XXX(FILV)XXXX(FILW) (Fig. 6A). The binding of CaM to Src was characterized using recombinant Src and CaM proteins. As shown in Fig. 6B, GST-Src protein strongly bound to CaM-Sepharose but not to control Sepharose in the presence of Ca2+, whereas the calcium chelator EGTA largely decreased the binding. With the use of recombinant CaM, we further confirmed the direct binding of CaM to GST-Src, which was decreased by EGTA (Fig. 6C). GST protein alone did not interact with CaM (data not shown), confirming the direct binding of CaM to Src. Direct binding of Src and Fas was not detected with the use of recombinant proteins of Fas and Src (Fig. 6D).
FIGURE 6.
Direct interaction of Src and CaM. A, prediction of a CaM-binding site in Src amino acids 190–230. The amino acid sequence of Src was analyzed to predict CaM-binding sites, using a CaM target data base. The numbers below the sequences indicate normalized scores (0–9) based on the evaluation criteria for CaM-binding sites. The higher the number, the more likely it is to bind CaM. The predicted CaM-binding motif is underlined. Arrows with solid lines indicate hydrophobic residues, and arrows with dotted lines indicate positively charged residues. B, recombinant Src protein (GST-Src) (100 ng) was incubated with CaM-Sepharose (CaM-S) or control Sepharose (CS) overnight in 0.5 ml of binding buffer supplemented with Ca2+ (2.5 mm) or EGTA (2.5 mm). Western blot analysis was performed to determine the proteins pulled down by the beads. C, left panel, GST-Src (100 ng) was incubated with purified CaM (25 ng) (1:1 in molar ratio) in binding buffer with Ca2+ (2.5 mm) or EGTA (2.5 mm); right panel, GST or GST-Src was incubated with purified CaM protein for 2 h and then followed by incubation with glutathione-Sepharose (GST beads) overnight at 4 °C. Proteins pulled down by GST beads were analyzed by Western blot with antibodies for CaM, Src, or GST as indicated. D, GST-Src (100 ng) was incubated with purified His-Fas or CaM protein for 2 h and then followed by incubation with glutathione-Sepharose (GST beads) overnight at 4 °C. Proteins pulled down by GST beads were analyzed by Western blot with antibodies for His, CaM, or GST. Representative blots from three independent experiments are shown.
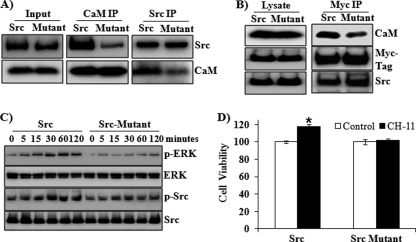
The direct binding of CaM and Src was further characterized using a mutant Src protein with mutations in the predicted amino acids 204–214 region (Fig. 7). Compared with wild-type Src protein, the mutant Src protein reduced binding to CaM-Sepharose beads (Fig. 7A). To assess the effects of the mutation on the binding of CaM and Src in vivo and thus their roles in regulating Fas-induced survival signals, the mutant or wild-type Src protein were overexpressed in the FADD knockdown BxPC-3 cells. Consistently, reduced CaM/Src binding was found in cells overexpressing the mutant Src compared with those with wild-type Src (Fig. 7B). Furthermore, overexpression of the mutant Src resulted in decreased activation of Src and ERK in response to Fas stimulation, compared with the wild-type Src (Fig. 7C). Fas-induced proliferation was blocked in the cells overexpressing the mutant Src protein (Fig. 7D).
FIGURE 7.
Effect of mutations of Src in the predicted CaM-binding site on CaM binding and ERK activation. A, wild-type Src and Src protein with mutations in the CaM binding domain 203–212 (KHYKIRKLDS mutated to alanine) was produced as a His-SUMO fusion protein. Recombinant Src protein (100 ng) was incubated with purified CaM (25 ng) protein (1:1 in molar ratio) in 500 μl of binding buffer for 2 h at 4 °C. Immunoprecipitation (IP) was performed with antibodies against Src (left panel) or CaM (middle panel). Protein complexes pulled down by immunoprecipitation were analyzed by Western blot with antibodies for Src and CaM. Western blot was also performed to show Src and CaM protein levels in 1/30 of the lysate used in protein pull down (left panel, indicated as input). Representative blots from three independent experiments are shown. B, BxPC-3 cells with FADD knockdown were transfected with Myc-Src or Myc-Src mutant plasmids for 24 h. Immunoprecipitation was performed with antibodies against Myc tag. Western blot analysis was performed to analyze Myc tag, Src, and CaM in the immunoprecipitated protein complexes. C, BxPC-3 cells with FADD knockdown were transfected with Src or Src mutant plasmids for 24 h. Cells then were exposed to CH-11 (0.5 μg/ml) for 0, 5, 15, 30, 60, and 120 min. Western blot analysis was performed to determine the expression of phospho-Src, Src, phospho-ERK, and ERK. Representative blots from three independent experiments are shown. D, BxPC-3 cells with FADD knockdown were transfected with Src or Src mutant plasmids for 24 h. Cells were treated with or without CH-11 (0.5 μg/ml) for 48 h. The accumulation of cells in each condition was compared with that in control condition (1st column, defined as 100%). Results shown are from three independent experiments. Statistical significance is as follows: *, p ≤ 0.05 compared with control group.
DISCUSSION
The Fas/FasL system is generally thought of primarily as an inducer of apoptosis. Consistently, we found that Fas activation induced apoptosis in MiaPaCa-2 and BxPC-3 pancreatic cancer cells. MiaPaCa-2 cells were less sensitive to CH-11-induced apoptosis compared with BxPC-3 cells, despite similar levels of expression of the Fas receptor on these two cell types. This may be attributed to MiaPaCa-2 cells expressing relatively higher levels of c-FLIP, an enzymatically inactive homologue of caspase-8 that inhibits Fas-induced apoptosis.
In addition to Fas-induced apoptotic signaling pathways, compelling evidence demonstrates that Fas activates nonapoptotic signaling pathways that induce cellular activation, proliferation, differentiation, and migration (24, 38). Fas-enhanced proliferation has been shown to be mediated by a FADD-dependent signaling pathway in T cell receptor-stimulated T cells and thymocytes (30, 39). In mice with partial hepatectomy, Fas stimulation accelerates liver regeneration via FADD-independent signaling pathways (17). In this study, we characterized the function of FADD in Fas-activated signaling pathways in MiaPaCa-2 and BxPC-3 pancreatic cells with FADD knockdown by lentivirus-mediated shRNA for FADD.
Knockdown of FADD blocked Fas-induced activation of caspase-8 and caspase-3, thus rendering them resistant to Fas-induced apoptosis. In the FADD knockdown of MiaPaCa-2 and BxPC-3 cells, Fas stimulation failed to recruit caspase-8 and c-FLIP into the Fas-induced signaling complex, which excludes the role of FLIP in mediating Fas-activated survival signaling pathways in these cells. Consistent with this, there was no Fas-induced activation of NF-κB, which has been demonstrated to be activated by Fas via FLIP-mediated signaling (26, 28, 30, 31, 40). Therefore, the Fas-activated survival signaling in the pancreatic cells with FADD knockdown was independent of FLIP-mediated NFκB activation.
We found that Fas induced the activation of ERK that regulated survival of the FADD knockdown MiaPaCa-2 and BxPC-3 pancreatic cells. Fas-stimulated ERK activation has been reported to regulate cell proliferation and survival in many cell lines, including lymphocytes, leukemia, glioma, and primary neuron cells (25, 29, 41–43). We confirmed that Fas activation induced activation of ERK in wild-type MiaPaCa-2 and BxPc-3 cells (data not shown). These results demonstrate that Fas activated ERK by a FADD-independent pathway and are consistent with previous observations in Fas-induced FADD-independent neuron cell proliferation (25), as well as Fas-induced liver regeneration in mice with mutated Fas that lacked the binding site for FADD (17).
Activation of ERK by Fas stimulation was found to be rapid and maximized at 1 h. This observation is consistent with many previous reports of a biphasic activation of ERK1/2, with a rapid and strong burst of ERK activity peaking at 5–10 min followed by a second wave of lower but sustained activity up to 6 h (44–46). Activated ERK1 and ERK2 translocate to the nucleus where they activate multiple transcription factors ultimately resulting in protein synthesis that affects cell viability (47). An ERK inhibitor, PD98059, was found to inhibit Fas-induced cell proliferation, demonstrating an important role of ERK activation in mediating Fas-induced cell proliferation.
In search of a Fas-activated kinase that would be responsible for Fas-induced phosphorylation/activation of ERK, we demonstrated activation of Src upon Fas stimulation. The Src family of kinases has a critical role in cell proliferation during tumor development. The Src family of kinases can affect cell proliferation via the Ras/ERK/MAPK pathway (48). Several reports have demonstrated that Fas activation induces phosphorylation of Src family kinases (49–51). In addition, Fas stimulation induces the activation of the Src family member, Yes, by recruiting Yes into the Fas-activated signaling complex (36, 52). At the cellular level, Fas-induced activation of ERK or Src was found to increase cancer cell motility and invasion (36, 53). In this study, we demonstrated that Fas stimulation induced the recruitment and activation of Src into the Fas-activated signal complex, which was responsible for Fas-induced ERK activation and promotion of cell survival.
Our studies also demonstrate for the first time that Src was associated with Fas by binding to CaM. The observation that Src phosphorylates CaM at tyrosine 99 implies an interaction between Src and CaM (54). We have previously reported that CaM directly binds to Fas, which is increased upon Fas stimulation and inhibited by the CaM antagonist TFP (14). Blocking the CaM/Fas binding by TFP also inhibited Fas-activated recruitment and activation of Src, implying that Src association with Fas is mediated by CaM. The importance of CaM in Fas-induced Src activation is further underlined by the observation that the CaM antagonist, TFP, decreased Fas-induced Src phosphorylation and Fas-induced cell survival. These findings strongly imply that CaM may work as a docking protein, which mediates Fas-induced downstream signaling pathways that promote cell proliferation and survival.
More direct evidence of the interaction between CaM and Src was determined with the use of purified recombinant CaM and Src proteins (Figs. 6 and 7). We found that the binding of CaM with Src was partially calcium-dependent, as the calcium chelator EGTA decreased but did not abolish the interaction. CaM interacts with a large number of proteins in a calcium-dependent or -independent manner. For instance, the binding of CaM to neurogranin protein does not require Ca2+ (55), although binding of CaM to calcineurin is Ca2+-dependent (56). The binding of CaM to myosin, however, requires Ca2+ for maximal binding (57), because the NH2-terminal motif binds to CaM in the absence of Ca2+, whereas the COOH-terminal motif binds to CaM in the presence of Ca2+. Based on the sequence of the CaM-binding proteins, three CaM recognition motifs have been classified (58). These include a modified variant of the IQ motif as a consensus for Ca2+-independent binding and two related motifs for Ca2+-dependent binding, termed 1-8-14 ((FILVW)XXXXXX(FAILVW)XXXXX(FILVW)) and 1-5-10 (XXX(FILVW)XXX(FILV)XXXX(FILVW)) according to the conserved hydrophobic residues within these motifs. The CaM-binding motif on the Src protein, KHYKIRKLDSGGF, fits the basic 1-5-10 motif (RK)(RK)(RK)(FILVW)XXX(FILV)XXXX(FILW). We have previously shown that Fas induces an increased intracellular Ca2+ within 5 min of Fas stimulation (14), which may contribute to the rapid increases of CaM/Src binding upon Fas stimulation (Fig. 4).
The CaM binding domain of Src between amino acids 204 and 214 overlaps the Src homology 2 (SH2) domain. Src contains a phosphorylated tyrosine at amino acid 530 that interacts with its own SH2 domain in the inactive conformation, which positions the SH3 domain to interact with the proline-rich linker domain and keeps Src in a tightly bound inactive state. Upon stimulation, the tyrosine 530 becomes dephosphorylated, which destabilizes the intramolecular interactions and ultimately results in autophosphorylation of tyrosine 416 and thus increases its activity (59). Accordingly, we found that Fas stimulation induced phosphorylation of Src at tyrosine 416. Inhibition of CaM binding to the Src SH2 domain by the Src mutant decreased Fas-activated Src phosphorylation, which inhibited Fas-induced activation of ERK and cell survival. Therefore, these results support the role of CaM binding to the Src SH2 domain in mediating Fas-activated Src, ERK, and survival signals.
In summary, we have demonstrated that Fas promoted cell survival signaling in FADD-independent pancreatic cancer cells via activation of Src/ERK. Fas stimulation induces the recruitment of Src into the Fas-induced signaling complex, by direct binding of the Src SH2 domain to CaM, which induces Src phosphorylation. Activated Src mediates Fas-induced activation of ERK, thus promoting cell survival. These studies provide novel insights into the molecular mechanism underlying Fas-regulated cell survival signaling in pancreatic cells. Fundamental understanding of the interaction between CaM, Src, and Fas may provide new therapeutic modalities with CaM antagonist targeting the CaM/Src interaction.
This work was supported in part by start-up funds from the Department of Pathology, University of Alabama at Birmingham (to Y. C.), and a Veterans Affairs merit review award (to J. M. M.).
- DISC
- death-inducing signaling complex
- TFP
- trifluoperazine
- SH
- Src homology.
REFERENCES
- 1. Maitra A., Hruban R. H. (2008) Annu. Rev. Pathol. 3, 157–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Park M. A., Mitchell C., Zhang G., Yacoub A., Allegood J., Häussinger D., Reinehr R., Larner A., Spiegel S., Fisher P. B., Voelkel-Johnson C., Ogretmen B., Grant S., Dent P. (2010) Cancer Res. 70, 6313–6324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walker T., Mitchell C., Park M. A., Yacoub A., Graf M., Rahmani M., Houghton P. J., Voelkel-Johnson C., Grant S., Dent P. (2009) Mol. Pharmacol. 76, 342–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Park M. A., Zhang G., Mitchell C., Rahmani M., Hamed H., Hagan M. P., Yacoub A., Curiel D. T., Fisher P. B., Grant S., Dent P. (2008) Mol. Cancer Ther. 7, 2633–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Krammer P. H. (2000) Nature 407, 789–795 [DOI] [PubMed] [Google Scholar]
- 6. Wallach D., Kovalenko A. V., Varfolomeev E. E., Boldin M. P. (1998) Curr. Opin. Immunol. 10, 279–288 [DOI] [PubMed] [Google Scholar]
- 7. Siegel R. M., Frederiksen J. K., Zacharias D. A., Chan F. K., Johnson M., Lynch D., Tsien R. Y., Lenardo M. J. (2000) Science 288, 2354–2357 [DOI] [PubMed] [Google Scholar]
- 8. Schneider P., Holler N., Bodmer J. L., Hahne M., Frei K., Fontana A., Tschopp J. (1998) J. Exp. Med. 187, 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Strasser A., Jost P. J., Nagata S. (2009) Immunity 30, 180–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nagata S. (1997) Cell 88, 355–365 [DOI] [PubMed] [Google Scholar]
- 11. Kischkel F. C., Hellbardt S., Behrmann I., Germer M., Pawlita M., Krammer P. H., Peter M. E. (1995) EMBO J. 14, 5579–5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clapham D. E. (2007) Cell 131, 1047–1058 [DOI] [PubMed] [Google Scholar]
- 13. Kahl C. R., Means A. R. (2003) Endocr. Rev. 24, 719–736 [DOI] [PubMed] [Google Scholar]
- 14. Chen Y., Pawar P., Pan G., Ma L., Liu H., McDonald J. M. (2008) J. Cell. Biochem. 103, 788–799 [DOI] [PubMed] [Google Scholar]
- 15. Ahn E. Y., Lim S. T., Cook W. J., McDonald J. M. (2004) J. Biol. Chem. 279, 5661–5666 [DOI] [PubMed] [Google Scholar]
- 16. Reinehr R., Sommerfeld A., Häussinger D. (2008) Gastroenterology 134, 1494–1506 [DOI] [PubMed] [Google Scholar]
- 17. Desbarats J., Newell M. K. (2000) Nat. Med. 6, 920–923 [DOI] [PubMed] [Google Scholar]
- 18. Suzuki I., Martin S., Boursalian T. E., Beers C., Fink P. J. (2000) J. Immunol. 165, 5537–5543 [DOI] [PubMed] [Google Scholar]
- 19. Ametller E., García-Recio S., Costamagna D., Mayordomo C., Fernández-Nogueira P., Carbó N., Pastor-Arroyo E. M., Gascón P., Almendro V. (2010) Mol. Cancer 9, 161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen L., Park S. M., Tumanov A. V., Hau A., Sawada K., Feig C., Turner J. R., Fu Y. X., Romero I. L., Lengyel E., Peter M. E. (2010) Nature 465, 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Christgen M., Schniewind B., Jueschke A., Ungefroren H., Kalthoff H. (2005) Cancer Lett. 227, 193–200 [DOI] [PubMed] [Google Scholar]
- 22. Kornmann M., Ishiwata T., Maruyama H., Beger H. G., Korc M. (2000) Pancreas 20, 123–128 [DOI] [PubMed] [Google Scholar]
- 23. Ungefroren H., Voss M., Jansen M., Roeder C., Henne-Bruns D., Kremer B., Kalthoff H. (1998) Cancer Res. 58, 1741–1749 [PubMed] [Google Scholar]
- 24. Peter M. E., Budd R. C., Desbarats J., Hedrick S. M., Hueber A. O., Newell M. K., Owen L. B., Pope R. M., Tschopp J., Wajant H., Wallach D., Wiltrout R. H., Zörnig M., Lynch D. H. (2007) Cell 129, 447–450 [DOI] [PubMed] [Google Scholar]
- 25. Desbarats J., Birge R. B., Mimouni-Rongy M., Weinstein D. E., Palerme J. S., Newell M. K. (2003) Nat. Cell Biol. 5, 118–125 [DOI] [PubMed] [Google Scholar]
- 26. Imtiyaz H. Z., Rosenberg S., Zhang Y., Rahman Z. S., Hou Y. J., Manser T., Zhang J. (2006) J. Immunol. 176, 6852–6861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dohrman A., Kataoka T., Cuenin S., Russell J. Q., Tschopp J., Budd R. C. (2005) J. Immunol. 174, 5270–5278 [DOI] [PubMed] [Google Scholar]
- 28. Kataoka T., Tschopp J. (2004) Mol. Cell. Biol. 24, 2627–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kataoka T., Budd R. C., Holler N., Thome M., Martinon F., Irmler M., Burns K., Hahne M., Kennedy N., Kovacsovics M., Tschopp J. (2000) Curr. Biol. 10, 640–648 [DOI] [PubMed] [Google Scholar]
- 30. Kennedy N. J., Kataoka T., Tschopp J., Budd R. C. (1999) J. Exp. Med. 190, 1891–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang J., Cado D., Chen A., Kabra N. H., Winoto A. (1998) Nature 392, 296–300 [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y., Rosenberg S., Wang H., Imtiyaz H. Z., Hou Y. J., Zhang J. (2005) J. Immunol. 175, 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Byon C. H., Javed A., Dai Q., Kappes J. C., Clemens T. L., Darley-Usmar V. M., McDonald J. M., Chen Y. (2008) J. Biol. Chem. 283, 15319–15327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pawar P., Ma L., Byon C. H., Liu H., Ahn E. Y., Jhala N., Arnoletti J. P., McDonald J. M., Chen Y. (2009) Clin. Cancer Res. 15, 1288–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pawar P. S., Micoli K. J., Ding H., Cook W. J., Kappes J. C., Chen Y., McDonald J. M. (2008) Biochem. J. 412, 459–468 [DOI] [PubMed] [Google Scholar]
- 36. Kleber S., Sancho-Martinez I., Wiestler B., Beisel A., Gieffers C., Hill O., Thiemann M., Mueller W., Sykora J., Kuhn A., Schreglmann N., Letellier E., Zuliani C., Klussmann S., Teodorczyk M., Gröne H. J., Ganten T. M., Sültmann H., Tüttenberg J., von Deimling A., Regnier-Vigouroux A., Herold-Mende C., Martin-Villalba A. (2008) Cancer Cell 13, 235–248 [DOI] [PubMed] [Google Scholar]
- 37. Griffiths G. J., Koh M. Y., Brunton V. G., Cawthorne C., Reeves N. A., Greaves M., Tilby M. J., Pearson D. G., Ottley C. J., Workman P., Frame M. C., Dive C. (2004) J. Biol. Chem. 279, 46113–46121 [DOI] [PubMed] [Google Scholar]
- 38. Bouillet P., O'Reilly L. A. (2009) Nat. Rev. Immunol. 9, 514–519 [DOI] [PubMed] [Google Scholar]
- 39. Newton K., Harris A. W., Bath M. L., Smith K. G., Strasser A. (1998) EMBO J. 17, 706–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen Y., Xu J., Jhala N., Pawar P., Zhu Z. B., Ma L., Byon C. H., McDonald J. M. (2006) Am. J. Pathol. 169, 1833–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nabissi M., Morelli M. B., Amantini C., Farfariello V., Ricci-Vitiani L., Caprodossi S., Arcella A., Santoni M., Giangaspero F., De Maria R., Santoni G. (2010) Carcinogenesis 31, 794–803 [DOI] [PubMed] [Google Scholar]
- 42. Chen K. C., Liu W. H., Chang L. S. (2010) J. Cell. Physiol. 222, 625–634 [DOI] [PubMed] [Google Scholar]
- 43. Cuvillier O., Pirianov G., Kleuser B., Vanek P. G., Coso O. A., Gutkind S., Spiegel S. (1996) Nature 381, 800–803 [DOI] [PubMed] [Google Scholar]
- 44. Kahan C., Seuwen K., Meloche S., Pouysségur J. (1992) J. Biol. Chem. 267, 13369–13375 [PubMed] [Google Scholar]
- 45. Meloche S. (1995) J. Cell. Physiol. 163, 577–588 [DOI] [PubMed] [Google Scholar]
- 46. Meloche S., Seuwen K., Pagès G., Pouysségur J. (1992) Mol. Endocrinol. 6, 845–854 [DOI] [PubMed] [Google Scholar]
- 47. Mebratu Y., Tesfaigzi Y. (2009) Cell Cycle 8, 1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim L. C., Song L., Haura E. B. (2009) Nat. Rev. Clin. Oncol. 6, 587–595 [DOI] [PubMed] [Google Scholar]
- 49. Reinehr R., Becker S., Eberle A., Grether-Beck S., Häussinger D. (2005) J. Biol. Chem. 280, 27179–27194 [DOI] [PubMed] [Google Scholar]
- 50. Schlottmann K. E., Gulbins E., Lau S. M., Coggeshall K. M. (1996) J. Leukocyte Biol. 60, 546–554 [DOI] [PubMed] [Google Scholar]
- 51. Eischen C. M., Dick C. J., Leibson P. J. (1994) J. Immunol. 153, 1947–1954 [PubMed] [Google Scholar]
- 52. Cursi S., Rufini A., Stagni V., Condò I., Matafora V., Bachi A., Bonifazi A. P., Coppola L., Superti-Furga G., Testi R., Barilà D. (2006) EMBO J. 25, 1895–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barnhart B. C., Legembre P., Pietras E., Bubici C., Franzoso G., Peter M. E. (2004) EMBO J. 23, 3175–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Salas V., Sánchez-Torres J., Cusidó-Hita D. M., García-Marchan Y., Sojo F., Benaim G., Villalobo A. (2005) Protein Expr. Purif. 41, 384–392 [DOI] [PubMed] [Google Scholar]
- 55. Baudier J., Deloulme J. C., Van Dorsselaer A., Black D., Matthes H. W. (1991) J. Biol. Chem. 266, 229–237 [PubMed] [Google Scholar]
- 56. Rusnak F., Mertz P. (2000) Physiol. Rev. 80, 1483–1521 [DOI] [PubMed] [Google Scholar]
- 57. Bähler M., Kroschewski R., Stöffler H. E., Behrmann T. (1994) J. Cell Biol. 126, 375–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rhoads A. R., Friedberg F. (1997) FASEB J. 11, 331–340 [DOI] [PubMed] [Google Scholar]
- 59. Wheeler D. L., Iida M., Dunn E. F. (2009) Oncologist 14, 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]