Abstract
Photosystem II (PSII) is a large membrane bound molecular machine that catalyzes light-driven oxygen evolution from water. PSII constantly undergoes assembly and disassembly because of the unavoidable damage that results from its normal photochemistry. Thus, under physiological conditions, in addition to the active PSII complexes, there are always PSII subpopulations incompetent of oxygen evolution, but are in the process of undergoing elaborate biogenesis and repair. These transient complexes are difficult to characterize because of their low abundance, structural heterogeneity, and thermodynamic instability. In this study, we show that a genetically tagged Psb27 protein allows for the biochemical purification of two monomeric PSII assembly intermediates, one with an unprocessed form of D1 (His27ΔctpAPSII) and a second one with a mature form of D1 (His27PSII). Both forms were capable of light-induced charge separation, but unable to photooxidize water, largely because of the absence of a functional tetramanganese cluster. Unexpectedly, there was a significant amount of the extrinsic lumenal PsbO protein in the His27PSII, but not in the His27ΔctpAPSII complex. In contrast, two other lumenal proteins, PsbU and PsbV, were absent in both of these PSII intermediate complexes. Additionally, the only cytoplasmic extrinsic protein, Psb28 was detected in His27PSII complex. Based on these data, we have presented a refined model of PSII biogenesis, illustrating an important role of Psb27 as a gate-keeper during the complex assembly process of the oxygen-evolving centers in PSII.
Keywords: Membrane Proteins, Photosynthesis, Protein Assembly, Protein Purification, Protein Turnover
Introduction
Photosystem II (PSII)3 is a large membrane protein complex found in the thylakoids membranes of cyanobacteria, algae, and higher plants. It catalyzes oxidation of water and reduction of plastoquinone (PQ). These activities are essential for the conversion of solar energy to the chemical energy used by the vast majority of life on earth and for the production of molecular oxygen.
Recent structural studies have provided a detailed yet static view of PSII (1–3). Accordingly, a PSII monomer is comprised of 20 protein subunits, which form a proteinaceous scaffold holding two pheophytin a (Pheo), 35 chlorophyll a (Chla), one non-heme iron, two hemes (Cyt b559 and Cyt c550), three PQ, and 12 β-carotene (4, 5) molecules as cofactors. The light driven electron transfer reactions through the PSII complex requires precise positioning of all these cofactors and an inorganic (Mn4-Ca) cluster on lumenal side, the latter of which is also called water-oxidizing complex (WOC). However, the cost of water oxidation is high: photooxidative damage inevitably occurs to PSII, in which P680+, the strongest biological oxidant, is generated and finally leads to the splitting of water to molecular oxygen, protons, and electrons. Under physiological conditions, an elaborate and well-orchestrated repair process repairs such damages, so that PSII can function optimally.
Because PSII is under constant repair and biogenesis, elucidation of the details of the PSII assembly pathway has been the focus of intensive research. Many studies have been conducted to define the order of association of intrinsic and extrinsic PSII polypeptide subunits (6, 7). During the sequential assembly of the PSII complex, cofactors such as chlorophylls, carotenoids, non-heme iron, Pheo, and (Mn4-Ca) center must bind to the proteinaceous environment and be oriented properly. The final step leading to functional PSII is thought to be the photoassembly of the (Mn4-Ca) cluster on the electron transfer donor side (8, 9) and the association of the lumenally localized extrinsic proteins, which enhance the stability of the (Mn4-Ca) cluster. However, the detailed order in which the dynamic recruitment of such extrinsic proteins during photoactivation in vivo remains unclear.
One essential step before the photoassembly of (Mn4-Ca) cluster can take place is the proteolytic processing of the D1 protein by a C-terminal processing enzyme (CtpA). The D1 protein is synthesized as a precursor protein (pD1) with a short (8–16 residues) amino acid extension at its C terminus (10, 11). The extension must be cleaved before (Mn4-Ca) cluster assembly, because the exposed terminal carboxylate group on D1 appears to provide a coordination ligand for a manganese atom in the catalytic (Mn4-Ca) cluster of PSII (12–14). Following the proteolytic processing of pD1, assembly of the (Mn4-Ca) center takes place in a light-dependent manner. This process occurs very rapidly in vivo, and thus transient assembly intermediates have been difficult to capture experimentally.
In this context, Psb27, an 11 kDa lumenal protein, has been found to be a component of PSII during its assembly process, but not a component in the mature and functional form of PSII (15–17). Here, we report two intermediate PSII complexes, isolated using a genetically tagged form of the Psb27 protein in the cyanobacterium Synechocystis 6803. Both of these PSII preparations could undergo charge separation, but could not catalyze water oxidation. Thus, these isolated complexes represent consecutive assembly intermediates before the photoactivation of PSII. His27PSII complex contains mature D1 protein, while His27ΔctpAPSII complex contains pD1 protein due to the genetic deletion of the ctpA gene. Analysis of these two isolated membrane protein complexes has allowed us to develop a more detailed model of PSII biogenesis in cyanobacteria.
EXPERIMENTAL PROCEDURES
Growth of Synechocystis sp. PCC 6803 Strains
HT3, His27, and His27ΔctpA strains of Synechocystis 6803 were grown in BG11 medium (18) at 30 °C under 30 μmol photons m−2·s−1. These strains were grown in BG11 supplemented with antibiotics as follows: 5 μg/ml kanamycin (HT3); 5 μg/ml gentamicin (His27); 5 μg/ml gentamicin, 5 μg/ml erythromycin, 10 μm DCMU (3-(3′,4′-dichlorophenyl)-1,1-dimethylurea), and 5 mm glucose (His27ΔctpA). The HT3 strain was provided by Dr. Terry M. Bricker (19). Because the His27ΔctpA cells are light sensitive, they were cultured at a lower light intensity by wrapping culture bottles with Kimwipes.
Mutant Construction
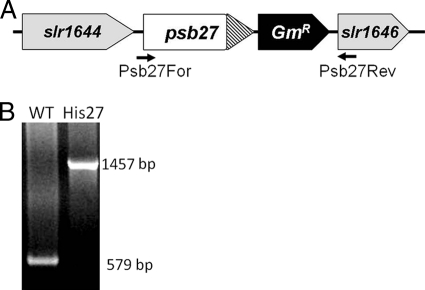
To generate the His27 strain, the full-length psb27 gene without its stop codon was amplified using the 5pBADF27 and 3pBAD27 primers (Table 1) and cloned into the pBAD/Myc-His vector (Invitrogen, Carlsbad, CA). The PCR fragment containing the affinity tag (both Myc and His6) derived from the pBAD/Myc-His vector and psb27 gene was amplified using 27HisFor and 27HisRev primers and then cloned into pUC18 vector to yield the 27His/pUC18 plasmid. In a separate step, the DNA fragment corresponding to the 489 base pair (bp) downstream of the psb27 gene was amplified using the 27dsFor and 27dsRev primers, and was cloned into pUC18 to yield 27ds/pUC18 plasmid. The KpnI fragment containing the gentamicin resistance gene from the pUCGM vector (20) was cloned into the 27ds/pUC18 plasmid to yield the GM/27ds/pUC18 plasmid. Finally, the BamHI/EcoRI fragment from the GM/27ds/pUC18 plasmid containing both the gentamicin resistance gene and the psb27 gene downstream fragment was cloned into the 27His/pUC18 plasmid to yield the Tag27 plasmid. The Tag27 plasmid was used to transform wild type Synechocystis cells. Segregation of the modified psb27 locus was verified by PCR analysis with Psb27For and Psb27Rev primers. All primer sequences are listed in Table 1.
TABLE 1.
Primers used in the construction and segregation analysis
| Name | Sequence (5′ to 3′) |
|---|---|
| 5pBADF27 | CCCGCCGCTCGAGATCCTTTTTGAAAAATCAG |
| 3pBAD27 | CCCAAGCTTCACGCCCCGTTCAATGGATC |
| 27HisFor | ACATGCATGCATGGATCCGAGCTCGAGATCCT |
| 27HisRev | CTAGTCTAGAAGCTGGAGACCGTTTAAACTCA |
| 27dsFor | CGAGCTCGAACCATCCCGATTTCCC |
| 27dsRev | CGGAATTCATGGTCGGCACTAGTTTTGG |
| Psb27For | AGCCGGTCATAGGAAGGAGT |
| Psb27Rev | CCTGGCCAACTGGTAGGATA |
After successful isolation of the His27PSII assembly intermediate, we replaced the ctpA gene with a 1.5-kb erythromycin resistance marker in the genetic background of the His27 strain to generate the double mutant His27ΔctpA. Complete segregation of the ΔctpA mutation was confirmed by PCR analysis of the ctpA locus (data not shown).
PSII Purification
PSII complexes from the HT3, His27, and His27ΔctpA strains were purified as previously described (19) with minor modifications for an AKTA FPLC system (GE Healthcare, Little Chalfont, Buckinghamshire, UK). Solubilized membranes were injected onto a nickel-nitrilotriacetic acid agarose column (Qiagen, Valencia, CA) pre-equilibrated with buffer containing 50 mm Mes-NaOH, pH 6.0/5 mm CaCl2/10 mm MgCl2/25% glycerol/0.04% β-dodecylmaltoside (β-DDM). PSII was eluted with buffer supplemented with 50 mm l-histidine. The elution peak containing PSII was precipitated in the presence of polyethylene glycol.
Polypeptide Profile and BN-gel Analysis
Electrophoresis and immunodetection were performed using an 18–24% gradient acrylamide 6 m urea SDS/PAGE system (21, 22). PSII subunits were identified by using specific antisera after transferring onto PVDF membranes (Millipore). Bands were visualized by using enhanced chemiluminescence reagents (WestPico; Pierce) on a Fujifilm LAS-1000 Plus imager (Fujifilm, Stamford, CT). Monomer and dimer analysis of PSII assembly intermediates were performed by Blue-Native gel (BN-gel) electrophoresis (23).
Protein Identification by Mass Spectrometry
After SDS-PAGE of purified PSII preparations, samples of specific protein bands were analyzed on a QSTAR XL (AB/MDS Sciex) mass spectrometer at the Danforth Plant Science Center proteomics facility (St. Louis).
Cofactor Analysis
Chla concentration in PSII complexes was determined spectrophotometrically (24). Simultaneous quantification of Chla and Pheo was performed using HPLC. Pigments were extracted from purified PSII complexes in 7:2 of acetone: methanol (v:v) at 4 °C and immediately analyzed. Pigments were resolved on a ZORBAX XDB-C18 (4.6 × 250 mm, 5 μm) column on an Agilent 1200 series HPLC using a diode array detector (Agilent Technologies, Santa Clara, CA). The mobile phase consisted ethanol in methanol and separation was achieved using a linear gradient. Chla and Pheo were detected at 408 nm and characteristic absorption spectra of the corresponding peak confirmed the identities of the compounds. Quantification of Chla and Pheo was performed by comparison of peak area to standards of known concentration. Pheophytin a standards were prepared by acidification of Chla with 2.5 mm HCl. Concentration of Mn was determined on an AA600 atomic absorption spectrophotometer (PerkinElmer Life Sciences, Wellesley, MA). The Mn: PSII ratio was calculated based on 41 molecules of Chla/PSII (21).
Oxygen Evolution Measurements
Steady state rates of oxygen evolution of PSII were measured on a Clark-type electrode in the presence of 1 mm potassium ferricyanide and 0.5 mm 2, 6-dichloro-p-benzoquinone as electron acceptors at 5 μg of Chla/ml in 50 mm Mes-NaOH (pH 6.0)/20 mm CaCl2/0.5 m sucrose. PSII samples were incubated for 1 min in the dark at 30 °C before the assay under 8,250 μmol photons m−2 · s−1.
Fluorescence Analysis
PSII samples were diluted to 0.1 mg Chla/ml and fluorescence emission spectra at 77K were measured on a Fluoromax-2 fluorometer (Jobin Yvon, Longjumeau, France) with excitation at 435 nm (25). Variable Chla fluorescence relaxation kinetics at room temperature were recorded on a double-modulation fluorometer FL-200 (Photon System Instruments, Brno, Czech Republic), with a built-in analyzing program, FluorWin. The sample concentration was adjusted to 5 μg/ml of Chla in the buffer used for oxygen evolution with 0.04% of β-dodecylmaltoside added additionally. All samples were dark adapted for 5 min at room temperature before the measurements. DCMU was added to 10 μm, when needed.
RESULTS
Mutant Construction
To facilitate PSII purification, a hexahistidine tag was designed at the C terminus of the Psb27 protein, so as not to disrupt lipid modification of the N-terminal cysteine residue of the mature protein (17). The His27 strain construct contains the native promoter of the psb27 gene. Following the coding sequence of Psb27 protein is a sequence encoding for a c-Myc epitope and for a hexahistidine tag (Fig. 1A). A gentamicin resistance marker was introduced between the polyhistidine tag and the downstream slr1646 gene (encoding ribonuclease III) for selection purpose. Complete segregation of the His27 strain with the resultant larger PCR product was confirmed by PCR analysis (Fig. 1B). Psb27 is not absolutely essential for photosynthesis in Synechocystis 6803 cells (26), as well as in the vascular plant Arabidopsis thaliana (27). Similarly, when grown in nutrient replete BG11 medium, the His27 strain exhibits normal photoautotrophic growth and photochemical activities (data not shown).
FIGURE 1.
Construction of His27 mutant. A, psb27 locus in the His27 mutant which expresses a C-terminally Myc/His6-tagged Psb27 protein. A gentamicin resistance gene cassette is located between the psb27 locus and the downstream gene slr1646. B, PCR segregation analysis using the Psb27For and Psb27Rev primers (Table 1); positions of primers are labeled in A.
PSII Isolation
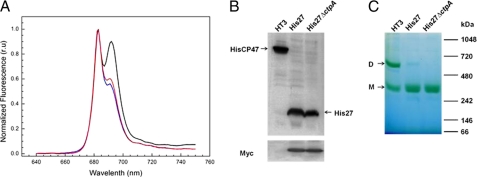
As reported before, the Psb27 protein is present in substoichiometric amount in PSII complex isolated using a polyhistidine tag on the CP47 protein of Synechocystis 6803 (21). To investigate the nature of the protein complex that has a stoichiometric amount of Psb27, the tagged Psb27 protein was used to isolate PSII complexes from His27 cells. We have used a similar strategy in the past to purify a highly active PSII preparation using a C-terminally polyhistidine tagged extrinsic PsbQ protein (28). Isolation of His27PSII using FPLC chromatography resulted in a single small peak (data not shown). 77 K fluorescence measurements confirmed that the elution peak contained PSII complexes (Fig. 2A), which show a specific PSII peak at 685 nm (F685) and a much decreased peak at 695 nm (F695). Notably, no characteristic peak from PSI was observed. We named the corresponding purified PSII complex as His27PSII. The yield of this PSII complex was about 10–15 fold lower than PSII from the HT3 strain (Table 2), consistent with the notion that the Psb27 protein is present in a PSII assembly intermediate pigment-protein complex, but not in the mature and functional PSII complex.
FIGURE 2.
Characterization of purified His27PSII and His27ΔctpAPSII protein complexes. A, 77 K fluorescence emission spectra of the His27PSII (red) and His27ΔctpAPSII (blue) complexes are shown with that of HT3PSII (black) complexes as a reference. Samples were excited at 435 nm and fluorescence emission was normalized as (F-F640)/(F683 − F640). B, immunodetection of the polyhistidine tag and c-Myc tag epitopes in HT3PSII, His27PSII, and His27ΔctpAPSII. C, BN-gel analysis of HT3PSII, His27PSII, His27ΔctpAPSII preparation. Arrows indicate the dimeric (D) and monomeric (M) forms. For details, see “Experimental Procedures.”
TABLE 2.
PSII function and cofactor analysis
| PSII Yield | O2 evolutiona | Mn/PSII | Chla/Pheo | |
|---|---|---|---|---|
| HT3PSII | 4–6% | 1394 ± 103b | 3.6 ± 0.1 | 16.9 ± 1.4 |
| His27PSII | 0.4% | 115 ± 34 | 0.76 ± 0.1 | 20.6 ± 1.8 |
| His27ΔctpAPSII | 1.25% | NDc | 0.91 ± 0.08 | 20.3 ± 2.0 |
a Oxygen evolution rates in μmol of O2·mg−1 Chl·h−1.
b Standard error of the mean (n = 3).
c ND, not detected.
As expected, the His27ΔctpA strain could not grow photoautotrophically (data not shown), since the CtpA protein is essential for one of the terminal maturation steps in the assembly of functional PSII complex (11). We isolated pD1-containing PSII complex (named His27ΔctpAPSII) from this strain grown under photoheterotrophic conditions. Interestingly, the yield of PSII complex preparation from this strain is significantly higher than that from the His27 strain (Table 2).
Fig. 2B shows the immunodetection of the polyhistidine and c-Myc tags in the purified PSII complexes from strains under consideration. As expected, the HT3 strain contained only polyhistidine-tagged CP47, whereas both His27 and His27ΔctpA strains contained polyhistidine-tagged Psb27. Similarly, the Psb27 protein in the His27 and His27ΔctpA strains exhibits the presence of the c-Myc epitope (Fig. 2B, lower panel), whereas no c-Myc protein is present in HT3PSII.
Fig. 2C shows the results of blue-native gel (BN-gel) analysis of these three PSII preparations. The HT3PSII complex could be resolved into two major green bands corresponding to PSII dimers and PSII monomers, respectively. Interestingly, both His27PSII and His27ΔctpAPSII are present mostly as monomeric form. Recently, a new dimeric Psb27-PSII complex has been observed, which accumulates under specific stress conditions and is apparently involved in the replacement of damaged D1 (29). However, we only detected monomeric Psb27-containing PSII complexes with either processed or unprocessed D1 protein. Further experiments are needed to characterize the functional and structural differences between monomeric and dimeric Psb27-containing PSII complexes.
Activities of PSII from HT3, His27, and His27ΔctpA Strains
Table 2 documents the photochemical activities of the three PSII preparations. Compared with HT3PSII, the His27PSII preparation has marginal oxygen evolution activity, whereas the His27ΔctpAPSII sample does not exhibit any such activity. Table 2 also shows manganese content and Chla/Pheo ratios of these PSII samples. All three samples have nearly similar Chla/Pheo ratios. In contrast, His27PSII and His27ΔctpAPSII complexes show less than 1 Mn per PSII center, consistent with their inabilities to oxidize water. This observation is consistent with the finding that in a Psb27-containing PSII complex from the thermophilic cyanobacterium Thermosynechococcus elongatus, no observable EPR signal from Mn4-Ca cluster was observed (30).
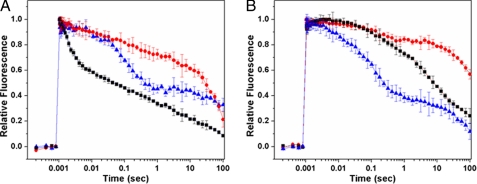
Electron transfer in the two PSII assembly intermediates was further investigated and compared with that of HT3PSII by using flash-induced variable fluorescence decay analysis. Fig. 3 shows the fluorescence decay kinetics measured in the absence or presence of DCMU in the three PSII preparations. In the absence of DCMU, after a single turnover saturating flash, the fluorescence decay occurs mainly as a consequence of forward electron transfer from QA− to QB. On the other hand, in the presence of DCMU, which prevents electron transfer from QA− to QB, the decay of fluorescence following a saturating flash is dominated by charge recombination between QA− and the oxidizing-side components of PSII. As shown in Fig. 3A, the decay kinetics of the flash-induced fluorescence from His27PSII and His27ΔctpAPSII preparations in the absence of DCMU are significantly slower than that from HT3PSII, indicating that the QA− reoxidation by forward electron transfer to QB has been severely affected in both of these preparations. In fact, the fluorescence decay trend in both His27PSII and His27ΔctpAPSII samples without DCMU treatment are very similar to those treated with DCMU. It seems that compared with HT3PSII, forward electron transfer from QA− to QB in both His27PSII and His27ΔctpAPSII is blocked. These findings are consistent with those from the studies on monomeric PSII complexes from T. elongatus containing Psb27 (30).
FIGURE 3.
Fluorescence kinetic analysis. Chlorophyll a fluorescence decay in the absence (A) and presence (B) of 10 μm DCMU, following a single saturating flash on HT3PSII (black), His27PSII (red), and His27ΔctpAPSII (blue) samples. Error bars represent the standard error of the mean (n = 3).
Polypeptide Profiles of the His27PSII and His27ΔctpAPSII Complexes
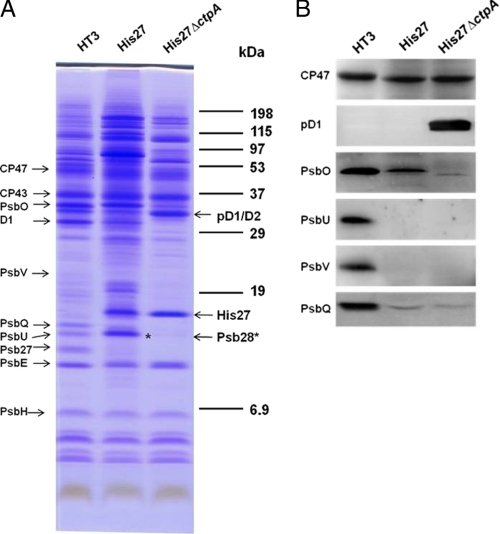
Fig. 4A shows a comparison of the polypeptide profiles of PSII preparations from HT3PSII, His27PSII, and His27ΔctpAPSII. Protein components were identified based on their migration on the SDS-PAGE system (31) or by immunodetection (Fig. 4B). As expected, tagged Psb27 protein migrated more slowly in the His27PSII and His27ΔctpAPSII samples, due to the additon of the c-Myc and polyhistidine tags (Fig. 2B).
FIGURE 4.
A, polypeptide profiles of isolated HT3PSII, His27PSII, and His27ΔctpAPSII complexes. The positions of major PSII protein components are indicated on the left. The Psb28 protein in His27PSII is indicated by an asterisk. 5 μg of Chla containing sample was loaded in each lane. B, immunodetection of PSII polypeptides in the isolated complexes. 0.2 μg of Chla sample was fractionated by SDS-PAGE, transferred to PVDF membrane, and probed for the indicated PSII proteins. Specific antibodies against CP47, D1 extension, PsbO, PsbU, PsbV, and PsbQ were used for immunodetection of the corresponding proteins, respectively.
In both His27PSII and His27ΔctpAPSII preparations, most PSII intrinsic core subunits, such as CP47, CP43, D1, D2, and PsbE, are observed. As expected, His27ΔctpAPSII contains only the pD1 protein, which migrates at the same location of D2 protein. An unexpected finding is the detection of a significant amount of the PsbO protein in the His27PSII preparation (Fig. 4, A and B). In contrast, PsbU and PsbV are not observed (Fig. 4B), whereas PsbQ is observed only as a very weak band in both the His27PSII and His27ΔctpAPSII samples.
Another notable finding is the enriched presence of the Psb28 protein (the gene product of sll1398) in His27PSII preparation. There was a distinct band in the region where PsbU migrates in HT3PSII and His27PSII, but imunological methods showed PsbU was not detected in His27PSII via Western blot (Fig. 4, A and B). Psb28 (11 kDa) migrates in the same region of PsbU (12 kDa) and thus they are difficult to separate using SDS-PAGE. An excised band corresponding to the general location of PsbU from the His27PSII lane (Fig. 4A, asterisk) was analyzed by mass spectrometry analysis and the results indicated no PsbU was identified, consistent with our Western blot analysis (Fig. 4B). MS/MS analysis identified three Psb28 protein peptide fragments corresponding to residues 9–19 (GVAETVVPEVR), 33–43 (FYFLEPTILAK), and 100–112 (GAENGLGFSKSE) of the 112 amino acid residues long Psb28 protein in Synechocystis 6803 (supplemental Fig. S1). Psb28 is the only cytoplasmically (stromal) exposed extrinsic protein in PSII (32). This protein is absent in the His27ΔctpAPSII preparation, suggesting it becomes associated to PSII after the processing of the C-terminal extension of the D1 protein.
In addition to Psb28, Sll1130, a hypothetical 11 kDa protein was identified. Also identified were the carbon dioxide concentrating mechanism protein (CcmK homolog2, 11 kDa); 30 S ribosomal protein S10 (12 kDa); ribulose bisphosphate carboxylase small subunit (13 kDa). Because of the high sensitivity of modern mass spectrometry, it is highly possible that these proteins are contaminants rather than PSII components. Sll1130 has been previously reported in PSII preparations (21).
DISCUSSION
In summary, using genetic and biochemical methods, we have isolated and characterized two PSII assembly intermediates, which represent two stages immediately preceding and following the C-terminal processing of the pD1 protein, respectively. The ability to biochemically purify such complexes using C-terminally polyhistidine-tagged Psb27 protein indicates that Psb27 is a tightly bound subunit of these PSII assembly intermediates.
The yield of the fraction of Psb27-containing PSII assembly intermediates from the His27 cells is around 7–10% of that from HT3 cells (Table 2). This provides strong evidence to that there is always a subpopulation of PSII, which undergoes dynamic assembly. This fraction is characterized by the presence of Psb27, the absence of (Mn4-Ca) cluster, and absence of oxygen evolution activity. Absence of the (Mn4-Ca) cluster in PSII assembly intermediates particularly may have profound physiological and ecological adaptive effects. It has recently been reported that coupling of phycobilisome, the large light harvesting antenna complex of cyanobacteria, to PSII is dependent on the integrity of the (Mn4-Ca) cluster on the donor side of PSII (33). Thus, it is conceivable that without phycobilisomes functionally attaching to Psb27 containing PSII intermediates; no energy coupling can take place, resulting in less damage to the complex. This is particularly important, since protection of PSII assembly intermediates from photodamage before the functional state of PSII is achieved is essential for efficient PSII biogenesis under physiological conditions.
The presence of the PsbO protein in the His27PSII but not in the His27ΔctpAPSII assembly intermediate prompts us to propose that PsbO is the first extrinsic protein recruited to the binding interface of PSII during donor side assembly. Absence of PsbO in His27ΔctpAPSII is consistent with the data from PSII complexes isolated from our earlier studies on a ΔctpAHT3 strain (16). As it is the only difference between the two PSII assembly intermediates in our current study, it is reasonable to speculate that it is either the presence of the C-terminal extension of pD1, or the subsequent conformational changes in CP47 or CP43 that blocks the interactions between PsbO and CP47 and CP43 (4, 34). The absence of functional (Mn4-Ca) clusters (Table 2) in both PSII intermediates may also contribute substantially to the reduced extrinsic protein binding affinity to PSII, as has been observed in in vitro experiments (35, 36). Our results argue against the hypothesis that Psb27 facilitates the (Mn4-Ca) clusters assembly by preventing the PsbO protein from binding to PSII (37).
A novel finding in this report is the presence of Psb28 in His27PSII, but not in His27ΔctpAPSII. The functional role of this extrinsic protein, with a unique location in the cytoplasm side of thylakoids membrane, remains poorly understood. A recent study has suggested that this protein is somehow involved in the biosynthesis of chlorophyll, and also is closely associated with the CP47 protein. Our data indicate that the binding of Psb28 to the cytoplasmic side of the His27-containing PSII assembly intermediate occurs after the C-terminal processing of the pD1 protein on the lumenal side of PSII. In addition, accumulation of Psb28 has only been found in PSII monomers from dgdA mutant cells (38). The authors hypothesized that Psb28 may have an important function in the assembly of subunits in PSII. It seems that in His27PSII both donor and acceptor sides are structurally compromised and different from those of functional PSII. The functional implications of such events deserve further detailed investigation.
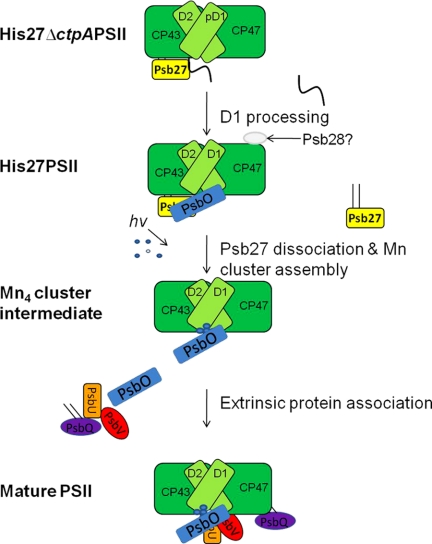
We propose a refined schematic model for the biogenesis of the PSII complex (Fig. 5), combining our data with previous studies, especially recent reports (30, 39, 40). Psb27 is the first lumenal protein that binds to the lumenal interface of PSII assembly intermediates containing a number of intrinsic membrane proteins such as CP47, CP43, Cyt b559, D2, and pD1. Next, the CtpA enzyme cleaves the C-terminal extension of pD1 to form mature D1. At this stage, due to the presence of Psb27 and potential effects on lumenal domains of intrinsic subunits, PsbO is only able to weakly bind to PSII. However, the presence of Psb27 completely prevents PsbU, PsbV, and PsbQ from binding to intrinsic PSII subunits and prevents the (Mn4-Ca) cluster from assembling. Finally, the detachment of Psb27 allows photoactivation of the (Mn4-Ca) cluster as well as the binding of other extrinsic proteins, to form stable PSII complexes with normal water oxidation capacity.
FIGURE 5.
A schematic model for PSII assembly. The core membrane proteins are shown in green, whereas the extrinsic proteins are Psb27 (yellow), Psb28 (gray), PsbO (blue), PsbU (orange), PsbV (red), and PsbQ (purple). The Psb27 and PsbQ lipoproteins are depicted with their lipid modifications. The atoms of Mn cluster are shown in blue and white. The different assembly events are labeled.
In conclusion, we have demonstrated that Psb27 is tightly bound to two PSII assembly intermediates, and that PsbO, but not PsbV and PsbU, can bind to the Psb27 containing PSII assembly intermediates after pD1 processing. However, the binding partner(s) of Psb27 in such PSII complex remain poorly defined. For example, it has been hypothesized that Psb27 binds to the lumenal area of CP47 (37, 41) or CP43 (42). We are currently investigating the structural location of Psb27 in the PSII assembly intermediate complexes described in this report.
Supplementary Material
Acknowledgments
We thank Dr. Terry M. Bricker for the HT3 strain and other members of the Pakrasi laboratory for collegial discussions.
This work was supported by National Science Foundation Grant NSF-MCB0745611 (to H. B. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PSII
- photosystem II
- CtpA
- C-terminal processing enzyme
- DCMU
- (3-(3′,4′-dichlorophenyl)-1,1-dimethylurea)
- PQ
- plastoquinone
- Pheo
- pheophytin a.
REFERENCES
- 1. Zouni A., Witt H. T., Kern J., Fromme P., Krauss N., Saenger W., Orth P. (2001) Nature 409, 739–743 [DOI] [PubMed] [Google Scholar]
- 2. Kamiya N., Shen J. R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 98–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. (2005) Nature 438, 1040–1044 [DOI] [PubMed] [Google Scholar]
- 4. Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., Iwata S. (2004) Science 303, 1831–1838 [DOI] [PubMed] [Google Scholar]
- 5. Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009) Nat. Struct. Mol. Biol. 16, 334–342 [DOI] [PubMed] [Google Scholar]
- 6. Rokka A., Suorsa M., Saleem A., Battchikova N., Aro E. M. (2005) Biochem. J. 388, 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nixon P. J., Michoux F., Yu J., Boehm M., Komenda J. (2010) Ann. Bot. 106, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheniae G. M., Martin I. F. (1971) Biochim. Biophys. Acta. 253, 167–181 [DOI] [PubMed] [Google Scholar]
- 9. Frasch W. D., Sayre R. T. (2001) Photosynth. Res. 70, 245–247 [DOI] [PubMed] [Google Scholar]
- 10. Seibert M., Tamura N., Inoue Y. (1989) Biochim. Biophys. Acta 974, 185–191 [Google Scholar]
- 11. Metz J. G., Pakrasi H. B., Seibert M., Arntzen C. J. (1986) FEBS. Letts. 205, 269–274 [Google Scholar]
- 12. Chu H. A., Hillier W., Debus R. J. (2004) Biochemistry. 43, 3152–3166 [DOI] [PubMed] [Google Scholar]
- 13. Stull J. A., Stich T. A., Service R. J., Debus R. J., Mandal S. K., Armstrong W. H., Britt R. D. (2010) J. Am. Chem. Soc. 132, 446–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nixon P. J., Diner B. A. (1992) Biochemistry. 31, 942–948 [DOI] [PubMed] [Google Scholar]
- 15. Ikeuchi M., Inoue Y., Vermaas W. (1995) in Photosynthesis: From Light to Biosphere (Mathis P. ed) pp. 297–300, vol. III, Kluwer Academic Publishers [Google Scholar]
- 16. Roose J. L., Pakrasi H. B. (2004) J. Biol. Chem. 279, 45417–45422 [DOI] [PubMed] [Google Scholar]
- 17. Nowaczyk M. M., Hebeler R., Schlodder E., Meyer H. E., Warscheid B., Rögner M. (2006) Plant. Cell. 18, 3121–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Allen M. M. (1968) J. Phycol. 4, 1–4 [DOI] [PubMed] [Google Scholar]
- 19. Bricker T. M., Morvant J., Masri N., Sutton H. M., Frankel L. K. (1998) Biochim. Biophys. Acta. 1409, 50–57 [DOI] [PubMed] [Google Scholar]
- 20. Schweizer H. D. (1993) BioTechniques 15, 831–834 [PubMed] [Google Scholar]
- 21. Kashino Y., Lauber W. M., Carroll J. A., Wang Q., Whitmarsh J., Satoh K., Pakrasi H. B. (2002) Biochemistry 41, 8004–8012 [DOI] [PubMed] [Google Scholar]
- 22. Kashino Y., Koike H., Satoh K. (2001) Electrophoresis 22, 1004–1007 [DOI] [PubMed] [Google Scholar]
- 23. Shägger H., von Jagow G. (1991) Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 24. Porra R. J., Kriedemann P. E. (1989) Biochim. Biophys. Acta. 975, 384–394 [Google Scholar]
- 25. Zak E., Norling B., Maitra R., Huang F., Andersson B., Pakrasi H. B. (2001) Proc. Natl. Acad. Sci. U. S. A. 98, 13443–13448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roose J. L., Pakrasi H. B. (2008) J. Biol. Chem. 283, 4044–4050 [DOI] [PubMed] [Google Scholar]
- 27. Chen H., Zhang D., Guo J., Wu H., Jin M., Lu Q., Lu C., Zhang L. (2006) Plant. Mol. Biol. 61, 567–575 [DOI] [PubMed] [Google Scholar]
- 28. Roose J. L., Kashino Y., Pakrasi H. B. (2007) Proc. Natl. Acad. Sci. U. S. A. 104, 2548–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nowaczyk M. M., Sander J., Grasse N., Cormann K. U., Rexroth D., Bernát G., Rögner M. (2010) Eur. J. Cell Biol. 89, 974–982 [DOI] [PubMed] [Google Scholar]
- 30. Mamedov F., Nowaczyk M. M., Thapper A., Rögner M., Styring S. (2007) Biochemistry 46, 5542–5551 [DOI] [PubMed] [Google Scholar]
- 31. Kashino Y., Koike H., Yoshio M., Egashira H., Ikeuchi M., Pakrasi H. B., Satoh K. (2002) Plant Cell Physiol. 43, 1366–1373 [DOI] [PubMed] [Google Scholar]
- 32. Dobáková M., Sobotka R., Tichý M., Komenda J. (2009) Plant Physiol. 149, 1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hwang H. J., Nagarajan A., McLain A., Burnap R. L. (2008) Biochemistry 47, 9747–9755 [DOI] [PubMed] [Google Scholar]
- 34. Odom W. R., Bricker T. M. (1992) Biochemistry 31, 5616–5620 [DOI] [PubMed] [Google Scholar]
- 35. Bricker T. M., Frankel L. K. (1987) Arch. Biochem. Biophys. 256, 295–301 [DOI] [PubMed] [Google Scholar]
- 36. Bricker T. M. (1990) Photosynth. Res. 24, 1–13 [DOI] [PubMed] [Google Scholar]
- 37. Cormann K. U., Bangert J. A., Ikeuchi M., Rögner M., Stoll R., Nowaczyk M. M. (2009) Biochemistry 48, 8768–8770 [DOI] [PubMed] [Google Scholar]
- 38. Sakurai I., Mizusawa N., Wada H., Sato N. (2007) Plant. Physiol. 145, 1361–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bentley F. K., Luo H., Dilbeck P., Burnap R. L., Eaton-Rye J. J. (2008) Biochemistry 47, 11637–11646 [DOI] [PubMed] [Google Scholar]
- 40. Wei L., Guo J., Ouyang M., Sun X., Ma J., Chi W., Lu C., Zhang L. (2010) J. Biol. Chem. 285, 21391–21398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Becker K., Cormann K. U., Nowaczyk M. M. (2011) J. Photochem. Photobiol. B. 104, 204–211 [DOI] [PubMed] [Google Scholar]
- 42. Fagerlund R. D., Eaton-Rye J. J. (2011) J Photochem Photobiol B. 104, 191–203 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.