Abstract
Alternative processing of parvovirus B19 (B19V) pre-mRNA is critical to generating appropriate levels of B19V mRNA transcripts encoding capsid proteins and small nonstructural proteins. Polyadenylation of the B19V pre-mRNA at the proximal polyadenylation site ((pA)p), which prevents generation of full-length capsid proteins encoding mRNA transcripts, has been suggested as a step that blocks B19V permissiveness. We report here that efficient splicing of the B19V pre-mRNA within the first intron (upstream of the (pA)p site) stimulated the polyadenylation; in contrast, splicing of the B19V pre-mRNA within the second intron (in which the (pA)p site resides) interfered with the polyadenylation, leading to the generation of a sufficient number of B19V mRNA transcripts polyadenylated at the distal polyadenylation site ((pA)d). We also found that splicing within the second intron and polyadenylation at the (pA)p site compete during processing of the B19V pre-mRNA. Furthermore, we discovered that the U1 RNA that binds to the 5′ splice donor site of the second intron is fully responsible for inhibiting polyadenylation at the (pA)p site, whereas actual splicing, and perhaps assembly of the functional spliceosome, is not required. Finally, we demonstrated that inhibition of B19V pre-mRNA splicing within the second intron by targeting an intronic splicing enhancer using a Morpholino antisense oligonucleotide prevented B19V mRNA transcripts polyadenylated at the (pA)d site during B19V infection of human erythroid progenitors. Thus, our study reveals the mechanism by which alternative splicing coordinates alternative polyadenylation to generate full-length B19V mRNA transcripts at levels sufficient to support productive B19V infection.
Keywords: DNA Viruses, Polyadenylation, RNA Processing, RNA Splicing, Viral DNA, Viral Genetics, B19 Virus, Parvovirus, Viral DNA Replication
Introduction
Human parvovirus B19 (B19V)2 is a member of the genus Erythrovirus of the family Parvoviridae. B19V causes several diseases in humans (1), the most common of which is erythema infectiosum or Fifth disease. Other diseases include acute and chronic arthropathy, transient aplastic crisis (infection of patients with a high rate of red blood cell turnover), pure red cell aplasia (infection of immunocompromised patients), and hydrops fetalis (infection of pregnant women). Like other parvoviruses, B19V contains a linear single-stranded DNA genome of approximately (∼)5.6 kb, which is encapsidated by an icosahedrally symmetric capsid without an envelope (2, 3). In addition to infecting only humans, B19V infection shows a remarkable tropism for human erythroid progenitor cells (4–7).
Like members in the genus Bocavirus (8, 9) and Aleutian mink disease virus (10), also of the Parvoviridae family, the transcription profile of B19V is unusual for a DNA virus in that all of the mRNA transcripts are generated from a single precursor mRNA (pre-mRNA) transcribed from a single promoter at map unit 6 (P6) and feature alternative polyadenylation and splicing (see Fig. 1A) (11, 12). The only unspliced mRNA encoding the large nonstructural protein (NS1) is polyadenylated at the proximal poly(A) site ((pA)p) and retains the first intron. The B19V mRNA transcripts in which the first intron is spliced out, through the A1-1 site, and which are polyadenylated at the (pA)p site encode the small 7.5-kDa nonstructural protein, whose function remains unknown. The B19V mRNA transcripts in which the first intron is spliced out and in which read-through at the (pA)p site leads to polyadenylation at the (pA)d site (R4-R9) (see Fig. 1A) encode the structural proteins VP1 and VP2, as well as the small 11-kDa nonstructural protein, whose function is required for the induction of apoptosis during B19V infection (13). Therefore, read-through at the internal polyadenylation site is a key step in producing viable progeny virus.
FIGURE 1.
Schematic illustration of the B19V genome and the probes used in RNase protection assays. A, B19V genetic map (11, 12). The B19V genome (isolate J35, GenBankTM accession no. AY386330) (19) is shown to scale, with the major transcription units, terminal repeats (TR), P6 promoter, RNA initiation site, splice donor sites (D1 and D2) and splice acceptor sites (A1-1, A1-2, A2-1, and A2-2), and (pA)p and (pA)d sites indicated. The nine major transcripts are depicted below the map (designated R1 to R9), with their sizes and those of the encoded proteins shown on the right. B, probes used in RNase protection assays. Probe 8 (nt 2641–2960), probe 9 (nt 3121–3383), probe 10 (nt 4721–5040), and probe 11 (nt 2001–2560), which span the (pA)p site, the A2-1 site, the A2-2 site, and the central exon region, respectively, are diagrammed; and the individual bands they protected in the RNase protection assays and their sizes are shown. RT, read-through.
In B19V-permissive erythroid progenitor cells, production of VP1/VP2- and 11-kDa-encoding mRNA transcripts involves preventing polyadenylation of the B19V pre-mRNA at the (pA)p site (14). More than half of the B19V mRNAs are spliced at the second intron, which leads to the generation of sufficient VP2-encoding transcripts to enable viruses to assemble, as well as the 11-kDa-encoding transcripts. However, in B19V-nonpermissive cells, and even early in permissive infection of erythroid progenitor cells, the majority of B19V mRNA transcripts are polyadenylated at the (pA)p site and thus produce only the nonstructural proteins NS1 and the 7.5 kDa protein (14, 15). These facts indicate that alternative processing of the B19V pre-mRNA must be tightly controlled so that the amounts of B19V mRNA transcripts and proteins necessary to produce progeny virus are generated.
We and others (11, 12, 14, 16, 17) have analyzed the transcription profile obtained when COS-7 cells were transfected with a replication-competent, nearly full-length B19V genome and found that it resembles that observed during B19V infection of B19V-permissive cells. Using this system, we have identified multiple cis-acting elements that facilitate inclusion of the second exon (from the A1-1/A1-2 acceptor site to the D2 donor site), which was confirmed by monitoring transcription profiles during B19V infection of human primary erythroid progenitor cells (18). Here, we describe an extensive mutagenesis-based analysis of the first intron, second intron, and the (pA)p site in the context of a B19V replication-competent genome, and the implications of our findings for how alternative splicing and alternative polyadenylation influence one another during processing of the B19V pre-mRNA.
EXPERIMENTAL PROCEDURES
Plasmid Construction
All of the nucleotide numbers for the B19V genome refer to the J35 isolate of B19V (GenBankTM accession no. AY386330) (19). The parent plasmid C1NS1(−), which contains the SV40 replication origin and produces a prematurely terminated NS1 gene has been described previously (14, 17, 20). The parent plasmid pB19-N8 was described previously (19–21).
Reporter Constructs to Test Polyadenylation Strength
The parent polyadenylation reporter plasmid (pCI(pA)test) was constructed by inserting an internal ribosome entry site and a GFP expression cassette before the SV40 poly(A) signal, through the SalI and NotI sites of the pCI vector (Promega), after which a Renilla-encoding sequence was inserted between the intron and the internal ribosome entry site via the NheI and EcoRI sites (see Fig. 2A). The pCI(pA)p reporter plasmids, pCI(pA)p400, pCI(pA)p240, pCI(pA)p1248, pCI(pA)p3426, and pCI(pA)p4800 were constructed by inserting the B19V sequences of nt 2641–3040, nt 2721–2960, nt 1248–2960, nt 2641–3426, and nt 2641–4800, respectively, into the EcoRI and XbaI sites between the Renilla expression cassette and the internal ribosome entry site (see Fig. 2A). The pCI(pA)d reporter plasmids pCI(pA)d400 and pCI(pA)d240 were generated by inserting B19V nt 4961–5360 and nt 5041–5280, respectively, into the parent reporter within the EcoRI and XbaI sites. Replacing the SV40 poly(A) signal with the (pA)d signal in the construct pCI(pA)p400 generated pCI(pA)p(pA)d.
FIGURE 2.
An efficient poly(A) signal of the (pA)p site requires an extended upstream and downstream region but not that of the (pA)d site. A, C1NS1(−), pCI(pA)test-based (pA)p, and (pA)d test plasmids are illustrated schematically. The probes used are indicated, with the putative protected bands, their sizes, and their designations shown. B, COS-7 cells were transfected with pCI(pA)test reporter-based (pA)p and (pA)d plasmids. Total RNA was protected by probe p(pA)p + SV40 in the case of lanes 1–6, by probe (pA)d+SV40 in the case of lanes 7–9, and by both in the case of lane 10. Ratios of the mRNAs polyadenylated at (pA)SV40 versus (pA)p (lanes 1–6), at (pA)SV40 versus (pA)d (lanes 7–9), and at (pA)d versus (pA)p (lane 10) are shown as averages with the S.D. and reflect the results of at least three individual experiments. Total RNA from uninfected cells generated no protection products (data not shown). C1NS1(−) was transfected as a control. RT, read-through. TR, terminal repeats.
Constructs to Analyze Effect of Splicing of First Intron on Polyadenylation at (pA)p Site
All of the constructs used for this purpose were based on the C1NS1(−) and were diagrammed in Fig. 3A. Plasmids C1B19A1-1KOa and C1B19A1-2KO were constructed by mutating the A1-1 or the A1-2 sites from AG to AA. C1B19A1-1KOb was generated by mutating the A1-1 site from AG to AA and the purine-rich exonic splicing enhancer (ESE1) (nt 2158–2173) to a sequence of 5′-CCTTAGCCATATTGT-3′ between the A1-1 and A1-2 acceptor sites (18). Further mutation of the A1-2 site from AG to AA on the plasmid C1B19A1-1KO generated C1B19A1-1/2KO. The plasmid C1B19A1-1U2AF was constructed by mutating the A1-2 site from AG to AA and further mutating the polypyrimidine tract of the A1-1 site to a consensus U2AF65 (U2 auxiliary factor 65)-binding sequence (5′-TTCCCTTTTTTTTC-3′) (see Fig. 3A) (22).
FIGURE 3.
Splicing of the first intron is required for efficient internal polyadenylation of the B19V pre-mRNA at the (pA)p site. A, C1NS1(−) is schematically illustrated, and the nucleotide substitutions in each derivative mutant are shown. B and C, COS-7 cells were transfected with C1NS1(−) and its derivatives. Total RNA isolated was protected by homologous probe 11 (B) or probe 8 (C). A representative experiment is shown with the identities of the protected bands indicated on the right. Ratios of RNAs polyadenylated at (pA)p versus (pA)d are shown as averages with S.D. and were calculated from the results of at least three individual experiments. RT, read-through; TR, terminal repeats.
Constructs to Analyze Effects of Splicing of Second Intron on Polyadenylation at (pA)p Site
All of the plasmids were based on C1NS1(−) and B19-N8, as shown in Fig. 4A. The acceptor 3′ splice site knock-out constructs C1B19A2-1KO, C1B19A2-2KO, and C1B19A2-1/2KO were made by mutating the A2-1, A2-2, and both A2-1 and A2-2 sites, respectively, from AG to AA. Plasmids C1B19A2-1U2AF and C1B19A2-2U2AF were generated by mutating the polypyrimidine tract of the A2-1 and A2-2 sites to an U2AF65-binding sequence as described above. Further knock-out of the A2-2 site on C1B19A2-1U2AF resulted in plasmid C1B19A2-1U2AF/A2-2KO. Donor mutants C1B19D2KO and C1B19D2Cons were constructed by mutating the D2 site to 5′-CCG/CGGAAC-3′ and 5′-CAG/GTATGT-3′, respectively. B19-N8-based plasmids B19-N8A2-1U2AF and B19-N8A2-1/2KO were constructed by mutating the A2-1 site to an U2AF65-binding sequence and by knocking out both the A2-1 and A2-2 sites, respectively, as described above.
FIGURE 4.
Splicing of the second intron competes with internal polyadenylation of the B19V pre-mRNA at the (pA)p site. A, C1NS1(−) is schematically illustrated, and the nucleotide substitutions at the respective splice site in each derivative mutant are shown. B–D, COS-7 cells were transfected with C1NS1(−) and its derivatives, and UT7/Epo-S1 cells were electroporated with linearized B19V N8 DNA and its derivatives as indicated. Total RNA isolated from COS-7 and mRNA isolated from UT7/Epo-S1 cells were protected by homologous probe 9 (B), homologous probe 10 (C), or probe 8 (D). E, COS-7 cells were transfected with C1NS1(−), C1B19D2KO, or C1B19D2ISE2KO (18) as indicated. Total RNA isolated was protected by probe 8. A representative experiment is shown with the identities of the protected bands indicated on the right. Quantification of the ratios of RNAs spliced (Spl) versus unspliced (Unspl) at the A2-1 site (B), the ratios of RNAs spliced versus unspliced at the A2-2 site (C), and the ratios of RNAs polyadenylated at (pA)p versus (pA)d (D) are shown as averages with the S.D., and which were calculated from the results of at least three individual experiments. Asterisk indicates likely nonspecific hybridized products (14). RT, read-through; TR, terminal repeats.
Constructs to Analyze Significance of Polyadenylation at (pA)p in Splicing of Second Intron
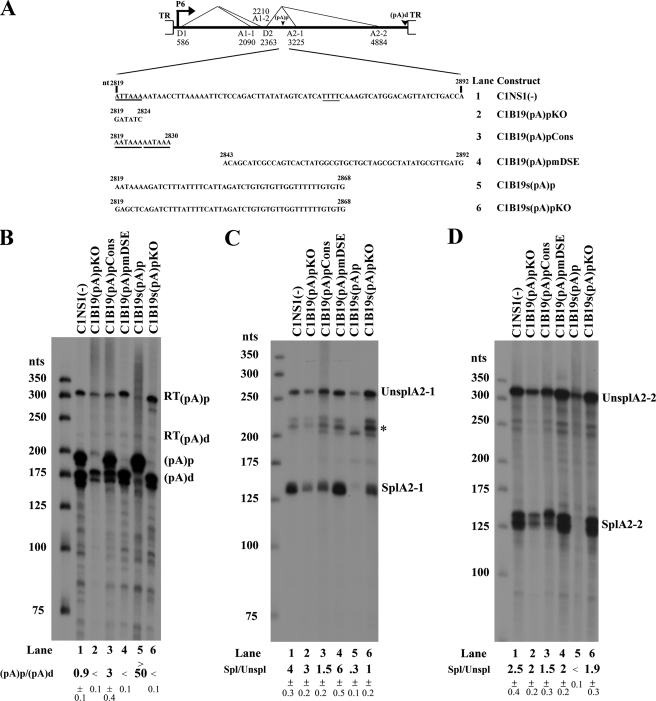
All of the (pA)p mutants were based on C1NS1(−) and diagrammed in Fig. 5A. C1B19(pA)pKO and pCIB19(pA)pCons were constructed by mutating the potential cleavage and polyadenylation specificity factor-binding hexanucleotides from 5′-ATTAAA/AATAAC-3′ to 5′GATATC/AATAAC-3′, and from 5′ATTAAA/AATAAC-3′ to 5′-AATAAA/AATAAA-3′, respectively. Plasmid C1B19(pA)pmDSE was generated by mutating the 50-nt downstream element (DSE) (17). Plasmid C1B19s(pA)p was made by replacing the B19V sequence nt 2819–2868 with a synthetic poly(A) signal (23), as shown in Fig. 5A. C1B19s(pA)pKO was generated by further mutating the synthetic poly(A) signal in the C1B19s(pA)p from 5′-AAUAAA-3′ to 5′-GAGCUC-3′ as described previously (18).
FIGURE 5.
When internal polyadenylation of the B19V pre-mRNA at the (pA)p site is strong, it competes with splicing at the second intron. A, C1NS1(−) is schematically illustrated, and its derivative mutants are shown with nucleotides changes at the (pA)p core element between nt 2819 and 2892 shown. B–D, COS-7 cells were transfected with C1NS1(−) and its derivative mutants. Total RNA isolated was protected by homologous probe 8 (B) or probe 9 (C), or probe 10 (D). A representative experiment is shown, with the identities of the protected bands indicated on the right. Ratios of RNAs polyadenylated at (pA)p versus (pA)d (B), of RNAs spliced (Spl) versus unspliced (Unspl) at the A2-1 site (C), and of RNAs spliced versus unspliced at the A2-2 site (D) are shown as averages with the S.D. and are the results of at least three individual experiments. RT, read-through; TR, terminal repeats. * indicates non-specific bands.
Constructs to Analyze Significance of U1 RNA Binding in Polyadenylation at (pA)p
Plasmid C1B19ISE2A2-1KO was constructed from the parent plasmid C1NS1(−) by mutating the G-rich intronic splicing enhancer (ISE) (nt 2371–2400) to 5′-CCGCGGCACGAGATCATCACGATCGAACAG-3′ as described previously (18) and the A2-1 site from AG to AA. The plasmid C1B19ISE2A2-1/2KO was generated by mutating the A2-2 site in the parent plasmid C1B19ISE2A2-1KO from AG to AA. Plasmid U1B19VD2 was made by mutating the donor site binding sequence in the U1 RNA sequence of the U1 RNA-expressing plasmid (24) such that it is complementary to the B19V D2 donor site (5′-ACT/GTTAGT-3′).
Constructs Used to Generate Probes for RNase Protection Assays
Probes 9 and 10 were constructed by inserting the B19V nt 3121–3383 or nt 4721–5040 sequences, respectively, into BamHI-HindIII-digested pGEM4Z (Promega). Probe 11 was constructed by cloning the corresponding B19V sequences of nt 2001–2560 into the C1NS1(−)-based constructs described above into BamHI-HindIII-digested pGEM3Z. Probe 8 has been described previously (14). All these probes and the bands they protect are shown in Fig. 1B. Probes p(pA)p + SV40 and p(pA)d+SV40 were generated by inserting the poly(A) signal of the SV40 virus of the pCI vector (nt 1114–1330) into pGEM4Z, through PstI and HindIII sites, and the B19V sequences of nt 2760–2940 and nt 5041–5280, respectively, were then inserted into EcoRI-KpnI-digested pGEM4Z. The probes and their protected bands are diagrammed in Fig. 2A.
Cells, Transfection, and Virus Infection
COS-7 cells (ATCC, CRL-1651) were maintained in DMEM with 10% FCS at 37 °C in 5% CO2. Cells were transfected with 2 μg of DNA per 60-mm plate using LipofectamineTM and PLUSTM reagent (Invitrogen) according to the manufacturer's instructions. UT7/Epo-S1 cells, which are permissive to B19V infection (20, 25, 26), were cultured in DMEM with 10% FCS and 2 units/ml of Epo (Epogen; Amgen, Thousand Oaks, CA) at 37 °C in 5% CO2. Plasmids B19-N8, B19-N8A2-1U2AF, and B19-N8A2-1/2KO were digested with XhoI/EcoRI to recover the excised B19V DNA fragments that were used for electroporation. UT7/Epo-S1 cells were electroporated with 2 μg of linearized B19V DNA per 2 × 106 cells with an Amaxa Nucleofector device (Lonza) as described previously (20). Primary human erythroid progenitor cells (CD36+ EPCs) were generated; large numbers of CD36+ EPCs, at day 8 of ex vivo expansion, were treated with Morpholino oligonucleotides M-ISE2 and M-ISE2 control (18) at 16 h prior to B19V infection, as described previously (13). The cells were infected with B19V (a gifted plasma sample from ViraCor-IBT, Lee's Summit, MO) at a multiplicity of infection of 1 (20, 27).
Quantification of Virus Particles (Packaged Viral Genomes)
Virus-containing cell lysates were digested with nuclease Benzonase® (2.5 units/μl) overnight, prior to viral DNA extraction using the QIAamp DNA Blood Mini Kit (Qiagen). Viral genomes were quantified by quantitative PCR, using a probe and a primer set targeting to the NS1 gene as described previously (20, 27).
RNA Isolation and RNase Protection Assays
Total RNA was isolated from transfected cells 2 days after transfection or virus infection using TRIzol reagent (Invitrogen), and in some experiments, where indicated, mRNA was purified from total RNA using FastTrack MAG mRNA kit (Invitrogen), following the protocols provided by the manufacturer. RNase protection assays were performed essentially as described previously (14, 28, 29). Briefly, probes were generated by in vitro transcription with T7 or SP6 polymerase, using the MAXIscript® (Ambion) and following the protocol supplied by the manufacturer. Ten μg of total RNA or mRNA purified from 100 μg of total RNA was hybridized with a substantial excess of probe, and RNase protection signals were quantified with the Storm 856 PhosphorImager and Image Quant TL software (version 2005, GE Healthcare). Relative molar ratios of individual species of RNAs were determined after adjustment for the number of 32P-labeled uridines (U) in each protected fragment, as described previously (28, 29).
RESULTS
Efficient Poly(A) Signal of (pA)p Site Requires an Extended Upstream and Downstream Region but Not That of (pA)d Site
To study the regulation of B19V internal polyadenylation, we first examined the strength of the core poly(A) signals at the (pA)p and the (pA)d sites. We constructed and used a poly(A) reporter plasmid pCI(pA)test, comparing the strength at each site with that at the SV40 poly(A) site ((pA)SV40) (Fig. 2A). We found that >95% of the mRNA transcripts generated from the pCI(pA)p400 and pCI(pA)p240 read through the (pA)p site (Fig. 2B, lanes 2 and 3). The ratio of (pA)SV40/(pA)p was ∼40, indicating that an efficient core poly(A) signal of the (pA)p requires extended sequences beyond this 400-nt B19V sequence. When the parallel experiment was carried out using probe p(pA)d+SV40, we found that nearly one-third of the pre-mRNAs were polyadenylated at the (pA)d site, indicating that the 240-nt core signal of the (pA)d (nt 5041–5280) is sufficient, and has a strength of a half of that at the SV40 poly(A) site (Fig. 2B, lane 9). Finally, analysis of the mRNA transcripts generated from transfection of the construct pCI(pA)p(pA)d showed that ∼95% of the mRNAs read through the core (pA)p site, and they were polyadenylated at the core (pA)d site (Fig. 2B, lane 10). The strength of polyadenylation at the 400-nt core (pA)d was found to be twenty times greater than that at the 400-nt core (pA)p.
We next tested functions of the sequences outside of the 400-nt core (pA)p signal, including sequences from either the first intron (pCI(pA)p1248)) or the second intron (pCI(pA)p3426 and pCI(pA)p4800). We found that ∼25% of the pre-mRNAs generated from transfection of the pCI(pA)p1248 were polyadenylated at the (pA)p site (Fig. 2B, lane 4) but that polyadenylation was higher when the core signal sequence was extended toward the second intron (Fig. 2B, lanes 5 and 6). Inclusion of the core poly(A) signal of B19V sequence nt 2641–4800 resulted in ∼50% of the pre-mRNAs being polyadenylated at the (pA)p site (Fig. 2B, lane 6). Taken together, these results show that efficient polyadenylation at the (pA)p site requires cis-acting signals located beyond the 400-nt core (pA)p signal within the first and second introns, whereas the 400-nt core (pA)d signal is sufficient for polyadenylation of B19V pre-mRNA at the (pA)d site.
Splicing of First Intron Is Essential for Efficient Polyadenylation at (pA)p Site
We next investigated whether splicing of the upstream intron of the site (pA)p affects polyadenylation at this site. First, we mutated the acceptor sites of the first intron to decrease splicing between the D1 site and the A1-1 or A1-2 site. Mutating the dinucleotide AG of the A1-1 acceptor to AA led to usage of a cryptic acceptor site at nt 2104, with splicing at the A1-1 site decreased ∼2-fold compared with that in wild type (Fig. 3B, lane 2). In this context, polyadenylation at the (pA)p site was slightly decreased compared with that in wild type, with ratio of (pA)p/(pA)d reduced from 0.9 to 0.7 (Fig. 3C, compare lane 2 with 1). Further mutation of ISE1 (an intronic splicing enhancer), which facilitates splicing at the A1-1 site (18), abrogated this splicing (Fig. 3B, lane 3) and reduced polyadenylation at the (pA)p site 3-fold compared with that in the wild type, with the (pA)p/(pA)d ratio decreasing from 0.9 to 0.3 (Fig. 3C, lane 3). Mutating the A1-2 acceptor site from AG to AA resulted in nearly complete loss of splicing from the D1 site to the A1-2 site (Fig. 3B, lane 4) and to a significant reduction of polyadenylation at the (pA)p site with the (pA)p/(pA)d reduced from 0.9 to 0.6 (Fig. 3C, lane 4). When all splicing from the D1 site (to both the A1-1 and A1-2 sites) was knocked out through combined mutations at the enhancer and both acceptor sites, polyadenylation at the (pA)p site was decreased ∼10-fold compared with that in wild type (Fig. 3C, lane 5) with the (pA)p/(pA)d ratio reduced from 0.9 to 0.1. Notably, when splicing of the first intron was enhanced by mutating the polypyrimidine tract of the A1-1 site to a consensus U2AF65-binding sequence (Fig. 3A, construct 6), polyadenylation at the (pA)p site was slightly increased compared with that seen in the wild type (Fig. 3C, lane 6). Splicing of B19V pre-mRNA at the A1-1 site was found to be increased significantly, but splicing at the A1-2 site was nearly abolished (Fig. 3B, lane 6). Taken together, these results suggest that efficient polyadenylation of the B19V pre-mRNA at the (pA)p site requires efficient splicing of the first intron upstream of the (pA)p site.
Splicing in Second Intron Leads to Competition for Polyadenylation at (pA)p Site
Because the B19V internal (pA)p site lies in a functional intron, we investigated whether splicing of the second intron influences polyadenylation at the (pA)p site. To this end, we mutated the A2-1 or A2-2 acceptor site, abolishing splicing at the respective site (Fig. 4, B and C, lanes 2 and 3). In both of these contexts, polyadenylation at the (pA)p site remained the same as that in the wild type (Fig. 4D, lanes 2 and 3). When both of the A2-1 and A2-2 sites were mutated, splicing in the second intron was completely abrogated (Fig. 4, B and C, lane 4). In this context, ∼35% of the mRNA transcripts were polyadenylated at the (pA)p site, which represents a decrease of ∼30% relative to that of the wild type genome (Fig. 4D, lane 4). When the D2 donor site and the A2-1 and A1-2 acceptor sites were all mutated, splicing within the second intron was likewise abrogated (Fig. 4, B and C, lane 5); yet, to our surprise, more than 85% of the B19V mRNA transcripts were polyadenylated at the (pA)p site (Fig. 4D, lane 5). Similarly, while splicing in the second intron was abrogated by mutating ISE2, located 3′ of the D2 donor site (18), >95% of the mRNAs produced were polyadenylated at the (pA)p site (Fig. 4E, lane 2).
Based on the findings presented above, we concluded that the presence of the D2 donor site and the ISE2 site, both of which are critical for U1 snRNP binding the D2 donor site (18), plays an important role in regulating polyadenylation of the B19V pre-mRNA at the (pA)p site. To test this possibility, we made a mutant affecting only the D2 donor site. Despite the fact that mutation of the D2 donor site resulted in splicing from a cryptic donor site and a decrease splicing in the second intron (as shown previously in Ref. 18 and confirmed in Fig. 4, B and C, lane 9), ∼85% of the B19V mRNA transcripts were polyadenylated at the (pA)p site (Fig. 4D, lane 9). Notably, when the D2 site was mutated to a consensus U1 RNA-binding sequence (5′-CAG/GTATGT-3′), splicing from the D2 to the A2-1 site was confirmed as being significantly increased (Fig. 4B, lane 10), and this resulted in nearly all B19V mRNAs being polyadenylated at the (pA)d site (Fig. 4D, lane 10). Accordingly, increasing splicing from the second intron as a consequence of conversion of the A2-1 site, A2-2 site, or both to consensus U2AF65-binding sequences (Fig. 4, B and C, lanes 6, 7, and 8, respectively) resulted in significantly decreased polyadenylation at the (pA)p site (Fig. 4D, lanes 6, 7, and 8, respectively).
To ensure this competition between splicing and polyadenylation within the second intron occurs in B19V-permissive cells, we transfected UT7/Epo-S1 cells with linearized B19V DNA N8, which has been shown to replicate in these cells (20), and its A2-1U2AF and A2-1/2KO mutants. Replication of these B19V DNAs in UT7/Epo-S1 cells was confirmed by Southern blot analysis (data not shown). Similar to what we observed from transfection of these mutants in the context of B19V replication-competent backbone in COS-7 cells, we found that an increase in splicing of the second intron at the A2-1 site improved B19V pre-mRNA read through the (pA)p site significantly (ratio of (pA)p/(pA)d was decreased from 1 to <0.1; Fig. 4D, lane 12) and that knock-out of splicing of the central intron prevented B19V pre-mRNA read through the (pA)p site. (The ratio of (pA)p/(pA)d was increased from 1 to 10; Fig. 4D, lane 13.)
Collectively, our results show that poor splicing of the B19V pre-mRNA from the second intron results in preferred polyadenylation at the (pA)p site and that conversely strong splicing from the second intron inhibits polyadenylation at the (pA)p site. Additionally, because the mutant with knock-out of both the acceptor sites remained a fair inhibition of polyadenylation at the (pA)p site, our results implicate that U1 RNA binding at the D2 donor site likely plays a key role in inhibiting this polyadenylation.
Strong Polyadenylation at (pA)p Site Leads to Competition for Splicing at D2 Donor Site
We next investigated whether polyadenylation at the (pA)p site influences splicing from the second intron. To this end, we mutated the cleavage and polyadenylation specificity factor-binding hexanucleotide site (AAUAAA) or the DSE (50 nt) (17) to abolish polyadenylation at the (pA)p site. Mutation of the (pA)p site significantly reduced polyadenylation at the (pA)p site (Fig. 5B, lane 2). Nevertheless, it did not lead to an apparent splicing increase involving the D2 donor site (Fig. 5, C and D, lane 2). In contrast, mutation of the DSE, which nearly abolished all polyadenylation at the (pA)p site (Fig. 5B, lane 4), resulted in a slight increase in splicing from the D2 site to the A2-1 site (Fig. 5C, lane 4) and a slight decrease in splicing from the D2 site to the A2-2 site (Fig. 5D, lane 4). However, when the poly(A) signal was strengthened by conversion to a consensus cleavage and polyadenylation specificity factor-binding site, polyadenylation at the (pA)p site was increased ∼3-fold (Fig. 5B, lane 3), and splicing at the D2 site was significantly decreased (Fig. 5, C and D, lane 3).
As a second part of this analysis, we replaced the (pA)p site with a strong synthetic poly(A) signal, (s(pA)p). As expected, polyadenylation of the B19V mRNAs from the s(pA)p site was significantly higher (>50-fold) than that from the (pA)p site (Fig. 5B, lane 5). In this context, splicing at the D2 site was nearly abolished (Fig. 5, C and D, lane 5). In a control construct, the AAUAAA poly(A) signal in pC1B19s(pA)p was mutated, and this resulted in failure of polyadenylation at the s(pA)p site (Fig. 5B, lane 6). These changes were accompanied by a significant increase in splicing from the D2 site to the A2-1 and A2-2 sites (Fig. 5, C and D, lane 6). Together, these results show that polyadenylation of the B19V pre-mRNA at the (pA)p site, when it is strong at a level sufficient to prevent polyadenylation at the (pA)d site in more than half of the pre-mRNAs, competes with splicing from the second intron.
Binding of U1 RNA to D2 Donor Site Inhibits Polyadenylation at (pA)p Site
In AAV5, binding of the U1 snRNP to the donor site of the AAV5 P41-generated pre-mRNA inhibits polyadenylation of this pre-mRNA at its (pA)p site (30). Thus, we tested the U1 snRNP for a role in inhibiting B19V pre-mRNA polyadenylation at the (pA)p site. We found that when both the ISE2 and the A2-1 acceptor were mutated (Fig. 6A), splicing of the B19V mRNAs at the D2 site was abolished (Fig. 6, B and C, lane 2), whereas >90% of the B19V mRNA transcripts were polyadenylated at the (pA)p site (Fig. 6D, lane 2). However, when an engineered U1 RNA that contains a binding site perfectly complementary to the B19V D2 donor site (Fig. 6A) was expressed, splicing from the D2 to A2-2 sites was restored (Fig. 6C, lane 3). This result suggested that the engineered U1 RNA binds to the B19V D2 donor site and is functional to initiate splicing. We also found that in this context significantly more B19V mRNA transcripts read through the (pA)p site and ultimately were polyadenylated at the (pA)d site (Fig. 6D, lane 3), suggesting that either splicing of the second intron or U1 RNA binding to the D2 donor inhibits polyadenylation at the (pA)p site.
FIGURE 6.
U1 RNA binding to the D2 donor site inhibits all the internal polyadenylation of the B19V pre-mRNA at the (pA)p site. A, top: C1NS1(−) is schematically illustrated, and its two derivatives are shown with nucleotide changes at the D2, A2-1, and A2-2 sites shown. Bottom: the mutant U1 RNA and the putative core elements that regulate the (pA)p site are indicated, as are the mutations in the sequence that normally binds to the D2 donor site. B–D, C1B19ISE2A2-1KO or C1B19ISE2A2-1/2KO was cotransfected into COS-7 cells with U1B19VD2 (U1B19VD2+) or pBluescript SK+ (U1B19VD2 (−)). C1NS1(−) was used as a control. Total RNA was protected by probe 9 (B), probe 10 (C), or probe 8 (D). A representative experiment is presented with the identities of the protected bands shown on the right. Quantification of the ratios of RNAs polyadenylated at (pA)p versus (pA)d are shown in D as averages with S.D. and were calculated from the results of at least three individual experiments. RT, read-through; TR, terminal repeats; Spl, spliced; Unspl, unspliced. * indicates non-specific bands.
To distinguish between these two possibilities, we mutated both the ISE2 and the acceptor sites in the second intron of B19V pre-mRNA (Fig. 6A). Because no acceptor sites were available, even in the presence of the engineered U1 RNA, splicing of the B19V mRNAs at the D2 donor site was not restored (Fig. 6C, lane 5); yet, the engineered U1 RNA bound to the D2 donor site, presumably without further assembly of an effective spliceosome (30, 31). In this context, polyadenylation at the (pA)p site was inhibited, and the ratio of (pA)p/(pA)d was restored to a level seen in the C1NS1(−) control (Fig. 6D, lane 5). Collectively, these results strongly support for the notion that it is U1 RNA binding rather than the splicing process per se that inhibits polyadenylation of the B19V pre-mRNA at the (pA)p site.
Inhibition of B19V Pre-mRNA Splicing within Second Intron Increases B19V mRNA Transcripts Polyadenylated at (pA)p during B19V Infection of CD36+ EPCs
To confirm our findings obtained from the above transfection experiments, we used a Morpholino antisense oligonucleotide (M-ISE2), which targets the ISE2 enhancer adjacent to the D2 donor site (18), to inhibit splicing of B19V pre-mRNA within the second intron. We have previously shown that the M-ISE2 is effective to inhibit splicing and alter capsid protein expression (18). Here, we confirmed that application of the M-ISE2 decreased splicing at the A2-1 and A2-2 site compared with M-ISE control (ratio of Spl versus Unspl decreased from 6 to 3 and 1 to 0.3, respectively; Fig. 7A, compare lane 1 with 2 and lane 3 with 4, respectively), which resulted in significantly more B19V mRNA transcripts polyadenylated at the (pA)site (the ratio of (pA)p/(pA)d was increased from 0.3 to 1.2; Fig. 7A, compare lane 4 with 6). Further quantification of progeny virus produced from B19V-infected CD36+ EPCs showed ∼6-fold reduction in progeny virus production from infected cells treated with M-ISE2 ((2.5 × 105 genomic copies (gc)/μl) compared with the counterpart treated with the control Morpholino (1.4 × 106 gc/μl) (Fig. 7B). These results suggest that splicing of the B19V pre-mRNA within the second intron regulates the B19V capsid proteins and 11-kDa-encoding mRNA transcripts that are polyadenylated at the (pA)d site and thereafter determines progeny virus production during B19V infection.
FIGURE 7.
Inhibition of B19V pre-mRNA splicing within the second intron increases B19V mRNA transcripts polyadenylated at the (pA)p site during B19V infection of CD36+ EPCs. CD36+ EPCs were treated with a Morpholino oligonucleotide (M-ISE2) or its control (Ctrl) followed by B19V infection. A, at 48 h postinfection, total RNA was isolated and protected by homologous probe 8, 9, or 10. A representative experiment is shown with the identities of the protected bands indicated on the right. Ratios of RNAs spliced (Spl) versus unspliced (Unspl) at the A2-1 site (lanes 1 and 2), of RNAs spliced versus unspliced at the A2-2 site (lanes 3 and 4), and of RNAs polyadenylated at (pA)p versus (pA)d (lanes 5 and 6), are shown as averages with the S.D. and are the results of three individual experiments. B, At 48 h postinfection, the cells were collected for quantification of virus particle production as genomic copies (gc)/μl. Results shown represent the averages and S.D. of data from at least three independent experiments. p < 0.05, as assessed based on the Student's t test.
DISCUSSION
In this study, we analyzed how alternative polyadenylation of the B19V pre-mRNA is regulated by its alternative splicing in the context of a replication-competent B19V genome in B19V nonpermissive COS-7 and B19V-permisisve UT7/Epo-S1 cells (20, 25, 26), as well as during B19V infection of ex vivo-expanded primary erythroid progenitor cells. Our results reveal that that alterative splicing at the D2 donor site is a dominate step in processing of the B19V pre-mRNA, attenuating polyadenylation at the (pA)p site and thereby facilitating read-through of the B19V pre-mRNA such that both the capsid and the 11-kDa proteins are produced. Processing of the B19V pre-mRNA during transfection of replicative constructs in COS-7 cells reproduces that seen in UT7/Epo-S1 cells and during B19V infection of human erythroid progenitor cells (11, 14, 16, 17). Due to the difficulty of transfecting UT7/Epo-S1 cells and human erythroid progenitor cells, we performed most of the experiments using COS-7 cells in the context of a replication-competent B19V genome in which replication is supported by an SV40 replication origin. We confirmed that splicing within the second intron was critical B19V mRNA transcripts to read through the (pA)p site in UT7/Epo-S1 cells and, more importantly, during B19V infection of primary human erythroid progenitor cells. Thus, we believe that the regulatory mechanism identified in the context of the replication-competent B19V genome in COS-7 cells accurately reflects that used during B19V infection of permissive cells.
Internal polyadenylation often regulates viral gene expression. A functional polyadenylation site within an intron plays a regulatory role in expression of papillomavirus (32), adenovirus (33), and AAV5 (34). In AAV5, different levels of internal polyadenylation are constitutively seen for transcripts generated from different promoters (34). The DSE of the internal polyadenylation site of AAV5 overlaps with the polypyrimidine tract of the 3′ acceptor site (A2) of the intron (34). The B19V system is unique in that all transcripts are processed from a single pre-mRNA transcribed from a single promoter (P6); the core internal (pA)p site is distinct from the 3′ acceptor of the intron in which the (pA)p site lies (Fig. 1A); and alternative splice elements are interspersed throughout the B19V genome. Therefore, the B19V system is more complex than its AAV5 counterpart. More importantly, usage of the internal polyadenylation site correlates negatively with the levels of capsid protein-encoding and 11-kDa protein-encoding mRNAs, which are critical to the production of progeny virus.
Alternative usage of the poly(A) signals in processing the B19V pre-mRNA is unique in that a functional intron is present upstream of the (pA)p site. This suggests that the exon definition model of intron recognition (Fig. 8) may play a role in establishing whether the long exon 2 (exon 2′), located between the upstream 3′ splice site A1-1 or A1-2 site and the poly(A) site, is produced and that this mechanism may involve an interaction with U2AF65, which binds to the upstream 3′ splice acceptor site and thereby links it to factors present in the polyadenylation machinery (35, 36) (Fig. 8A). We observed that definition of the long exon 2′ is balanced with definition of the central exon (exon 2), which extends from the A1-1 or A1-2 site to the D2 site (Fig. 8B). Therefore, we speculate that efficient splicing of the upstream intron is required for polyadenylation at the (pA)p site, and that polyadenylation at this (pA)p is a compromised outcome of defining exon 2′ and exon 2 (Fig. 8).
FIGURE 8.
A model for defining the central exon of B19V pre-mRNA. B19V mRNAs polyadenylated at the (pA)p (A) and at the (pA)d (B) sites are schematically illustrated. Splicing factors that define exon 2 (from the A1-1 or A1-2 site to the D2 site; A) and the exon 2′ (from the A1-1 or A1-2 site to (pA)p site; B) are diagrammed. Positive (+) and negative (−) interactions between splicing factors are indicated with arrowheads (see text for details).
During processing of eukaryotic pre-mRNA, U1 RNA binding to a functional donor site is required to suppress premature polyadenylation from cryptic poly(A) signals in introns (37). However, our discovery that strong splicing within the second intron reduces polyadenylation at the (pA)p site and vice versa indicates that splicing of the B19V pre-mRNA within the second intron and internal polyadenylation competes for the same pool of B19V pre-mRNA molecules and that this regulation is critical to production of the B19V mRNA transcripts encoding viral capsid proteins and the 11-kDa nonstructural protein. Although it is similar, to some extent, to that competition during processing of the pre-mRNA generated from AAV5 P41 promoter (34), in AAV5, the DSE of the (pA)p site and the polypyrimidine tract site overlap, and thus polyadenylation factors (e.g. CstF64) compete with splicing factors (i.e. U2AF65), and this results in the inhibition of polyadenylation at the (pA)p site in ∼50% of the molecules (34). Binding of U1 snRNP to the central donor site of the AAV5 pre-mRNA inhibits another internal polyadenylation in the other ∼50% of the molecules. These two inhibitory mechanisms cause most of the pre-mRNA molecules to read through the (pA)p site, they are ultimately polyadenylated at the (pA)d site such that AAV5 capsid proteins are encoded (34). Strikingly, in B19V pre-mRNA processing, binding of the U1 RNA, presumably via its U1 snRNP component, to the upstream D2 donor site accounted for all inhibition of polyadenylation at the (pA)p site, even when the D2 site and the polyadenylation site were ∼500 nt apart. However, at this distance the functional donor site in AAV5 fails to inhibit polyadenylation of the pre-mRNA at the AAV5 (pA)p site (30). In a study using a more general heterogeneous system, the longest distance from which an U1 snRNP binding site was found to inhibit downstream polyadenylation of the same RNA was 1180 nt (38). In this study, a high consensus U1 RNA binding site maximizing inhibition of the downstream poly(A) site was designed. Although the B19V D2 donor site is a nonconsensus U1 RNA binding site, splicing at the D2 site is greatly enhanced by its upstream and downstream splicing enhancers (ESE3 and ISE2) (18). These enhancers likely promote the affinity of the U1 RNA for the D2 donor site, to a level similar to that characteristic of the consensus U1 RNA binding site. Therefore, we hypothesize that it is the binding of the splice factors (presumably that of these serine-arginine-rich splicing factors) to the enhancer sites that promotes the affinity of the U1 snRNP binding to the intervening donor site and thereafter inhibits internal polyadenylation of the B19V pre-mRNA.
In our experiments in which the B19V nonreplicative genome was transfected into COS-7 cells and those examining early time points of B19V infection of erythroid progenitor cells (14), the majority of the B19V pre-mRNAs were cleaved and polyadenylated at the (pA)p site. In this situation, the stronger polyadenylation at the (pA)p site may compete with splicing from the second intron. In contrast, experiments in which the B19V replicative genome was transfected into COS-7 cells and those examining later time points of B19V infection of erythroid progenitor cells (14), approximately equal numbers of B19V pre-mRNA molecules were polyadenylated at both sites. Because reduction of polyadenylation at the (pA)p site in the context of replicative B19V genome did not significantly enhance splicing of the second intron (Fig. 5D, lanes 1 and 4), we hypothesize that replication of the B19V genome increases splicing of the B19V pre-mRNA at the second intron, rather than decreasing polyadenylation at the (pA)p site. Given that subsequent splicing of the B19V pre-mRNA inhibits polyadenylation at the (pA)p site, it is possible that replication of the viral genome at particular foci, i.e. the B19V replication centers,3 recruits the ESE3/ISE2-binding splice factors to the active spliceosome; alternatively, the viral replication centers may be already enriched for these factors (39).
In conclusion, our results have shown that polyadenylation of B19V pre-mRNA at the (pA)p site is facilitated by efficient splicing of the upstream intron and inhibited by splicing within the intron in which the (pA)p site is located. We propose an exon definition model of B19V pre-mRNA processing (Fig. 8), in which the long exon 2′ (from the A1-1 or A1-2 site to the (pA)p site; Fig. 8A) cannot be defined when the short exon 2 (from the A1-1 or A1-2 site to the D2 site; Fig. 8B) is defined. Binding of the U1 RNA to the D2 donor site, which promotes definition of the short Exon 2 through binding of the U2AF65 to the A2-1 or A2-2 site, is fully responsible for the inhibition of polyadenylation of B19V pre-mRNA at the (pA)p site (Fig. 8B). Because splicing of the first and second introns to define the short exon 2 is regulated by several splicing enhancers lie in this exon (18), we hypothesize that the internal polyadenylation efficiency of B19V pre-mRNA depends on these serine-arginine-rich splicing factors binding to the splicing enhancers. Thus, studying alternative processing of the B19V pre-mRNA has revealed the basic mechanism underlying how alternative splicing coordinates alternative polyadenylation to define an exon.
Acknowledgment
We are indebted to Dr. Steve Kleiboeker at ViraCor-IBT (Lee's Summit, MO) for providing B19V plasma samples.
This work was supported, in whole or in part, by National Institutes of Health USPHS Grant R01 AI070723 from NIAID and Grant P20 RR016443 from the National Center for Research Resources Centers of Biomedical Research Excellence Program.
Y. Luo and J. Qiu, unpublished observations.
- B19V
- human parvovirus B19
- (pA)p
- polyadenylation at the proximal site
- (pA)d
- polyadenylation at the distal site
- pre-mRNA
- precursor message RNA
- nt
- nucleotide(s)
- DSE
- downstream element
- EPC
- erythroid progenitor cell
- ISE
- intronic splicing enhancer.
REFERENCES
- 1. Young N. S., Brown K. E. (2004) N. Engl. J. Med. 350, 586–597 [DOI] [PubMed] [Google Scholar]
- 2. Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (2004) Virus Taxonomy, VIIIth Report of the ICTV, pp. 277–370, Elsevier/Academic Press, London [Google Scholar]
- 3. Cotmore S. F., Tattersall P. (2005) in Parvoviruses (Kerr J., Cotmore S. F., Bloom M. E., Linden R. M., Parrish C. R. eds) pp. 73–94, Hodder Arnold, London [Google Scholar]
- 4. Ozawa K., Kurtzman G., Young N. (1986) Science 233, 883–886 [DOI] [PubMed] [Google Scholar]
- 5. Srivastava A., Lu L. (1988) J. Virol. 62, 3059–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sol N., Le Junter J., Vassias I., Freyssinier J. M., Thomas A., Prigent A. F., Rudkin B. B., Fichelson S., Morinet F. (1999) J. Virol. 73, 8762–8770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wong S., Zhi N., Filippone C., Keyvanfar K., Kajigaya S., Brown K. E., Young N. S. (2008) J. Virol. 82, 2470–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qiu J., Cheng F., Johnson F. B., Pintel D. (2007) J. Virol. 81, 12080–12085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun Y., Chen A. Y., Cheng F., Guan W., Johnson F. B., Qiu J. (2009) J. Virol. 83, 3956–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qiu J., Cheng F., Burger L. R., Pintel D. (2006) J. Virol. 80, 654–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ozawa K., Ayub J., Hao Y. S., Kurtzman G., Shimada T., Young N. (1987) J. Virol. 61, 2395–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Z., Qiu J., Cheng F., Chu Y., Yoto Y., O'Sullivan M. G., Brown K. E., Pintel D. J. (2004) J. Virol. 78, 12929–12939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen A. Y., Zhang E. Y., Guan W., Cheng F., Kleiboeker S., Yankee T. M., Qiu J. (2010) Blood 115, 1070–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guan W., Cheng F., Yoto Y., Kleiboeker S., Wong S., Zhi N., Pintel D. J., Qiu J. (2008) J. Virol. 82, 9951–9963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu J. M., Green S. W., Shimada T., Young N. S. (1992) J. Virol. 66, 4686–4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. St Amand J., Beard C., Humphries K., Astell C. R. (1991) Virology 183, 133–142 [DOI] [PubMed] [Google Scholar]
- 17. Yoto Y., Qiu J., Pintel D. J. (2006) J. Virol. 80, 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guan W., Cheng F., Huang Q., Kleiboeker S., Qiu J. (2011) J. Virol. 85, 2463–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhi N., Zádori Z., Brown K. E., Tijssen P. (2004) Virology 318, 142–152 [DOI] [PubMed] [Google Scholar]
- 20. Guan W., Wong S., Zhi N., Qiu J. (2009) J. Virol. 83, 9541–9553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhi N., Mills I. P., Lu J., Wong S., Filippone C., Brown K. E. (2006) J. Virol. 80, 5941–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Valcárcel J., Gaur R. K., Singh R., Green M. R. (1996) Science. 273, 1706–1709 [DOI] [PubMed] [Google Scholar]
- 23. Levitt N., Briggs D., Gil A., Proudfoot N. J. (1989) Genes Dev. 3, 1019–1025 [DOI] [PubMed] [Google Scholar]
- 24. Zhuang Y., Weiner A. M. (1986) Cell. 46, 827–835 [DOI] [PubMed] [Google Scholar]
- 25. Morita E., Tada K., Chisaka H., Asao H., Sato H., Yaegashi N., Sugamura K. (2001) J. Virol. 75, 7555–7563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wong S., Brown K. E. (2006) J. Clin. Virol. 35, 407–413 [DOI] [PubMed] [Google Scholar]
- 27. Chen A. Y., Guan W., Lou S., Liu Z., Kleiboeker S., Qiu J. (2010) J. Virol. 84, 12385–12396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Naeger L. K., Schoborg R. V., Zhao Q., Tullis G. E., Pintel D. J. (1992) Genes Dev. 6, 1107–1119 [DOI] [PubMed] [Google Scholar]
- 29. Schoborg R. V., Pintel D. J. (1991) Virology. 181, 22–34 [DOI] [PubMed] [Google Scholar]
- 30. Qiu J., Pintel D. J. (2004) J. Biol. Chem. 279, 14889–14898 [DOI] [PubMed] [Google Scholar]
- 31. Ashe M. P., Griffin P., James W., Proudfoot N. J. (1995) Genes Dev. 9, 3008–3025 [DOI] [PubMed] [Google Scholar]
- 32. Barksdale S., Baker C. C. (1995) J. Virol. 69, 6553–6556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gilmartin G. M., Hung S. L., DeZazzo J. D., Fleming E. S., Imperiale M. J. (1996) J. Virol. 70, 1775–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qiu J., Nayak R., Pintel D. J. (2004) J. Virol. 78, 83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Millevoi S., Loulergue C., Dettwiler S., Karaa S. Z., Keller W., Antoniou M., Vagner S. (2006) EMBO J. 25, 4854–4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vagner S., Vagner C., Mattaj I. W. (2000) Genes Dev. 14, 403–413 [PMC free article] [PubMed] [Google Scholar]
- 37. Kaida D., Berg M. G., Younis I., Kasim M., Singh L. N., Wan L., Dreyfuss G. (2010) Nature. 468, 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fortes P., Cuevas Y., Guan F., Liu P., Pentlicky S., Jung S. P., Martínez-Chantar M. L., Prieto J., Rowe D., Gunderson S. I. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8264–8269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. de Jong L., Grande M. A., Mattern K. A., Schul W., van Driel R. (1996) Crit. Rev. Eukaryot. Gene Expr. 6, 215–246 [DOI] [PubMed] [Google Scholar]