Abstract
Krüppel-like factors (KLFs) control cell differentiation and embryonic development. KLF1 (erythroid Krüppel-like factor) plays essential roles in embryonic and adult erythropoiesis. KLF2 is a positive regulator of the mouse and human embryonic β-globin genes. KLF1 and KLF2 have highly homologous zinc finger DNA-binding domains. They have overlapping roles in embryonic erythropoiesis, as demonstrated using single and double KO mouse models. Ablation of the KLF1 or KLF2 gene causes embryonic lethality, but double KO embryos are more anemic and die sooner than either single KO. In this work, a dual human β-globin locus transgenic and KLF knockout mouse model was used. The results demonstrate that the human ϵ- (embryonic) and γ-globin (fetal) genes are positively regulated by KLF1 and KLF2 in embryos. Conditional KO mouse experiments indicate that the effect of KLF2 on embryonic globin gene regulation is at least partly erythroid cell-autonomous. KLF1 and KLF2 bind directly to the promoters of the human ϵ- and γ-globin genes, the mouse embryonic Ey- and βh1-globin genes, and also to the β-globin locus control region, as demonstrated by ChIP assays with mouse embryonic blood cells. H3K9Ac and H3K4me3 marks indicate open chromatin and active transcription, respectively. These marks are diminished at the Ey-, βh1-, ϵ- and γ-globin genes and locus control region in KLF1−/− embryos, correlating with reduced gene expression. Therefore, KLF1 and KLF2 positively regulate the embryonic and fetal β-globin genes through direct promoter binding. KLF1 is required for normal histone modifications in the β-globin locus in mouse embryos.
Keywords: Embryo, Erythropoiesis, Transcription Factors, Transgenic, Zinc Finger
Introduction
Erythroid cells are one of the first differentiated cell types in embryos (1). There are two unique processes in development: primitive and definitive erythropoiesis. Primitive erythropoiesis initiates from the extraembryonic mesoderm of the yolk sac as early as embryonic day 7.5 (E7.5)2 in mice (2, 3). Definitive erythropoiesis is detected in the mouse fetal liver by E11.5 (4). The human β-globins are encoded by four major genes, ϵ (embryonic), Gγ and Aγ (fetal), and β (adult), located on chromosome 11. The mouse β-globin locus contains four genes, two embryonic (Ey and βh1), and two adult (βmaj and βmin).
The expression of the β-globin genes is jointly regulated by elements in the promoter regions and an upstream enhancer region, the locus control region (LCR). The human β-globin LCR, located 6–22 kb upstream of the ϵ-globin gene, contains multiple erythroid-specific DNase I hypersensitive sites (HS) and plays a crucial role in maintaining β-globin gene expression (5, 6). Examples of regulatory elements within the promoters of all β-globin genes are TATA, CAAT, and CACCC (7).
Krüppel-like factors (KLFs) are a family of transcription factors that bind GC-rich sequences such as CACCC elements. The KLFs bind DNA via three carboxyl-terminal Cys-2/His-2 zinc fingers (8). Seventeen mammalian proteins have been identified in this family and are designated KLF1 to KLF17. KLFs are implicated in many cellular functions, such as erythropoiesis, cell differentiation, proliferation, and tissue development (9). The human and mouse KLF proteins are highly conserved. For example, KLF1 is 73% similar in the two species (although 90% similar within the zinc finger domain), and KLF2 is 90% similar in mouse and man.
KLF1, also known as erythroid Krüppel-like factor or EKLF, is expressed only in erythroid cells and plays essential roles in embryonic and adult β-globin gene expression (10–12). KLF1 is a master regulator of adult β-globin gene expression (10, 11). Semiquantitative ChIP assays revealed that HA-tagged KLF1 binds to the promoters of the embryonic β-like globin genes (Ey and βh1), to HS1, HS2, HS3, and HS5 in mouse primitive erythroid cells, and to the promoter of the mouse adult βmaj-globin gene in primitive and definitive cells (13). KLF1 interacts with CBP, p300, and PCAF, which have histone acetyltransferase activity (14). KLF1 is also implicated in erythroid processes other than β-globin gene regulation, such as cell maturation and cell membrane integrity (15, 16).
KLF2, originally known as lung KLF or LKLF, has important roles in T-cell differentiation and blood vessel development (9). KLF2 is a positive regulator of the mouse and human embryonic β-globin genes (17). KLF1 and KLF2 have high homology within their DNA-binding domains and reside close to each other on the same chromosome in human and mouse, suggesting that they originated from a gene duplication event (17). KLF1 and KLF2 can partially functionally compensate for each other in regulating the mouse embryonic β-globin genes. When both KLF1 and KLF2 are simultaneously ablated in mice, the amounts of Ey- and βh1-globin mRNA are reduced more than in KLF1 or KLF2 single knockouts (12). Like KLF1, KLF2 recruits proteins with histone acetyltransferase activity such as CBP, p300 and PCAF (18).
KLF1 and KLF2 coordinately regulate the mouse embryonic β-globin genes. In this study, we wished to determine whether KLF1 and KLF2 also control the human embryonic and fetal β-globin genes. This knowledge could facilitate therapeutic strategies to express these genes in adults with β-hemoglobinopathies. A second goal was to further establish the mechanistic roles of KLF1 and KLF2 in globin gene regulation. In this work, it was determined that KLF1 and KLF2 mRNA are expressed in similar amounts in mouse primitive erythroid cells. The expression of KLF1 but not KLF2 mRNA is greatly increased in definitive erythroid cells. Transgenic mice that harbor the complete human β-globin locus (19, 20), in conjunction with gene knockouts, were used to determine that KLF1 and KLF2 positively regulate human ϵ- and γ-globin gene expression in the embryo. KLF2 has an erythroid cell autonomous role in embryonic globin gene regulation, although it may also have non-cell-autonomous functions. In quantitative ChIP assays, KLF1 and KLF2 bind to the promoters of the mouse embryonic Ey- and βh1-globin and the human ϵ- and γ-globin genes in mouse primitive erythroid cells. KLF1, but not KLF2, is required to establish the normal histone modification status in the mouse and human β-globin loci in mouse primitive erythroid cells. Although both KLF1 and KLF2 directly affect globin gene regulation through the same DNA binding sites, their mechanisms of action appear to be somewhat different.
EXPERIMENTAL PROCEDURES
Generation of Knockout and Transgenic Mice
The KLF1 KO mouse model was developed by targeting the gene with the neomycin resistance gene (10). The KLF2 KO mouse model was developed by targeting the gene with the hypoxanthine phosphoribosyl-transferase (Hprt) gene (21). Because KLF1 and KLF2 are located in close proximity on mouse chromosome 8, recombination between the two genes is rare. Therefore, a mouse model that has the KLF1 and KLF2 KO alleles on the same DNA homolog was generated (12). Transgenic mice that carry the entire human β-globin locus (Tg-HBB) were described previously (19, 20). Tg-HBB mice were bred with KLF1+/− or KLF2+/− mice to obtain KLF1+/−Tg-HBB, KLF2+/−Tg-HBB, and KLF1+/−KLF2+/−Tg-HBB mice. Mouse embryonic yolk sacs and blood cells were collected as described (12).
For the conditional KO studies, mice with a floxed KLF2 allele were obtained from Dr. Jerry Lingrel (22). The ErGFP-Cre transgenic mice were described previously (23). Mice with a floxed KLF2 allele (KLF2F/+ mice) were mated with transgenic mice carrying an improved GFP-Cre fusion gene under the control of the endogenous erythropoietin receptor (EpoR) promoter (ErGFP-Cre mice) to obtain KLF2F/+, ErGFP-Cre mice.
RNA Preparation and cDNA Synthesis
Total RNA from yolk sacs, embryonic blood, or fetal liver cells was prepared using the TōTALLY RNAtm kit (Applied Biosystems, Foster City, CA). RNA concentrations were determined using a Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE) or an Agilent bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA). The integrity of RNA was assayed using gel electrophoresis or an Agilent bioanalyzer. cDNA was synthesized using the iScripttm cDNA synthesis kit (Bio-Rad).
Quantitative Reverse Transcriptase PCR (qRT-PCR)
Mouse KLF1, KLF2, cyclophilin A, glycophorin A (GPA), Ey- and βh1-globin, and human ϵ- and γ-globin mRNA amounts were quantified using qRT-PCR with SYBR Green or Taqman reagents (Applied Biosystems). Mouse cyclophilin A and GPA mRNA were used as internal standards for normalization as indicated in the figure legends. GPA is erythroid-specific and could therefore be used in the assays with yolk sac mRNA to correct for any reduction in the number of red blood cells in mutant compared with WT embryos. qRT-PCR was performed using an ABI Prism 7300 analyzer (Applied Biosystems). For quantification using SYBR Green chemistry, a dissociation step was performed, and it was verified that only one product was amplified. A standard curve from pooled cDNA samples was included in each run and used either to measure the relative amounts of unknown samples or to obtain the efficiency of specific primers (Efficiency [E] = 10^(-1/slope)). Fold change (relative expression) was calculated as E^Ctendogenous gene/E^Cttest gene (for Fig. 1). Primer and probe sequences are indicated in supplemental Table 1. Statistical significance was calculated using the Student's t test.
FIGURE 1.
Developmental expression patterns of KLF1 and KLF2 mRNA. Expression of KLF1 and KLF2 mRNA during primitive (E9.5 blood) and definitive (E12.5 fetal liver) erythropoiesis. Erythroid cells from E9.5 circulating blood and E12.5 fetal liver are in similar stages of differentiation. The amounts of mouse KLF1 and KLF2 mRNA were measured using qRT-PCR and normalized to cyclophilin A. Fold change was calculated using the 2ΔCT method after correcting for different primer efficiency. At least four biological replicates were tested at each time point. *, p <0.05). Error bars, S.D.
ChIP
ChIP assays were performed essentially as described previously (24). Briefly, for each biological replicate, ∼5 × 106 blood cells from Tg-HBB embryos were cross-linked with 1% formaldehyde for 10 min at room temperature. Cross-linking was stopped by adding glycine. Nuclei were purified and lysed to collect chromatin. Chromatin was sheared to ∼500 bp using a Bioruptor sonicator (Diagenode, Sparta, NJ). Chromatin was precleared using protein G (Millipore, catalog no. 16–266), and equal aliquots were incubated with either specific antibody (Ab) or nonspecific IgG. Precipitated chromatin was washed, and cross-links were reversed. DNA was purified and analyzed using quantitative PCR (qPCR) and SYBR Green chemistry. Fold enrichment was calculated as 2^(Ctinput − Cttest) and expressed relative to the IgG control. Antibodies used were anti-H3K9Ac (Abcam, catalog no. Ab4441), anti-H3K4me3 (Upstate-Millipore, catalog no. 07–473), anti-KLF1 (Abcam, catalog no. AB-2483), anti-KLF2 (KLF2_Ng (25), anti-KLF2 (KLF2-SC, Santa Cruz Biotechnology, catalog no. sc-18690), nonspecific IgG (rabbit, Abcam, catalog no. ab46540, or goat, Santa Cruz Biotechnology, catalog no. sc-2028). For optimal results, the KLF2_Ng antibody is recommended. Primer sequences for qPCR are indicated in supplemental Table 2.
RESULTS
KLF1 and KLF2 mRNA Amounts Are Similar in Primitive but Not Definitive Erythroid Cells
KLF1 is a major regulator of the human and mouse adult β-globin genes (10, 11), whereas KLF2 regulates embryonic but not adult β-globin gene expression (17). The relative amounts of KLF1 and KLF2 mRNA were compared in primitive and definitive erythroid cells matched for similar stages of differentiation. E9.5 blood (primitive) and E12.5 fetal liver (definitive) mRNA were used for the comparison because both contain mostly basophilic erythroblasts (26, 27). The vast majority of E12.5 fetal liver cells are erythroid (26, 27). qRT-PCR results indicate that KLF1 and KLF2 mRNAs are expressed in similar amounts at E9.5 (Fig. 1). The amount of KLF1 mRNA dramatically increases by E12.5, whereas KLF2 mRNA remains relatively unchanged. The ratio of KLF1 to KLF2 mRNA at E9.5 is less than 2, but it is more than 16 at E12.5. The developmental expression pattern of KLF1 and KLF2 mRNA correlates with globin gene expression. KLF1 and KLF2 regulate the mouse Ey- and βh1-globin genes that are expressed at E9.5, whereas KLF1, but not KLF2, regulates the adult β-globin genes expressed at E12.5 (12, 17).
KLF1 and KLF2 Regulate Human ϵ- and γ-Globin Gene Expression
KLF1 and KLF2 regulate the mouse embryonic β-globin genes, Ey and βh1, during primitive erythropoiesis (12). When KLF1 and KLF2 are simultaneously ablated, Ey- and βh1-globin mRNA amounts are reduced by severalfold to less than amounts measured in either single KO (12). The human embryonic and fetal β-globin genes, ϵ and γ, are both expressed at E10.5 in transgenic mouse models (28). To test whether KLF1 and KLF2 regulate human β-globin gene expression in the embryo, dual human β-globin locus transgenic (Tg-HBB) and KO mice were used. Dual Tg-HBB and heterozygous KO mice were crossed with heterozygous KO mice to obtain E10.5 KLF1−/− Tg-HBB, KLF2−/− Tg-HBB, and KLF1−/−KLF2−/− Tg-HBB embryos. qRT-PCR was used to measure the amounts of human ϵ- and γ-globin mRNA in mutant and WT E10.5 yolk sacs. GPA is a cell surface protein expressed exclusively on erythroid cells late in differentiation. GPA mRNA was used as an internal standard to which ϵ- and γ-globin mRNA amounts were normalized. GPA mRNA amounts in KLF1−/−KLF2−/− (double KO) and WT yolk sacs are not significantly different (12), but there is a modest elevation in GPA mRNA in KLF1−/− compared with WT yolk sacs (data not shown).
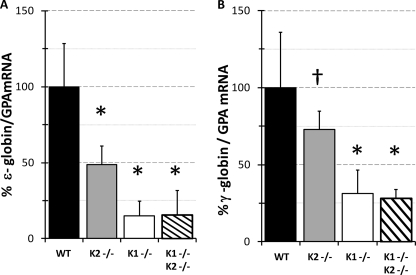
ϵ-globin mRNA is significantly reduced to 15 and 49% in KLF1−/− Tg-HBB and KLF2−/− Tg-HBB yolk sacs, respectively, compared with Tg-HBB (p < 0.025) (Fig. 2A). γ-globin mRNA was reduced to 31% of Tg-HBB in KLF1−/− Tg-HBB yolk sacs (p < 0.025) (Fig. 2B). KLF2 has a more modest effect on γ-globin gene expression (17), which is reduced to 73% of Tg-HBB in KLF2−/− Tg-HBB yolk sacs (p < 0.05) (Fig. 2B). KLF1 appears to have a greater effect on ϵ- and γ-globin gene expression, but KLF2 also contributes. The quantity of ϵ- and γ-globin mRNA in KLF1−/− Tg-HBB yolk sacs may be marginally underestimated compared with KLF1−/−KLF2−/− Tg-HBB. Thus, we cannot rule out possible synergistic regulation of the human ϵ- and γ-globin genes by KLF1 and KLF2 in the transgenic mouse model.
FIGURE 2.
KLF1 and KLF2 positively regulate human embryonic and fetal β-globin gene expression. Human embryonic and fetal β-globin gene expression in yolk sacs from KLF1−/−KLF2−/− at E10.5 compared with wild-type and single knockouts. A, ϵ-globin mRNA. B, γ-globin mRNA. GPA mRNA was used as an internal standard for quantitative RT-PCR. The globin-to-GPA mRNA ratio for WT was taken as 100% and for the other genotypes is expressed compared with WT. The KLF2−/− data comes from a previous study (17). †, p < 0.05; *, p < 0.025 compared with WT. Between four and six biological replicates were used. Error bars, S.D. K1−/−, KLF1−/−; K2−/−, KLF2−/−.
KLF2 Regulation of Murine Ey- and βh1-Globin Gene Expression Is Cell-autonomous
KLF1 has an erythroid-specific expression pattern, but KLF2 is expressed in numerous cell types, including erythroid and endothelial cells. To determine whether regulation of the mouse embryonic globin genes by KLF2 is erythroid cell-autonomous, a conditional KO mouse model was used in which the KLF2 gene is deleted in erythroid cells. Erythroid cell-autonomous effects are defined here as being due to loss of KLF2 in erythroid cells and not indirectly due to KLF2 ablation in another cell type. KLF2F/+, ErGFP-Cre and KLF2F/F mice were mated to obtain E10.5 KLF2F/F, ErGFP-Cre (test), and KLF2F/F (control) embryos. The ErGFP-Cre transgene is active in primitive erythroid cells (23, 29). RNA prepared from peripheral blood cells was used to quantify Ey- and βh1-globin expression by qRT-PCR. The amount of globin mRNA in KLF2F/F, ErGFP-Cre, and KLF2F/F (without Cre) was compared using five to eight biological replicates for each genotype. Cyclophilin A mRNA was used as an internal standard because it is expressed at the same level in test and control embryonic blood (data not shown). The use of peripheral blood rather than yolk sac in these experiments overcomes the need for an erythroid cell-specific internal standard.
As seen in Fig. 3, there is significantly less βh1-globin mRNA in KLF2F/F, ErGFP-Cre compared with KLF2F/F (without Cre) embryos (p < 0.05). The βh1-globin mRNA was reduced by ∼25% in KLF2F/F, ErGFP-Cre embryonic blood cells (Fig. 3A). There is a similar decreasing trend in Ey-globin mRNA in KLF2F/F, ErGFP-Cre compared with KLF2F/F embryos (Fig. 3B), although this difference is not statistically significant. These modest differences are consistent with the fact that there is ∼40% less KLF2 mRNA in KLF2F/F, ErGFP-Cre than in KLF2F/F embryos (Fig. 3C), making these embryos more comparable with KLF2+/− than to KLF2−/− (17). Although whole-mount staining of an ErGFP-Cre E13.5 embryo indicated transgene expression in a single blood vessel in the head, transgene expression is predominantly erythropoietic (23). The modest effect of the conditional knockout on globin gene regulation is likely due to inefficient excision of the KLF2 gene by the Cre recombinase. KLF2 very likely has an erythroid-cell autonomous role in the regulation of the βh1- and Ey-globin genes. However, the possibility that KLF2 also has non-cell-autonomous functions in globin gene expression cannot be excluded.
FIGURE 3.
Erythroid cell-autonomous regulation of mouse βh1- and Ey-globin expression by KLF2. Mouse embryonic globin mRNA was quantified in KLF2F/F, ErGFP-Cre (erythroid conditional knockout), and KLF2F/F (wild-type) mouse E10.5 erythroid cells. Cyclophilin A mRNA was used as an internal standard for qRT-PCR. The globin-to-cyclophilin mRNA ratio for KLF2F/F was taken as 100%. Shown are mouse embryonic βh1-globin (A), Ey-globin (B), and KLF2 mRNA (C). *, p < 0.05. For KLF2F/F, n = 5 or 6 and for KLF2F/F,ErGFP-Cre, n = 8. Error bars, S.D.
KLF1 and KLF2 Occupy the Embryonic and Fetal β-Globin Promoters and the LCR
KLF1 and KLF2 control human and mouse embryonic β-globin gene expression. To better understand the mechanism for this control, ChIP assays using polyclonal antibodies against KLF1 and KLF2 were performed on Tg-HBB transgenic mice, which have the human β-globin locus. Unlike definitive erythroid cells, which are enucleated, circulating primitive erythroid cells are nucleated at E10.5 and E11.5, making them amenable to ChIP analyses. The amounts of Ey-, γ-, and ϵ-globin mRNAs do not change appreciably between E10.5 and E11.5 in mouse embryos, although γ-globin is expressed more than ϵ-globin mRNA. However, the amount of βh1-globin mRNA at E11.5 is ∼25% lower than at E10.5 (20). To capture data for the βh1-globin gene, ChIP assays were performed at E10.5 where feasible, although less cells per embryo were available than at E11.5.
First, the specificity of the commercial KLF1 antibody was confirmed in ChIP assays using WT and KLF1−/− E13.5 fetal liver cells. The antibody is specific and binds to KLF1 in the promoters of the adult β-globin genes in the mouse and human β-globin loci in WT, but not in KLF1−/− fetal livers (supplemental Fig. S1). At E10.5, KLF1 is significantly enriched at the promoters of the Ey- and βh1-globin genes and at mouse 5′HS2 in the LCR (Fig. 4A, right panel). In the human β-globin locus, KLF1 is significantly enriched at the promoter of the γ-globin gene, 5′HS2, and 5′HS3 (Fig. 4A, left panel). At E11.5, the pattern of KLF1 enrichment at the mouse and human β-globin loci is similar to that at E10.5, except that binding of KLF1 to the ϵ-globin promoter is also evident at E11.5, perhaps because of increased sensitivity of the assay because of more available cells at this time point (Fig. 4B). As a negative control, KLF1 does not bind to the promoter of β-actin at E10.5 and E11.5. The ChIP assays indicate that KLF1 binds to all of the mouse and human embryonic and fetal β-globin gene promoters and to the LCR.
FIGURE 4.
KLF1 and KLF2 bind the mouse and human β-globin loci in primitive erythroid cells. ChIP assays were performed on E10.5 (A) or E11.5 (B and C) primitive erythroid cells of normal mice or transgenic mice that carry the entire human β-globin locus. Polyclonal antibodies specific for KLF1 (A and B) or KLF2 (C) and nonspecific IgG control antibody were used. The y axis represents the relative fold enrichment. The mean IgG enrichment was set as 1.0, and the enrichment of KLF1 was scaled appropriately. The x axis shows the location of the primers used for qPCR. Pr, promoter. The primers were specific to the DNase I hypersensitive sites 5′HS2 and 5′HS3, the promoters of the mouse (Ey-, βh1-, βmaj-) and the human β-globin genes (ϵ, γ, and β). Primers specific to β-actin were used as negative controls. *, significant enrichment compared with IgG (p <0.05). A, KLF1 ChIP on E10.5 erythroid cells, n = 3. B, KLF1 ChIP on E11.5 erythroid cells, n = 3. C, KLF2 ChIP on E11.5 erythroid cells. Two KLF2 antibodies were used on the mouse β-globin locus (one was from Santa Cruz, and the other was a gift from Dr. Ng). Mouse locus and β-actin, n = 4 for IgG and KLF2_Ng; n = 2 for KLF2_SC; human locus, n = 2. Error bars, S.E.
At E11.5, KLF2 is detected at the Ey- but not the βh1-globin promoter (Fig. 4C, right panel), which is consistent with higher expression of the Ey- than the βh1-globin gene at this time point (20). There was no evidence that KLF2 binds to 5′HS2 or 5′HS3 in the mouse β-globin LCR. In the human β-globin locus, KLF2 is enriched by about 2-fold at the γ-globin promoter and at 5′HS2 and 5′HS3 (Fig. 4C, left panel). Although the KLF2 binding measured at the ϵ-globin promoter was not statistically different from the negative control, it is approaching significance. The lower amount of expression of the ϵ- compared with the γ-globin gene in transgenic mice at E11.5 may have decreased the sensitivity of the ChIP assay. KLF2 binds to 5′HS2 and 5′HS3 in the human β-globin locus, which differs from data obtained by examining the mouse locus. These results strongly suggest that regulation of the embryonic and fetal β-globin genes by KLF1 and KLF2 is achieved by direct binding to the CACCC elements in the promoters and LCR. Binding of KLF1 and KLF2 to the LCR could be necessary for direct contact between the LCR and the β-globin gene promoters, as in adult erythroid cells (30).
KLF1 Affects Histone Modifications at the β-Globin Locus in Embryos
Histone modifications correlate with the state of gene transcription. In general, acetylated histones mark actively transcribed loci (31). More specifically, histone 3 lysine 9 acetylation (H3K9Ac) marks chromatin regions of open conformation, whereas histone 3 lysine 4 trimethylation (H3K4me3) marks chromatin regions of active transcription (31, 32). KLF1 and KLF2 interact with histone acetyltransferase cofactors such as CBP, p300, and PCAF (14, 18). To determine whether the absence of KLF1 and KLF2 in the blood cells from mutant embryos disrupts the normal deposition of histone marks at the mouse and human β-globin loci, ChIP assays were performed with antibodies specific to H3K9Ac and H3K4me3. These histone marks are mostly enriched within the second or third exons of the β-globin genes (33, 34).
At E10.5, the mouse Ey- and βh1-globin and human ϵ- and γ-globin mRNAs are expressed. As expected, in normal cells H3K9Ac (Fig. 5A) and H3K4me3 (B), marks were relatively enriched at the expressed genes compared with nonspecific antibodies. Surprisingly, H3K4me3 is also enriched in the adult mouse β-globin gene, βmaj, although it is not yet expressed (Fig. 5B). It is possible that H3K4me3 marks this gene prior to transcription. At the LCR, the amount of H3K9Ac and H3K4me3 is generally less than at the globin genes, but is enriched compared with IgG.
FIGURE 5.
Differential enrichment of H3K9Ac and H3K4me3 at the mouse and human β-globin loci in WT, KLF1−/− and KLF2−/− primitive erythroid cells. ChIP assays using anti-H3K9Ac (A and C) or anti-H3K4me3 (B and D) were performed on E10.5 erythroid cells from WT, KLF1−/− (A and B), or KLF2−/− (C and D) embryos with the human β-globin locus. H3K9Ac generally indicates open chromatin conformation, whereas H3K4me3 indicates active transcription. The mean IgG enrichment was set as 1.0, and the enrichment of H3K9Ac or H3K4me3 were scaled appropriately. Necdin was used as a negative control. Ex, exonic region. n = 3. Error bars, S.E.; *, significant enrichment compared with IgG (p <0.05).
When KLF1 is ablated, the enrichment of H3K9Ac is significantly reduced in the mouse Ey- and βh1-globin and human ϵ- and γ-globin genes (Fig. 5A). Similarly, the enrichment of H3K4me3 is significantly reduced in the Ey-, βh1-, and ϵ-globin genes when KLF1 is ablated (Fig. 5B). The amount of H3K4me3 in the γ-globin gene is modestly but not significantly reduced in the absence of KLF1, although γ-globin mRNA is significantly reduced in KLF1−/− compared with WT primitive erythroid cells. In the absence of KLF1, the amount of H3K9Ac and H3K4me3 at mouse and human 5′HS2 and 5′HS3 is also significantly decreased compared with WT (Fig. 5, A and B). The mouse necdin gene is a neuron-specific gene that is not expressed in erythroid cells (35). The amounts of H3K9Ac and H3K4me3 at the necdin gene are low and not different in WT and KLF1−/− primitive blood cells. The lower abundance of H3K9Ac and H3K4me3 at the mouse and human β-globin genes and LCR in KLF1−/− compared with WT cells correlates with reduced transcription.
KLF2 does not affect histone modifications to the same extent as KLF1. Most of the sites in the β-globin locus that were tested do not exhibit differences in H3K9Ac and H3K4me3 marks between WT and KLF2−/− primitive erythroid cells (Fig. 5, C and D). However, the H3K9Ac and H3K4me3 enrichment is reduced in KLF2−/− compared with WT at the adult human β-globin gene. Unexpectedly, H3K4me3 enrichment is higher at the Ey-globin gene in KLF2−/− compared with WT erythroid cells (Fig. 5D).
DISCUSSION
It was originally believed that KLF1 is required only for adult β-globin gene expression because KLF1 KO mice die just after the embryonic to adult β-globin gene switch (10, 11). However, more quantitative mRNA analyses in KLF1 KO mouse embryos established that KLF1 regulates mouse Ey- and βh1-globin gene expression in primitive erythroid cells (12). KLF2 also has a role in mouse Ey- and βh1-globin gene regulation (17). Furthermore, in transgenic mouse models, KLF2 regulates the human ϵ- and γ-globin genes in the E10.5 yolk sac (17). Here, we established that KLF1 is also required for normal expression of the human embryonic ϵ- and fetal γ-globin genes during mouse primitive erythropoiesis.
It is shown here that KLF2 has an erythroid cell autonomous role in mouse embryonic βh1-globin gene expression, as would be expected if it directly regulates the globin genes. Furthermore, KLF1 and KLF2 occupy the promoters of the mouse Ey- and βh1-globin and human ϵ- and γ-globin genes, supporting their direct roles in globin gene regulation. The ChIP data provides the first quantitative evidence that KLF1 binds to the β-globin promoters during primitive erythropoiesis. We did not detect KLF1 at the promoter of the βmaj-globin gene, as was reported in semiquantitative assays using a tagged KLF1 knockin in primitive erythroid cells (13). KLF2 directly binds to the promoters/enhancers of key regulators of stem cell pluripotency in ES cells (25). Our data shows that KLF2 is directly recruited to the promoters of the murine and human embryonic and fetal β-globin genes in native primitive erythroid cells.
Our work indicates that KLF1 acts directly as a positive regulator of the human ϵ- and γ-globin and mouse Ey- and βh1-globin genes during embryonic erythropoiesis. This is in contrast to increasing evidence that has recently emerged, supporting an indirect negative role for KLF1 in γ-globin gene regulation during adult erythropoiesis. Certain mutations in the human KLF1 gene have been correlated with hereditary persistence of fetal hemoglobin (36–39). An E325K mutation was detected in patients with congenital dyserythropoietic anemia who express increased amounts of HbF (36). A mutation in the analogous residue, E339D, occurs in the Nan mouse, which has an increase in βh1-globin expression in the fetal liver and adult spleen (37). A heterozygous K288X mutation is found in a Maltese family with hereditary persistence of fetal hemoglobin. The mutation eliminates the KLF1 zinc fingers and hence abrogates DNA binding (38). Interestingly, an S270X mutation in a Sardinian family does not cause an increase in HbF even though it eliminates the zinc fingers (39). A compound heterozygote with the S270X and a K332Q mutation does have hereditary persistence of fetal hemoglobin (39). The mechanism for the negative effect of KLF1 on γ-globin gene regulation in the adult is most probably indirect via up-regulation of BCL11A (38, 40). Apparently, KLF1 can positively or negatively affect γ-globin gene regulation, depending on the erythroid cell milieu.
The expression patterns of KLF1 and KLF2 in primitive and definitive erythroid cells were analyzed and reveal a possible explanation for the different milieu at the two stages. The ratio of KLF1 to KLF2 mRNA increases dramatically as erythroid cells switch from the primitive to the definitive stage. KLF1 acts as a repressor of megakaryocytic differentiation genes and therefore drives megakaryocyte-erythroid progenitor cells toward erythroid differentiation (41). It is possible that KLF1 is also involved in the switch from primitive to definitive erythropoiesis. It is plausible that KLF2 drives erythroid cells toward embryonic globin gene expression, opposing the role of KLF1.
KLF1 and KLF2 have a high degree of homology in their zinc finger domains and can partially compensate for each other in embryonic erythroid cells, probably because they regulate common target genes (12). Functional overlap between family members of transcription factors is common. GATA1 and GATA2, for example, can compensate fully or partially for each other during erythropoiesis (42, 43). Primitive erythroid cells are present in GATA1 KO embryos. GATA2 KO embryos show a modest reduction in the number of primitive erythroid precursors. However, when both GATA1 and GATA2 are ablated, no primitive erythroid cells are detected (42). When expressed under the control of the GATA1 hematopoietic regulatory domain, GATA2 can rescue the GATA1 KO phenotype (44). Moreover, cross-regulation can occur between transcription factor family members. KLF2, for example, positively regulates KLF1 in primitive erythroid cells (12). KLF8 is up-regulated by KLF1 and down-regulated by KLF3 (45). KLF1 also can activate KLF3 by directly binding to its promoter (46). GATA1 negatively regulates GATA2 in definitive erythroid cells and megakaryocytes (47–49).
Our data indicate that at E10.5, H3K9Ac, and H3K4me3 are enriched at the actively transcribed globin genes, mouse Ey and βh1 and human ϵ and γ. Previous reports have shown that H3K9Ac and H3K4me3 marks correlate with each other in the human β-globin locus during adult erythropoiesis (33). Our work provides the first demonstration in native embryonic erythroid cells that H3K9Ac is associated only with the actively transcribed globin genes. In contrast, in a previous study of mouse embryonic erythropoiesis using an antibody that detects both H3K9Ac and H3K14Ac (i.e. H3Ac), H3Ac and H3K4me2 enrichment was found at both the embryonic and adult β-globin genes (50). In the human β-globin locus, H3Ac and H3K4me2 are detected within the LCR, and only at the active β-globin genes (50, 51). In K562, a human cell line that expresses the fetal γ- but not the adult β-globin gene, H3Ac, H3K4me2, and H3K4me3 are enriched specifically at the γ-globin gene (52). H3K4me3 is enriched only in the actively transcribed fetal and adult β-globin genes in human and mouse tissue culture cell models, respectively (52, 53). Our results in native embryonic erythroid cells differ in that H3K4me3 is somewhat enriched at the mouse adult βmaj-globin gene at E10.5 prior to its activation.
KLF1 binds to histone-modifying proteins such as CBP, p300, and PCAF (14, 18, 54). Our data indicate for the first time that H3K9Ac and H3K4me3 are reduced at the mouse Ey- and βh1-globin and human ϵ- and γ-globin genes in primitive erythroid cells lacking KLF1. Transcription factors have been implicated previously in affecting histone modifications. The presence of GATA1 correlates with histone acetylation, H3K4me2, and H3K4me3 at the murine β-globin locus (55). Histone acetylation and H3K4me3 are significantly reduced in the adult β-globin gene by the absence of KLF1 in E13.5 fetal liver cells (56). It is difficult to dissect cause and effect in these cases. It is not known whether reduced transcription because of the absence of the transcription factor leads to decreased H3Ac and H3K4me3 or vice versa. However, there is a strong correlation between the presence of KLF1 and the occurrence of histone marks found in actively transcribed genes.
A simple model incorporating the available data is that KLF1 and KLF2 interchangeably (but not simultaneously) bind to the same CACCC elements in the LCR and promoters in the β-globin locus and promote gene expression in the embryo. This model is supported by the fact that KLF1 and KLF2 have very similar DNA binding domains, that there is no evidence that they can form dimers, and that they are likely to be present in similar amounts in embryonic blood cells. Individually, the effect of KLF1 on globin gene regulation tends to be greater than that of KLF2, based on qRT-PCR in the mouse KO models. Interestingly, this draws a parallel with our results studying histone marks in KLF1−/− and KLF2−/− embryonic blood cells. In these experiments, KLF1 binding correlates with histone marks that are associated with transcription, whereas KLF2 does not appear to dictate histone modifications in the β-globin locus. Yet, both KLF1 and KLF2 positively regulate globin gene transcription through direct binding to CACCC elements in the locus. Therefore, there are distinct yet overlapping mechanistic roles for KLF1 and KLF2 in embryonic red blood cells.
Supplementary Material
Acknowledgments
We thank Drs. Ann Dean and Chunhui Hou for technical advice on the ChIP assays, Jerry Lingrel for the KLF2 KO and KLF2 conditional KO mice, John Strouboulis for human β-globin locus transgenic mice, Huck-Hui Ng for the KLF2 custom antibody, and Gordon Ginder and Joseph Landry for frequent and invaluable advice.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK074694 (to J. A. L.). This work was also supported by the Ministry of Higher Education, Saudi Arabia (to Y. N. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables 1 and 2.
- E7.5
- embryonic day 7.5
- ϵ
- human embryonic globin gene
- γ
- human fetal globin gene
- β
- human adult globin gene
- CBP
- CREB-binding protein
- PCAF
- p300/CBP-associated factor
- LCR
- locus control region
- HS
- hypersensitive site(s)
- KLF
- Krüppel-like factor
- qRT-PCR
- quantitative RT-PCR
- GPA
- glycophorin A
- H3K9Ac
- histone 3 lysine 9 acetylation
- H3K4me3
- histone 3 lysine 4 trimethylation.
REFERENCES
- 1. Baron M. H., Fraser S. T. (2005) Curr. Opin. Hematol. 12, 217–221 [DOI] [PubMed] [Google Scholar]
- 2. Qiu C., Olivier E. N., Velho M., Bouhassira E. E. (2008) Blood 111, 2400–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McGrath K. E., Palis J. (2005) Exp. Hematol. 33, 1021–1028 [DOI] [PubMed] [Google Scholar]
- 4. Brotherton T. W., Chui D. H., Gauldie J., Patterson M. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 2853–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grosveld F., van Assendelft G. B., Greaves D. R., Kollias G. (1987) Cell 51, 975–985 [DOI] [PubMed] [Google Scholar]
- 6. Fraser P., Hurst J., Collis P., Grosveld F. (1990) Nucleic Acids Res. 18, 3503–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lloyd J. A., Lee R. F., Lingrel J. B. (1989) Nucleic Acids Res. 17, 4339–4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bieker J. J. (2001) J. Biol. Chem. 276, 34355–34358 [DOI] [PubMed] [Google Scholar]
- 9. McConnell B. B., Yang V. W. (2010) Physiol. Rev. 90, 1337–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perkins A. C., Sharpe A. H., Orkin S. H. (1995) Nature 375, 318–322 [DOI] [PubMed] [Google Scholar]
- 11. Nuez B., Michalovich D., Bygrave A., Ploemacher R., Grosveld F. (1995) Nature 375, 316–318 [DOI] [PubMed] [Google Scholar]
- 12. Basu P., Lung T. K., Lemsaddek W., Sargent T. G., Williams D. C., Jr., Basu M., Redmond L. C., Lingrel J. B., Haar J. L., Lloyd J. A. (2007) Blood 110, 3417–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou D., Pawlik K. M., Ren J., Sun C. W., Townes T. M. (2006) J. Biol. Chem. 281, 16052–16057 [DOI] [PubMed] [Google Scholar]
- 14. Zhang W., Bieker J. J. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9855–9860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hodge D., Coghill E., Keys J., Maguire T., Hartmann B., McDowall A., Weiss M., Grimmond S., Perkins A. (2006) Blood 107, 3359–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pilon A. M., Arcasoy M. O., Dressman H. K., Vayda S. E., Maksimova Y. D., Sangerman J. I., Gallagher P. G., Bodine D. M. (2008) Mol. Cell. Biol. 28, 7394–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Basu P., Morris P. E., Haar J. L., Wani M. A., Lingrel J. B., Gaensler K. M., Lloyd J. A. (2005) Blood 106, 2566–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. SenBanerjee S., Lin Z., Atkins G. B., Greif D. M., Rao R. M., Kumar A., Feinberg M. W., Chen Z., Simon D. I., Luscinskas F. W., Michel T. M., Gimbrone M. A., Jr., García-Cardeña G., Jain M. K. (2004) J. Exp. Med. 199, 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gaensler K. M., Kitamura M., Kan Y. W. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 11381–11385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Strouboulis J., Dillon N., Grosveld F. (1992) Genes Dev. 6, 1857–1864 [DOI] [PubMed] [Google Scholar]
- 21. Wani M. A., Means R. T., Jr., Lingrel J. B. (1998) Transgenic Res. 7, 229–238 [DOI] [PubMed] [Google Scholar]
- 22. Weinreich M. A., Takada K., Skon C., Reiner S. L., Jameson S. C., Hogquist K. A. (2009) Immunity 31, 122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heinrich A. C., Pelanda R., Klingmüller U. (2004) Blood 104, 659–666 [DOI] [PubMed] [Google Scholar]
- 24. Hou C., Zhao H., Tanimoto K., Dean A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20398–20403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang J., Chan Y. S., Loh Y. H., Cai J., Tong G. Q., Lim C. A., Robson P., Zhong S., Ng H. H. (2008) Nat. Cell Biol. 10, 353–360 [DOI] [PubMed] [Google Scholar]
- 26. Zhang J., Socolovsky M., Gross A. W., Lodish H. F. (2003) Blood 102, 3938–3946 [DOI] [PubMed] [Google Scholar]
- 27. Fraser S. T., Isern J., Baron M. H. (2007) Blood 109, 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Noordermeer D., de Laat W. (2008) IUBMB Life 60, 824–833 [DOI] [PubMed] [Google Scholar]
- 29. Maetens M., Doumont G., Clercq S. D., Francoz S., Froment P., Bellefroid E., Klingmuller U., Lozano G., Marine J. C. (2007) Blood 109, 2630–2633 [DOI] [PubMed] [Google Scholar]
- 30. Drissen R., Palstra R. J., Gillemans N., Splinter E., Grosveld F., Philipsen S., de Laat W. (2004) Genes Dev. 18, 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kiefer C. M., Hou C., Little J. A., Dean A. (2008) Mutat. Res. 647, 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hosey A. M., Chaturvedi C. P., Brand M. (2010) Epigenetics 5, 273–281 [DOI] [PubMed] [Google Scholar]
- 33. Demers C., Chaturvedi C. P., Ranish J. A., Juban G., Lai P., Morle F., Aebersold R., Dilworth F. J., Groudine M., Brand M. (2007) Mol. Cell 27, 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim A., Song S. H., Brand M., Dean A. (2007) Nucleic Acids Res. 35, 5831–5838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sengupta T., Chen K., Milot E., Bieker J. J. (2008) Mol. Cell. Biol. 28, 6160–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Arnaud L., Saison C., Helias V., Lucien N., Steschenko D., Giarratana M. C., Prehu C., Foliguet B., Montout L., de Brevern A. G., Francina A., Ripoche P., Fenneteau O., Da Costa L., Peyrard T., Coghlan G., Illum N., Birgens H., Tamary H., Iolascon A., Delaunay J., Tchernia G., Cartron J. P. (2010) Am. J. Hum. Genet. 87, 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Siatecka M., Sahr K. E., Andersen S. G., Mezei M., Bieker J. J., Peters L. L. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 15151–15156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Borg J., Papadopoulos P., Georgitsi M., Gutiérrez L., Grech G., Fanis P., Phylactides M., Verkerk A. J., van der Spek P. J., Scerri C. A., Cassar W., Galdies R., van Ijcken W., Ozgür Z., Gillemans N., Hou J., Bugeja M., Grosveld F. G., von Lindern M., Felice A. E., Patrinos G. P., Philipsen S. (2010) Nat. Genet. 42, 801–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Satta S., Perseu L., Moi P., Asunis I., Cabriolu A., Maccioni L., Demartis F. R., Manunza L., Cao A., Galanello R. (2011) Haematologica 96, 767–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou D., Liu K., Sun C. W., Pawlik K. M., Townes T. M. (2010) Nat. Genet. 42, 742–744 [DOI] [PubMed] [Google Scholar]
- 41. Isern J., Fraser S. T., He Z., Zhang H., Baron M. H. (2010) Blood 116, 3972–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fujiwara Y., Chang A. N., Williams A. M., Orkin S. H. (2004) Blood 103, 583–585 [DOI] [PubMed] [Google Scholar]
- 43. Takahashi S., Shimizu R., Suwabe N., Kuroha T., Yoh K., Ohta J., Nishimura S., Lim K. C., Engel J. D., Yamamoto M. (2000) Blood 96, 910–916 [PubMed] [Google Scholar]
- 44. Hosoya-Ohmura S., Mochizuki N., Suzuki M., Ohneda O., Ohneda K., Yamamoto M. (2006) J. Biol. Chem. 281, 32820–32830 [DOI] [PubMed] [Google Scholar]
- 45. Eaton S. A., Funnell A. P., Sue N., Nicholas H., Pearson R. C., Crossley M. (2008) J. Biol. Chem. 283, 26937–26947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Funnell A. P., Maloney C. A., Thompson L. J., Keys J., Tallack M., Perkins A. C., Crossley M. (2007) Mol. Cell. Biol. 27, 2777–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weiss M. J., Keller G., Orkin S. H. (1994) Genes Dev. 8, 1184–1197 [DOI] [PubMed] [Google Scholar]
- 48. Grass J. A., Boyer M. E., Pal S., Wu J., Weiss M. J., Bresnick E. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8811–8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Muntean A. G., Crispino J. D. (2005) Blood 106, 1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fromm G., Bulger M. (2009) Biochem. Cell Biol. 87, 781–790 [DOI] [PubMed] [Google Scholar]
- 51. Fang X., Yin W., Xiang P., Han H., Stamatoyannopoulos G., Li Q. (2009) J. Mol. Biol. 394, 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim A., Kiefer C. M., Dean A. (2007) Mol. Cell. Biol. 27, 1271–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fromm G., de Vries C., Byron R., Fields J., Fiering S., Groudine M., Bender M. A., Palis J., Bulger M. (2009) Blood 114, 3479–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kim S. I., Bresnick E. H., Bultman S. J. (2009) Nucleic Acids Res. 37, 6019–6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Im H., Grass J. A., Johnson K. D., Kim S. I., Boyer M. E., Imbalzano A. N., Bieker J. J., Bresnick E. H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17065–17070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bottardi S., Ross J., Pierre-Charles N., Blank V., Milot E. (2006) EMBO J. 25, 3586–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.